Linearization of the Kingman Coalescent
Abstract
1. Introduction
1.1. Coalescent Theory of Ancestral Processes
1.2. Coalescent Theory of Branching Processes
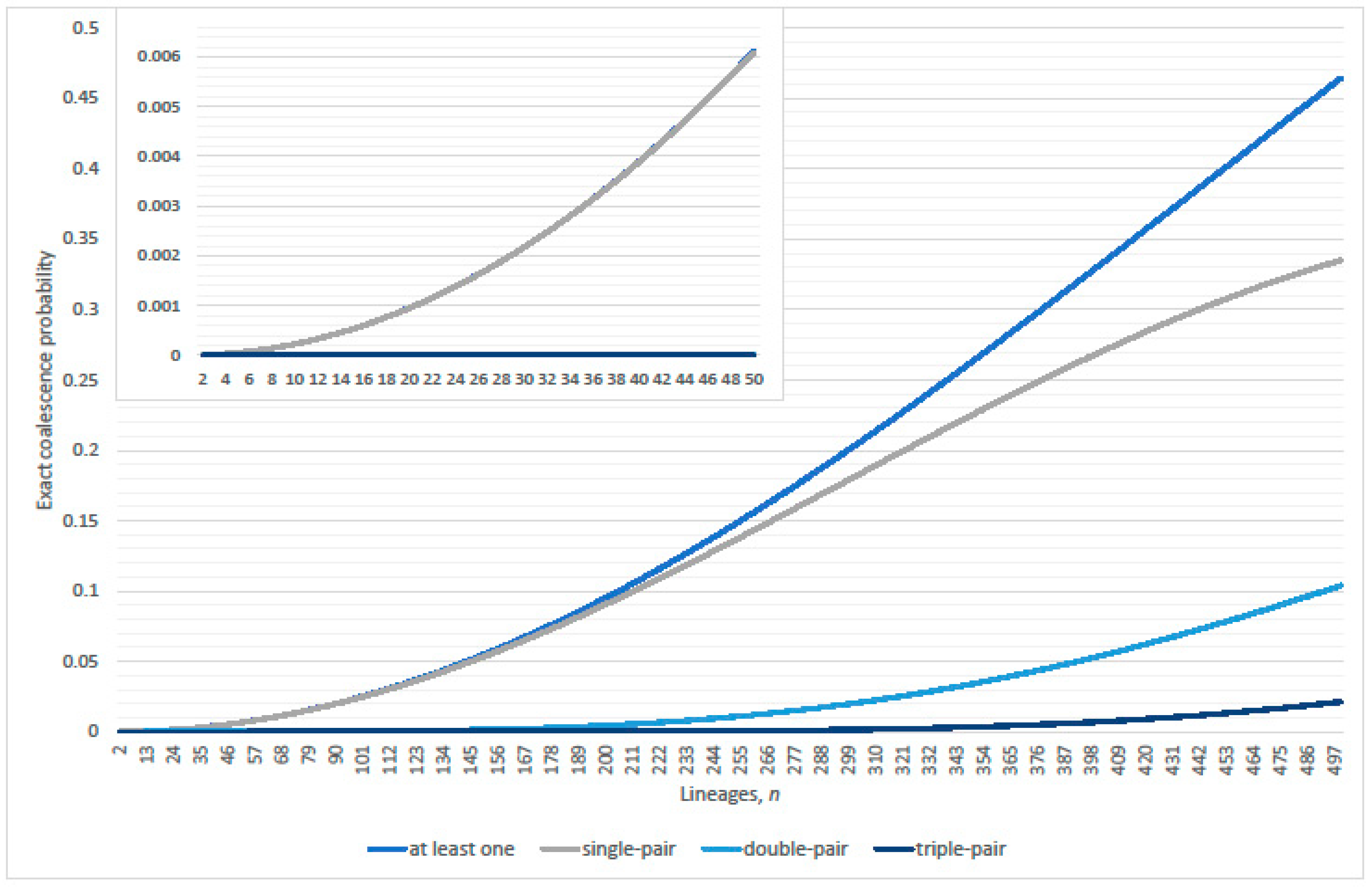
2. Ancestral Process, per Generation
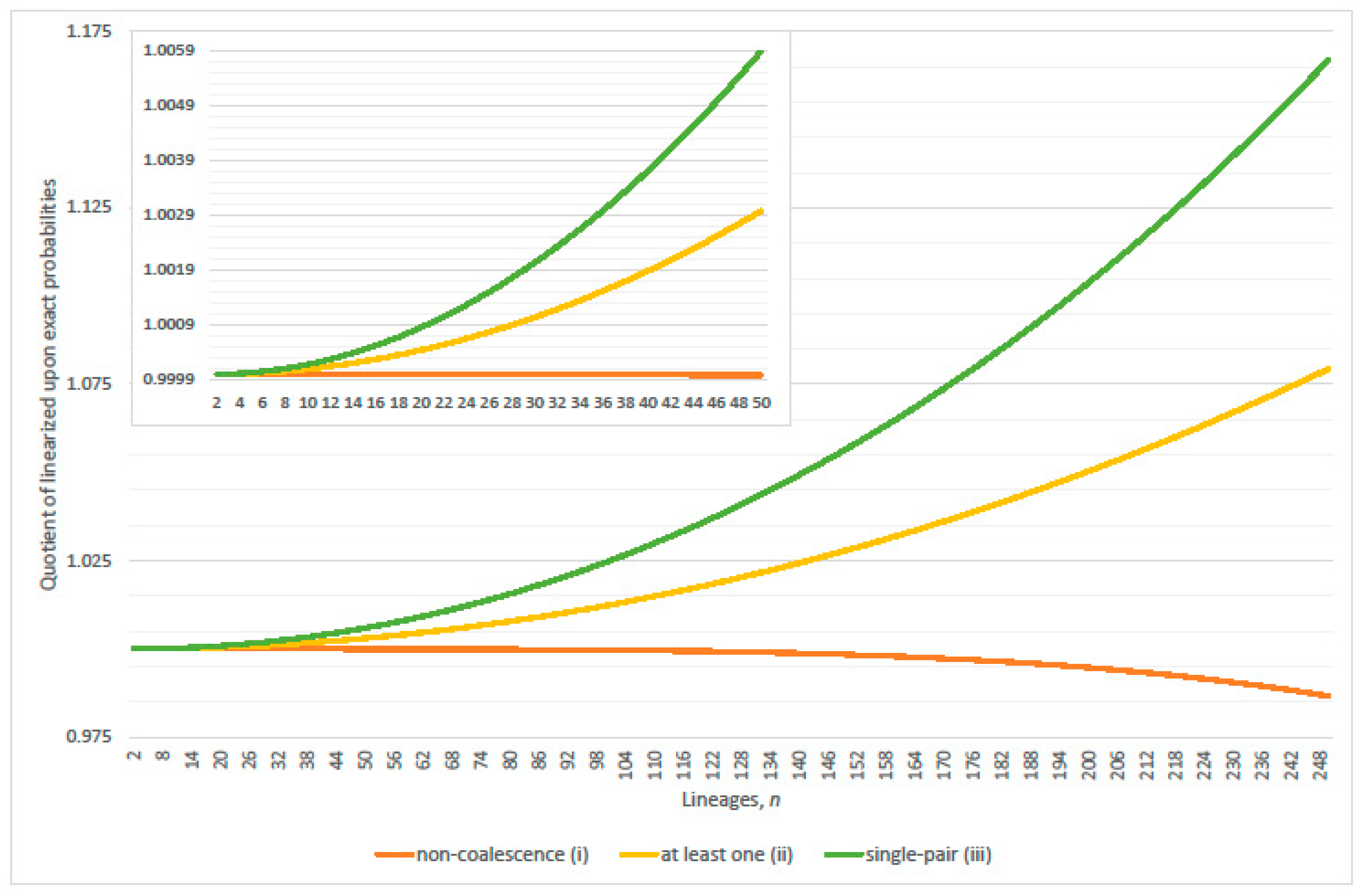
2.1. Zero Coalescence Events
2.2. Single Pair Coalescence Events
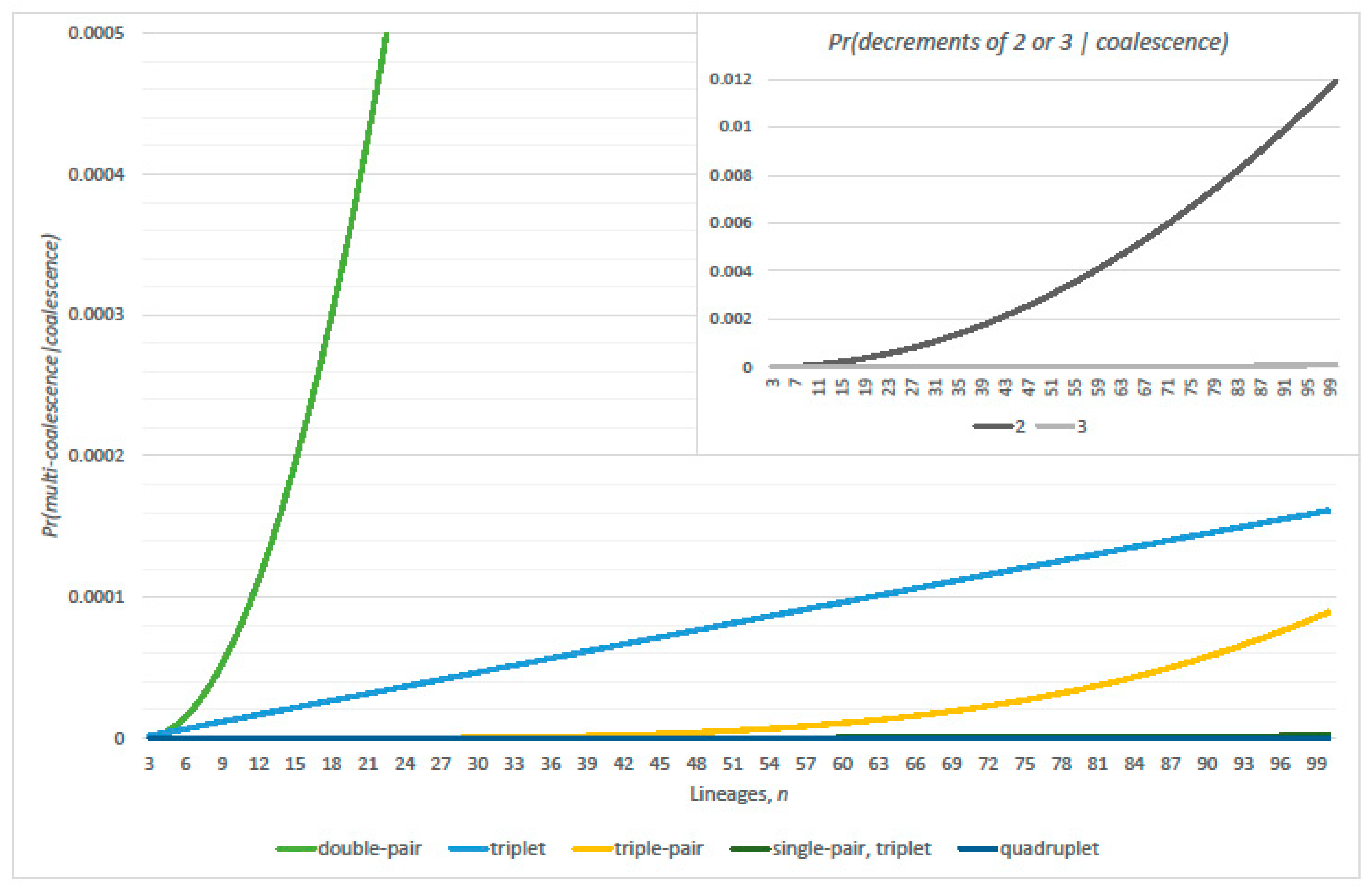
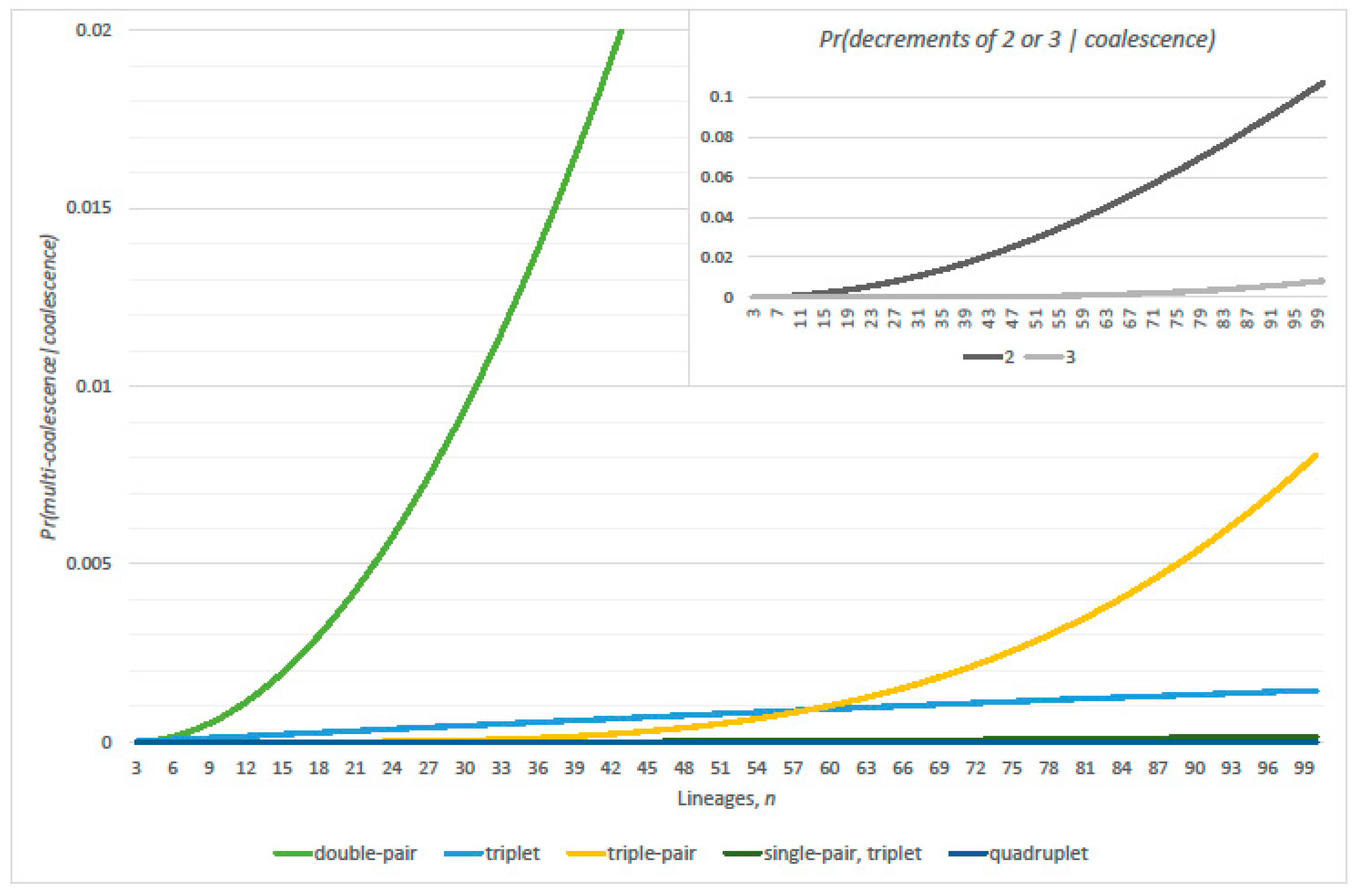
2.3. Multiple Coalescence Events
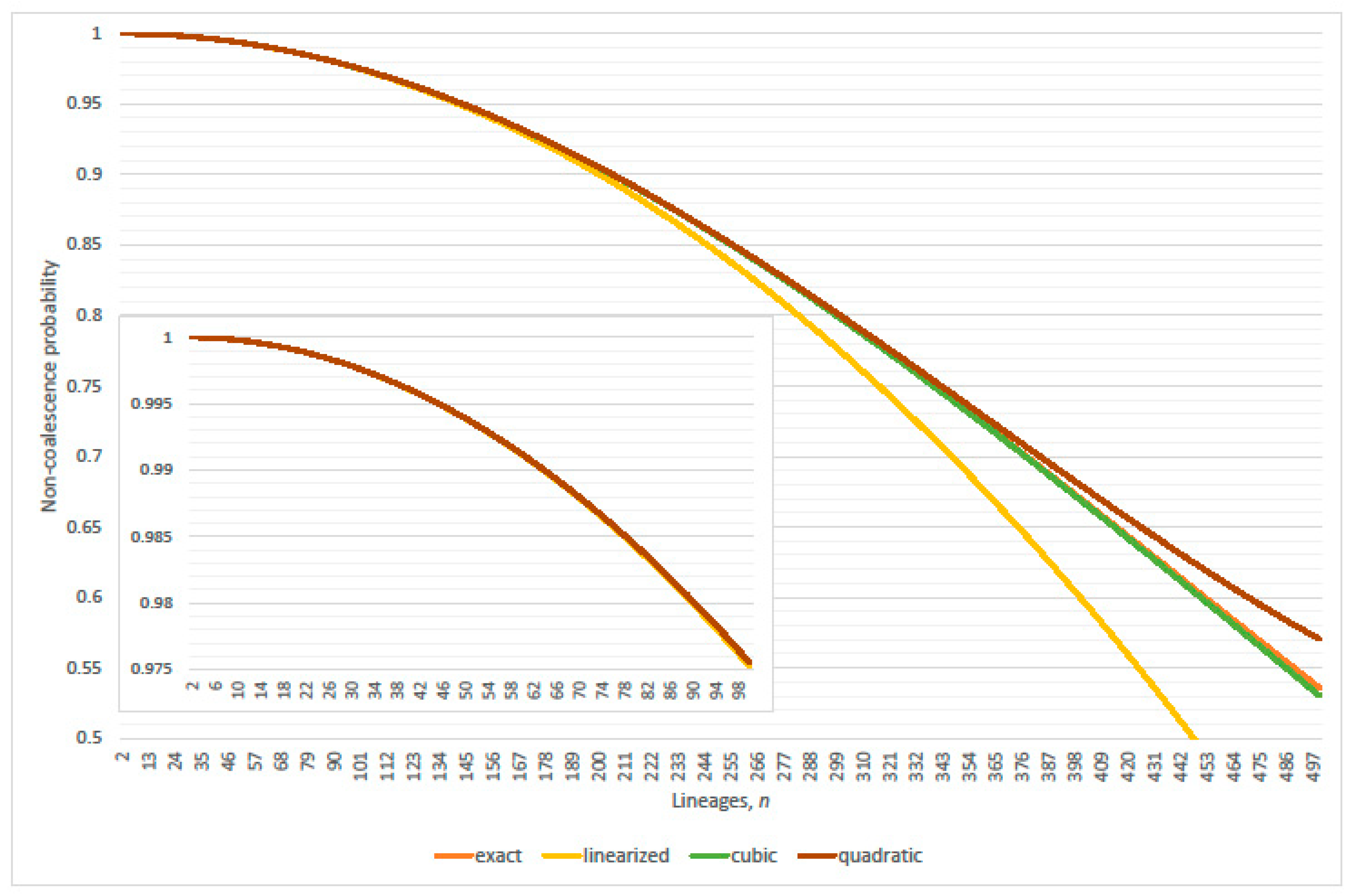
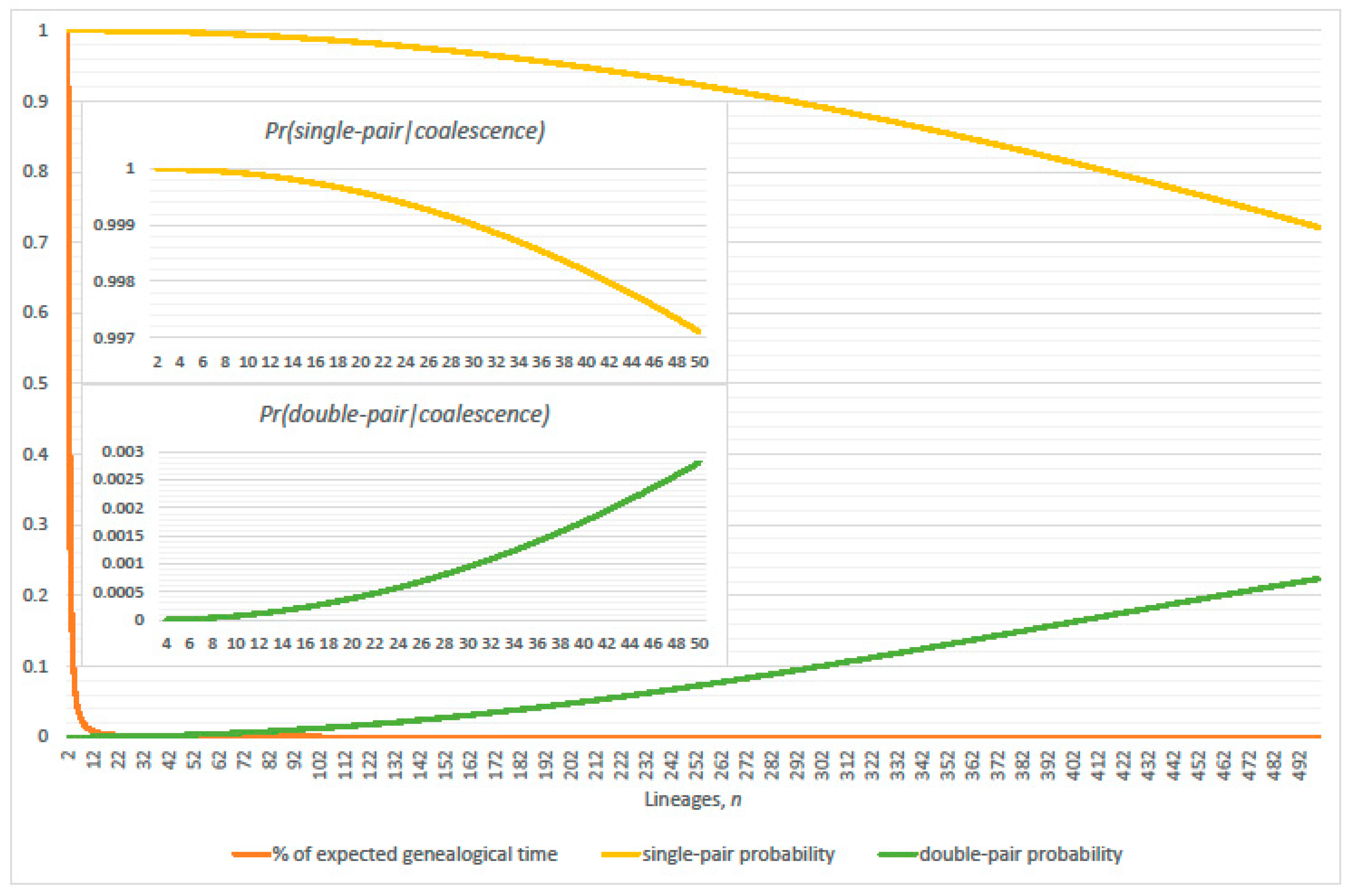
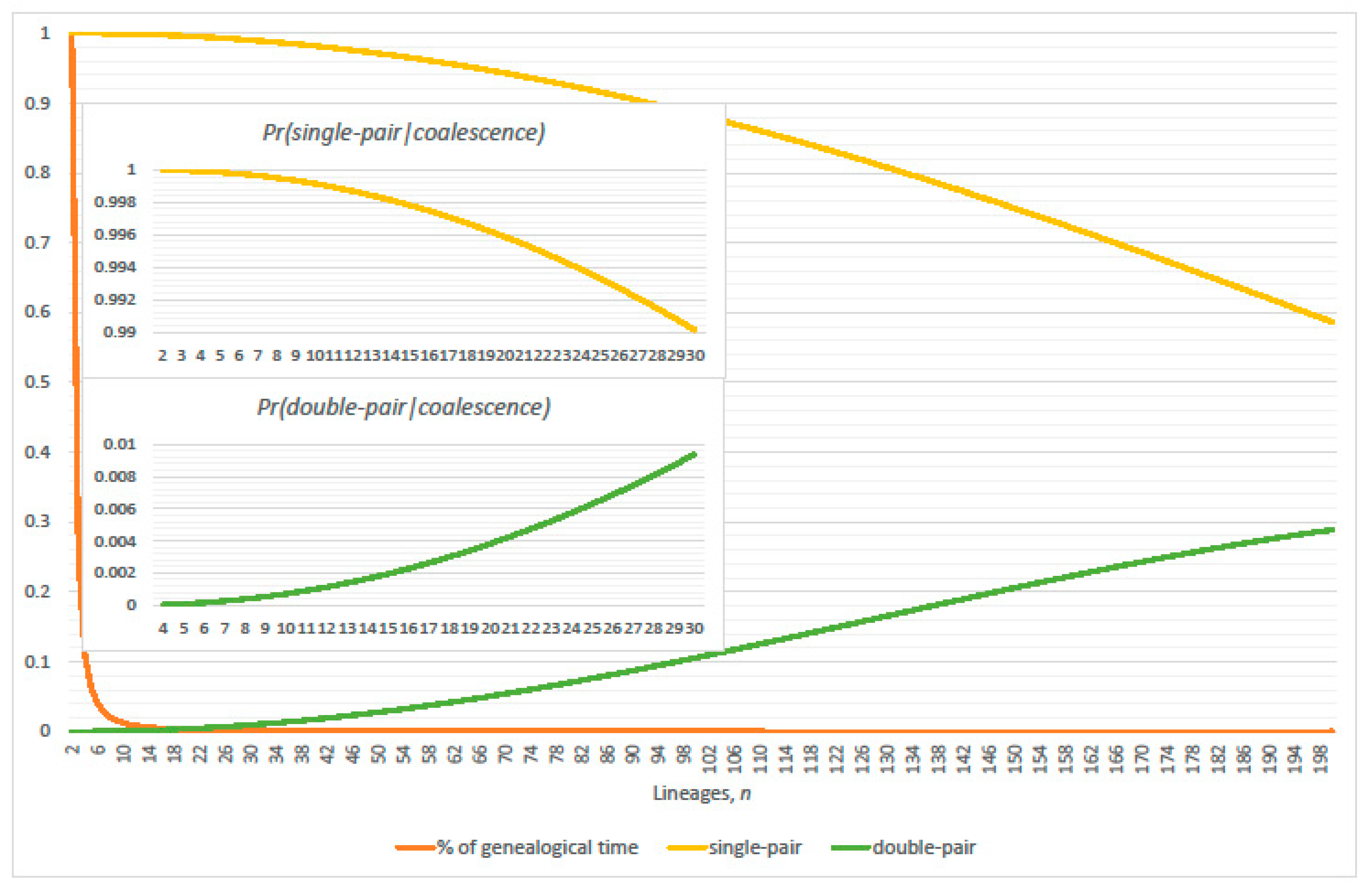
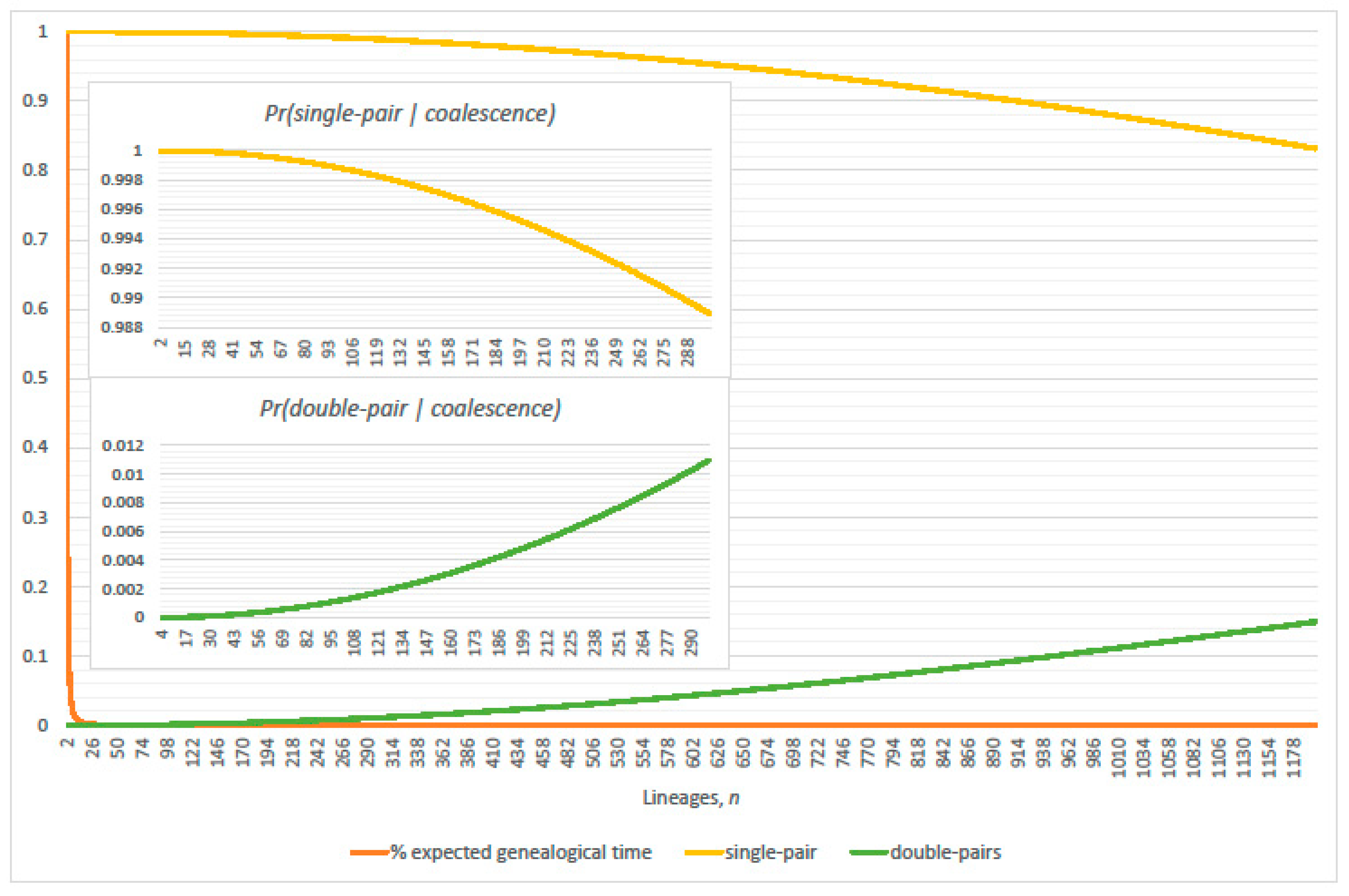
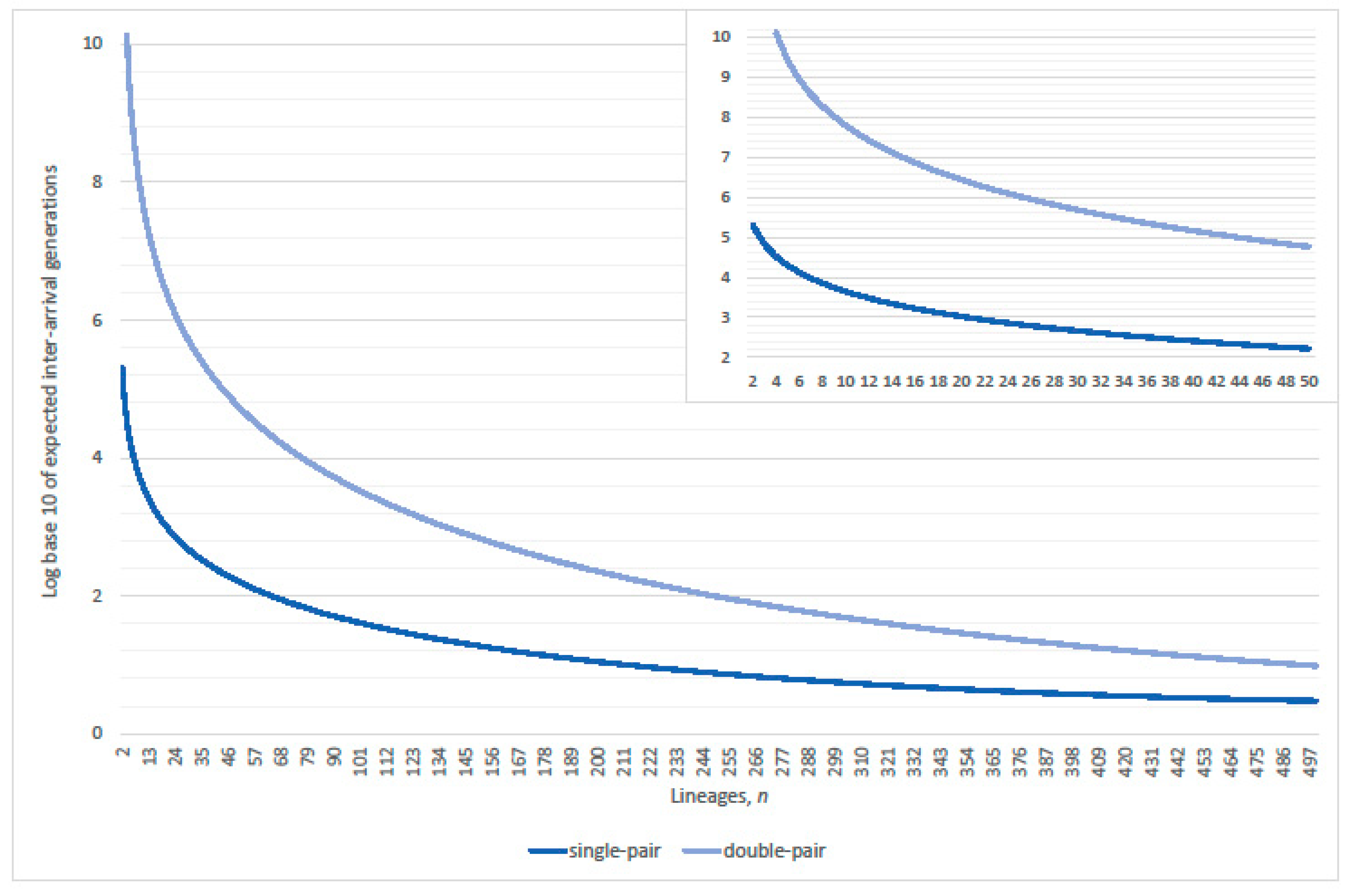
3. Genealogical Topology and Expected Inter-Arrival Generations
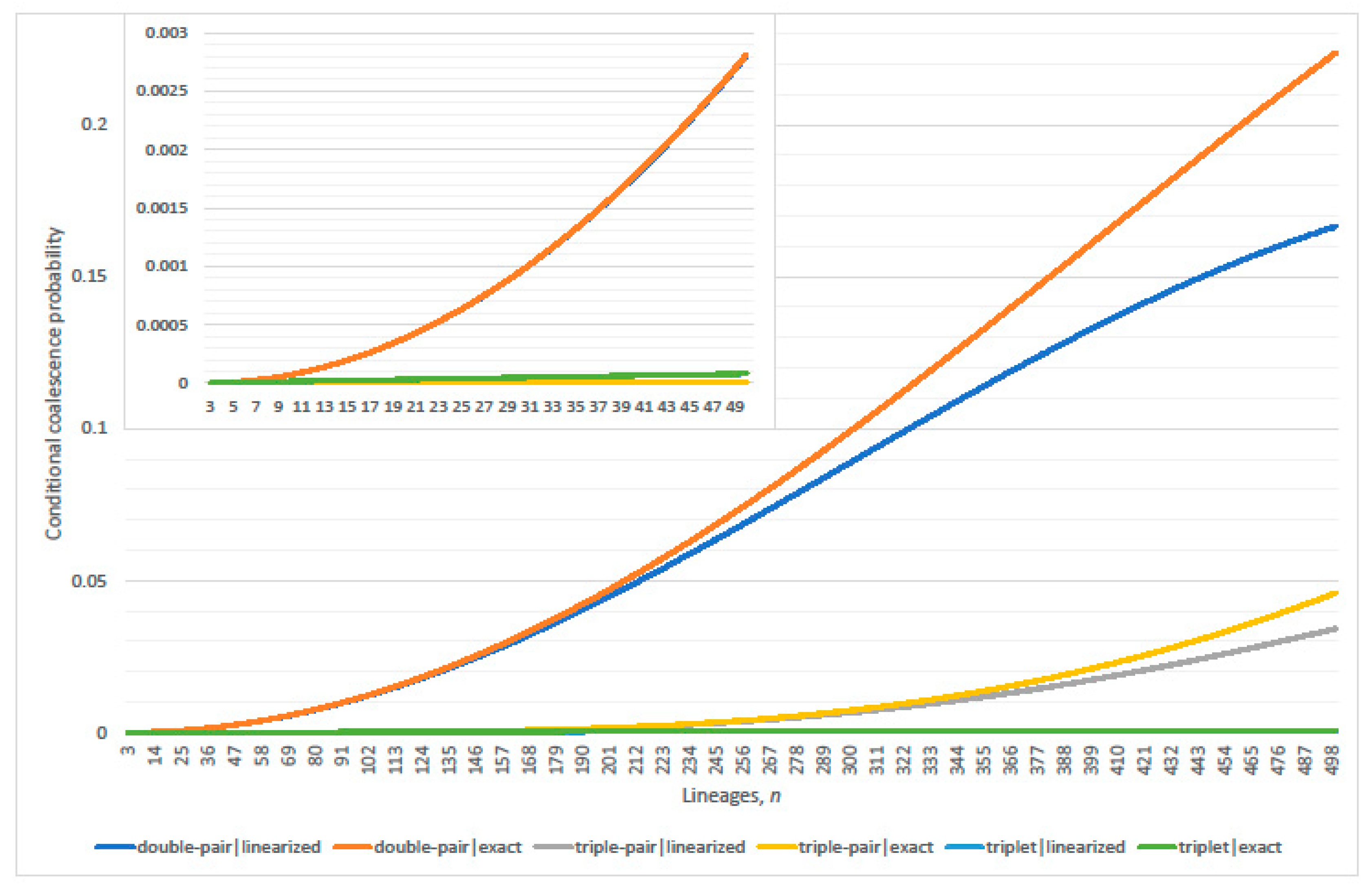
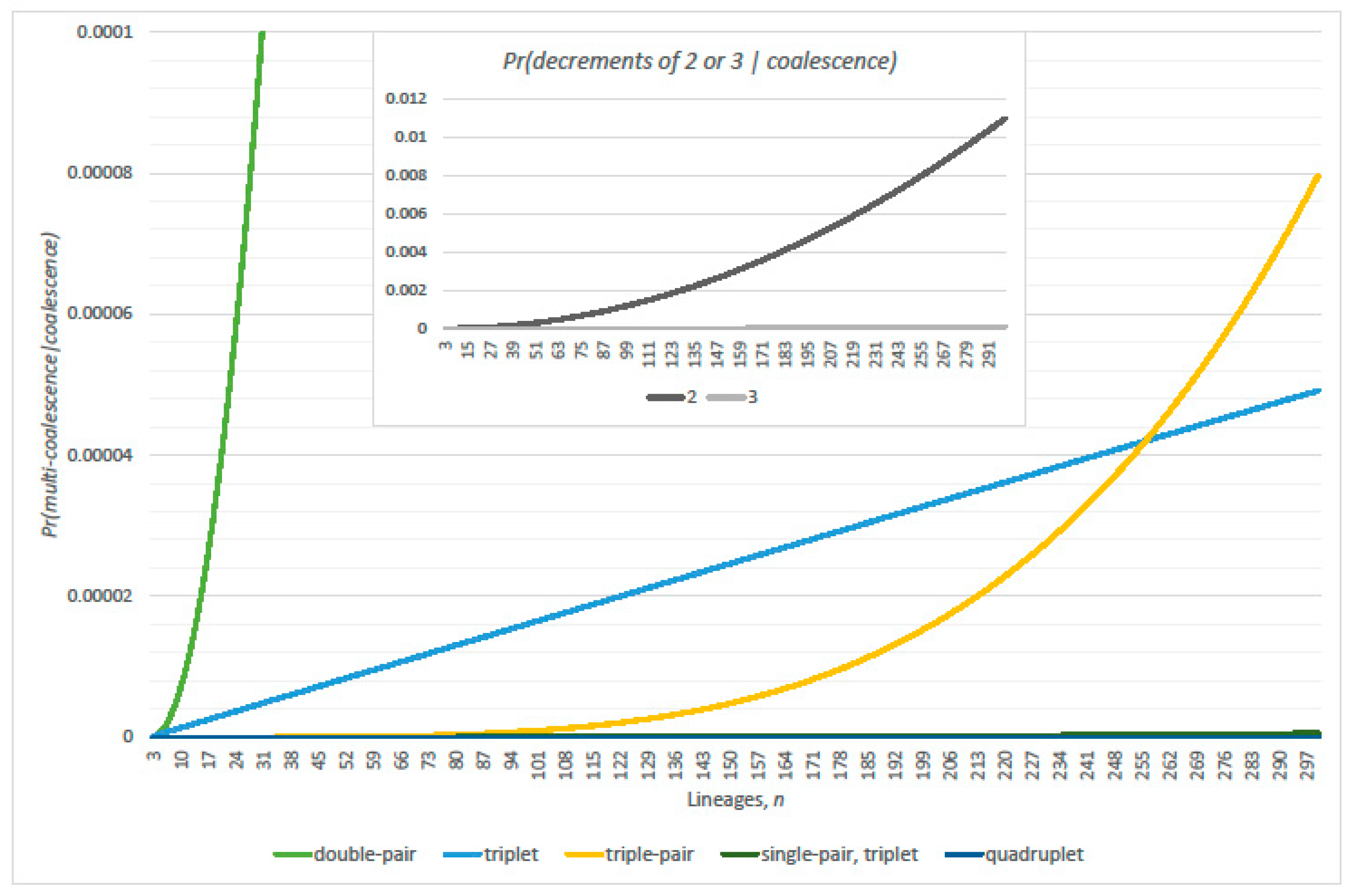
3.1. Conditional Probabilities of Multi-Coalescence
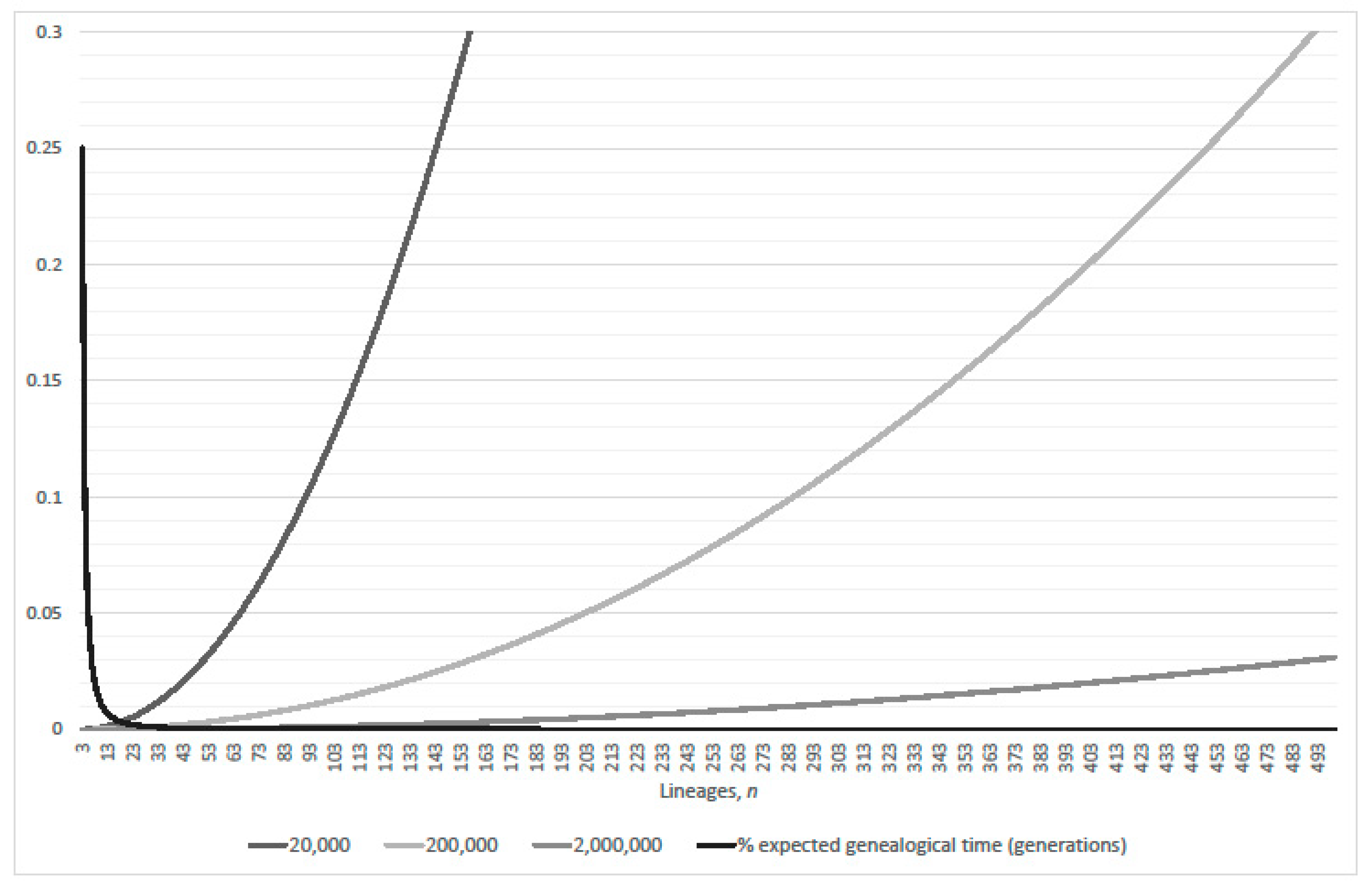
3.2. Single-Pairs Dominate Double-Pairs?
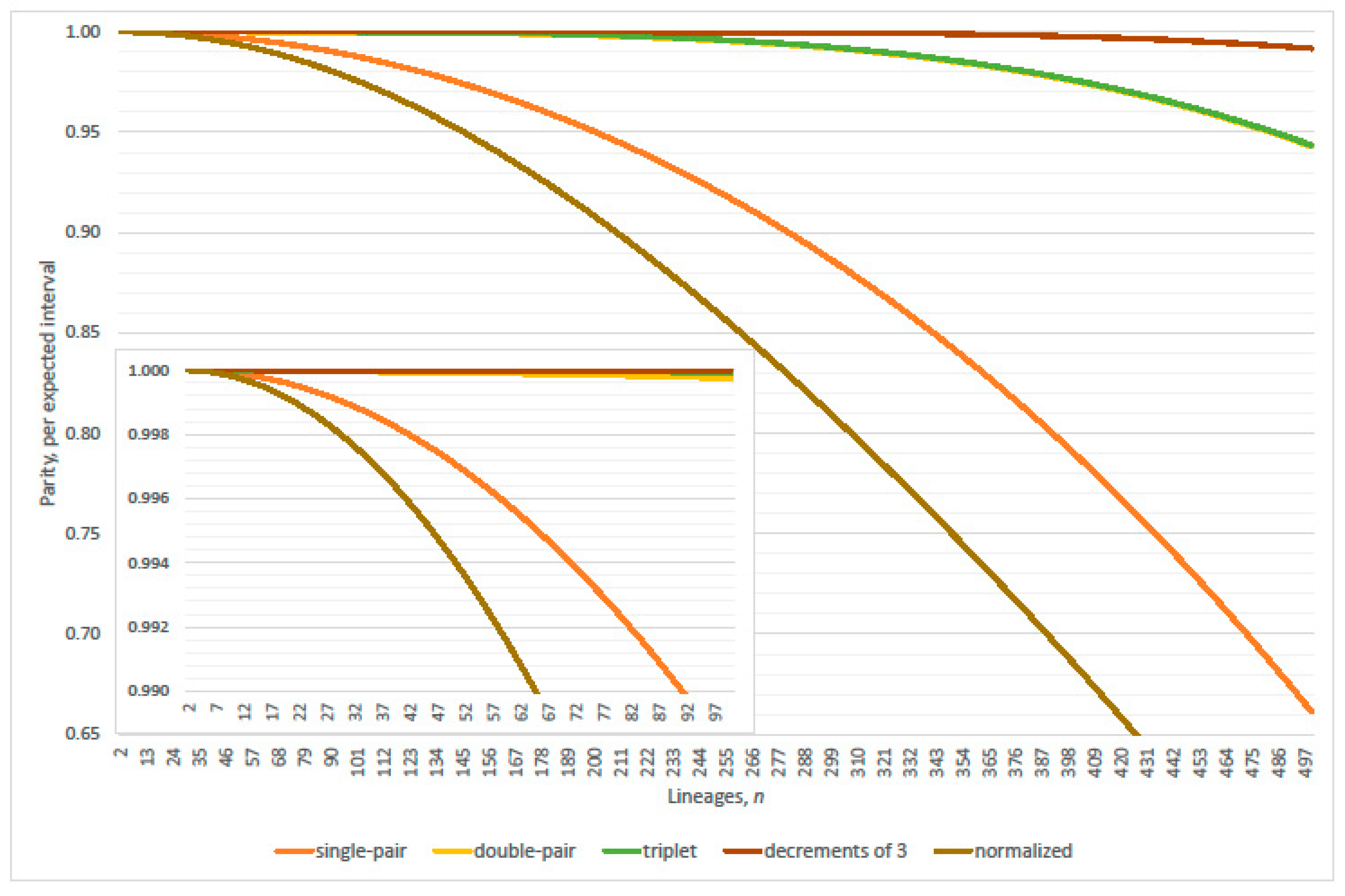
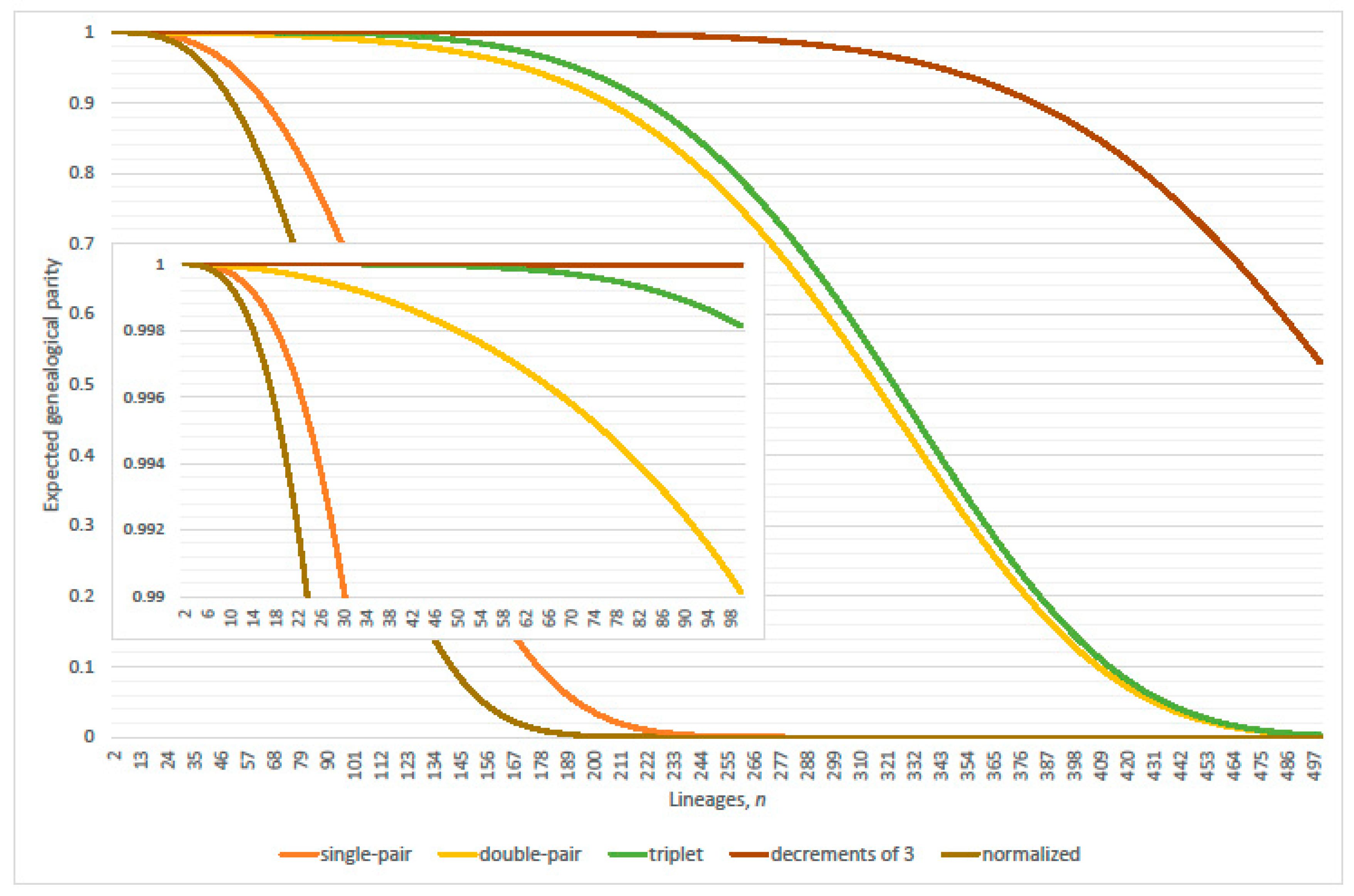
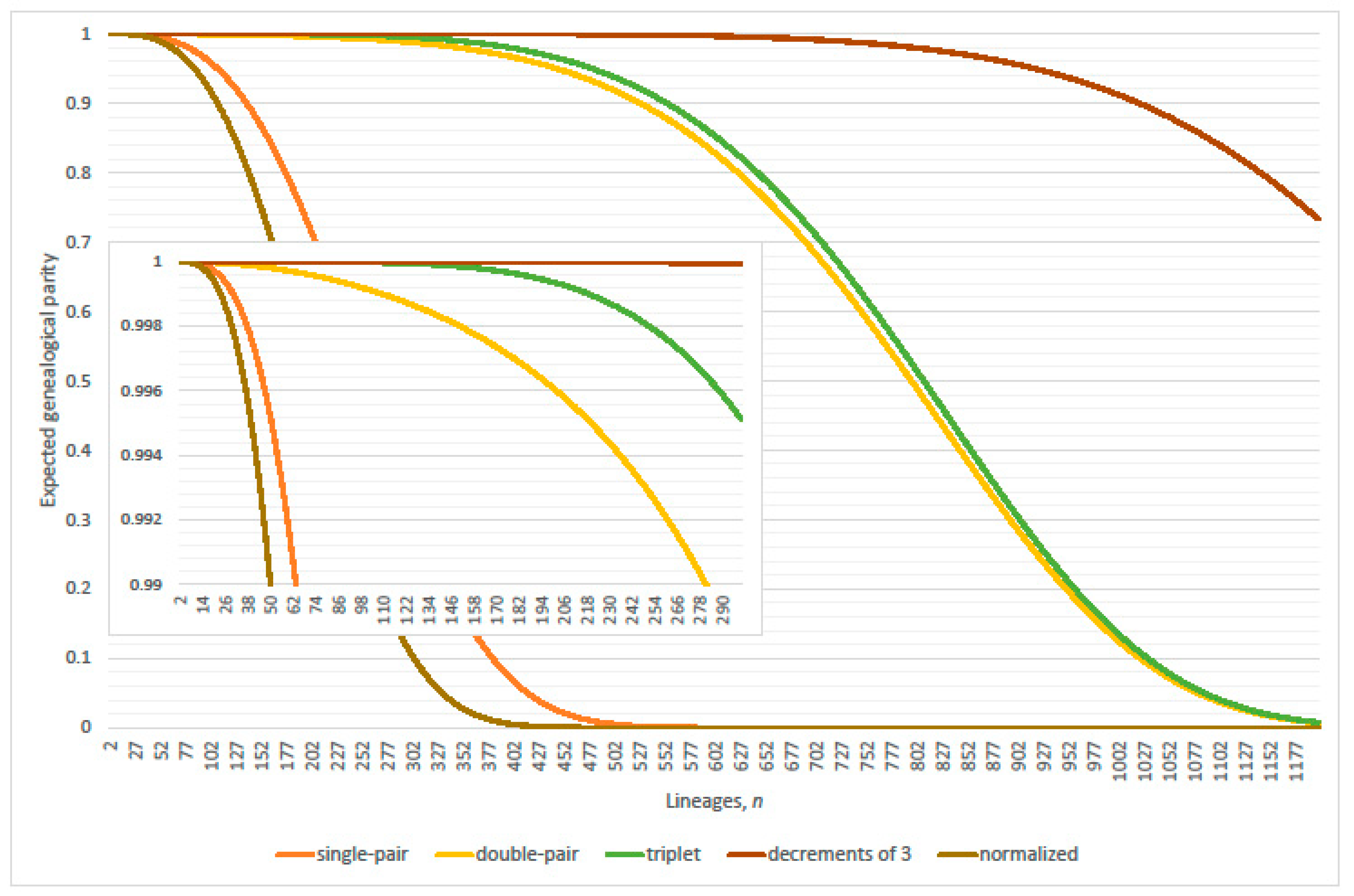
4. Parity of the Kingman Coalescent
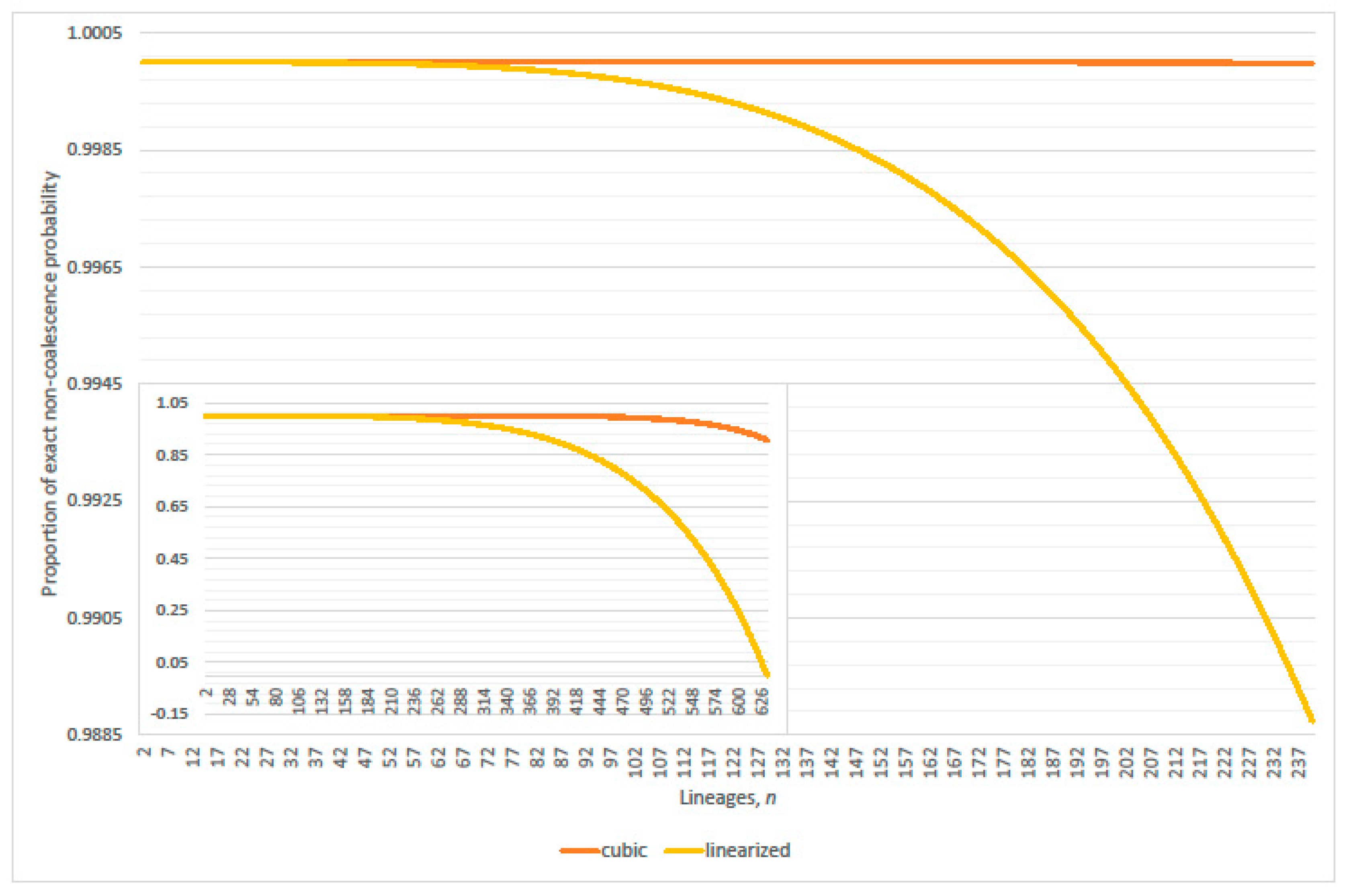
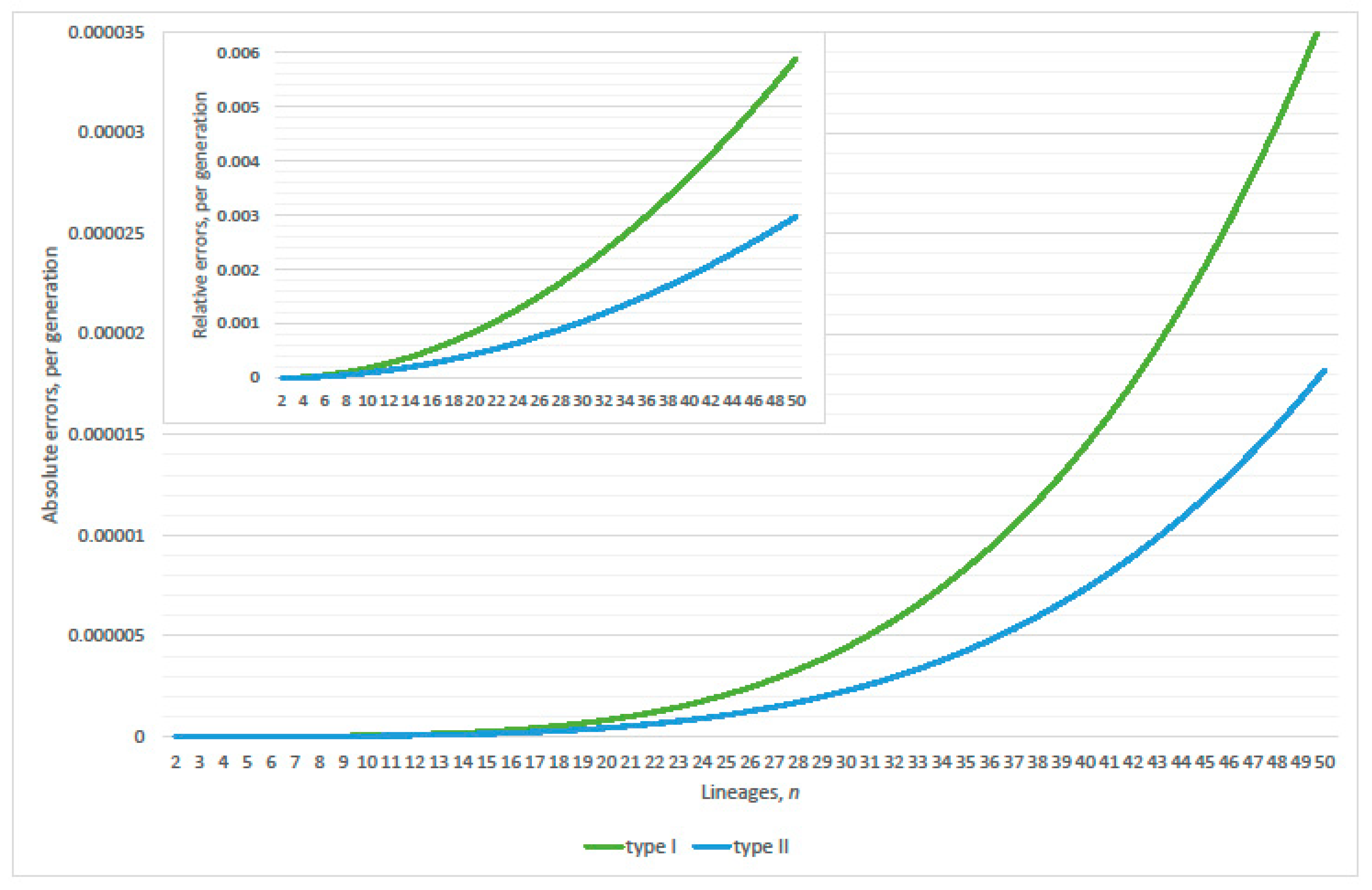
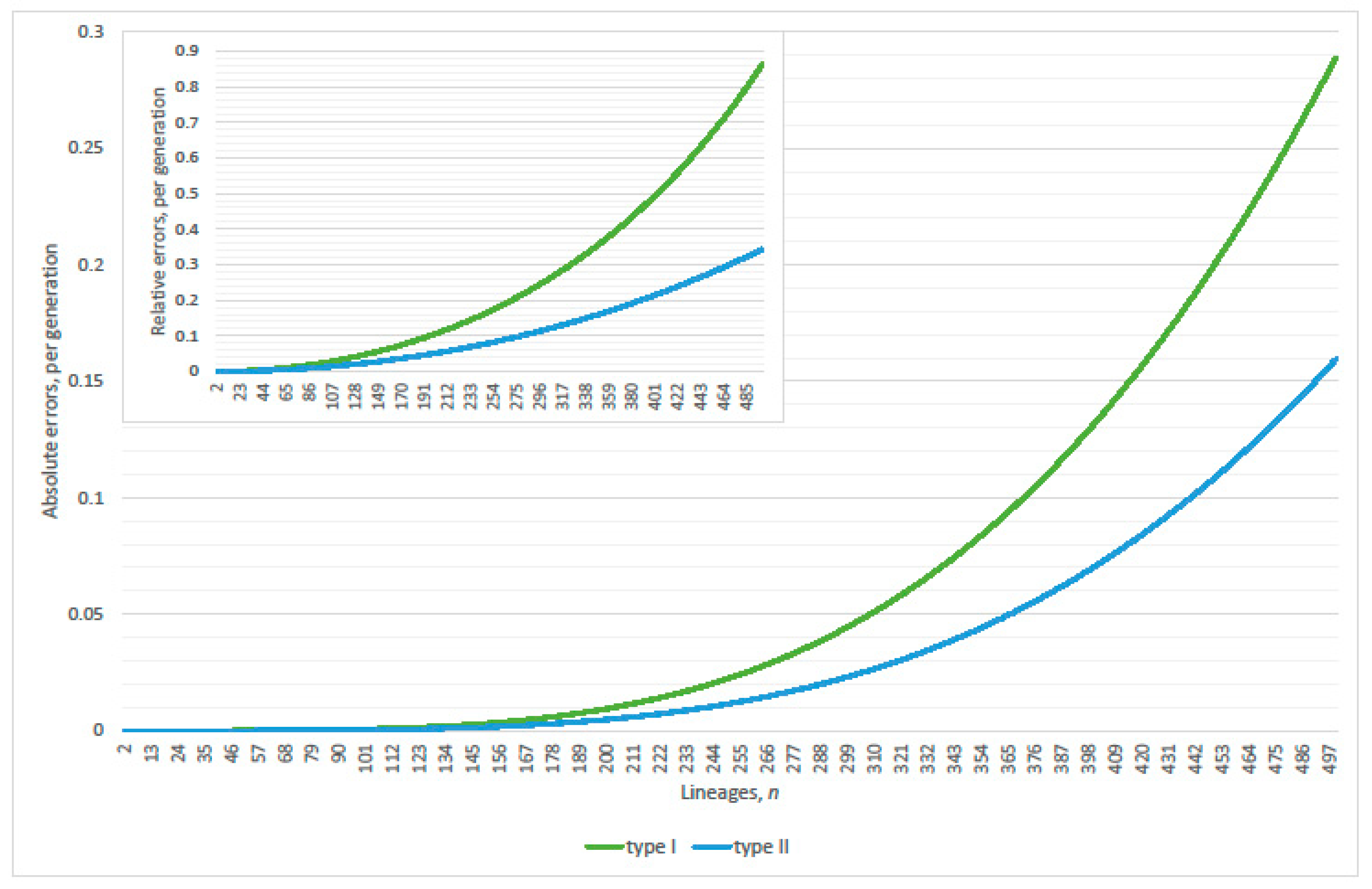
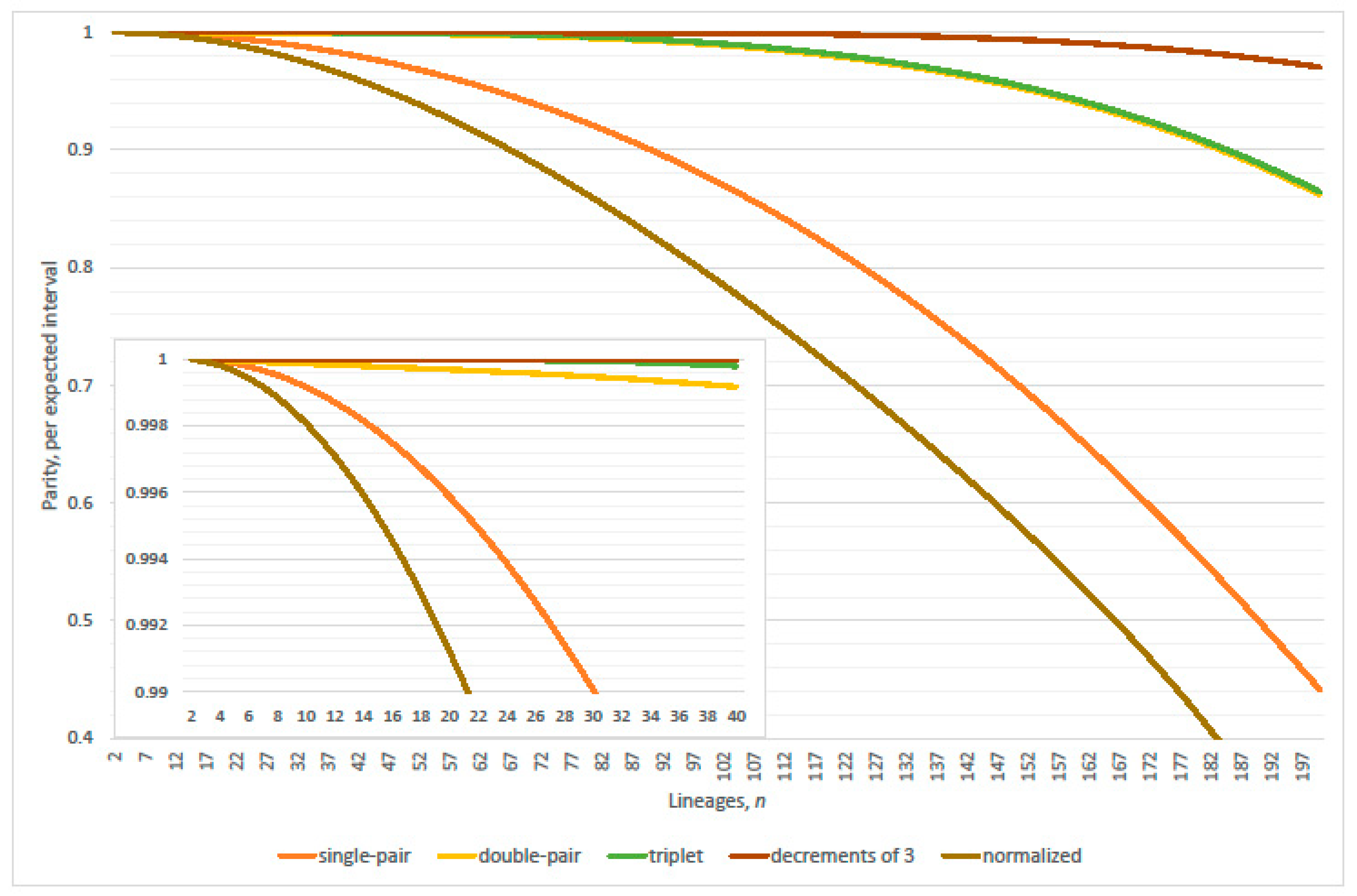
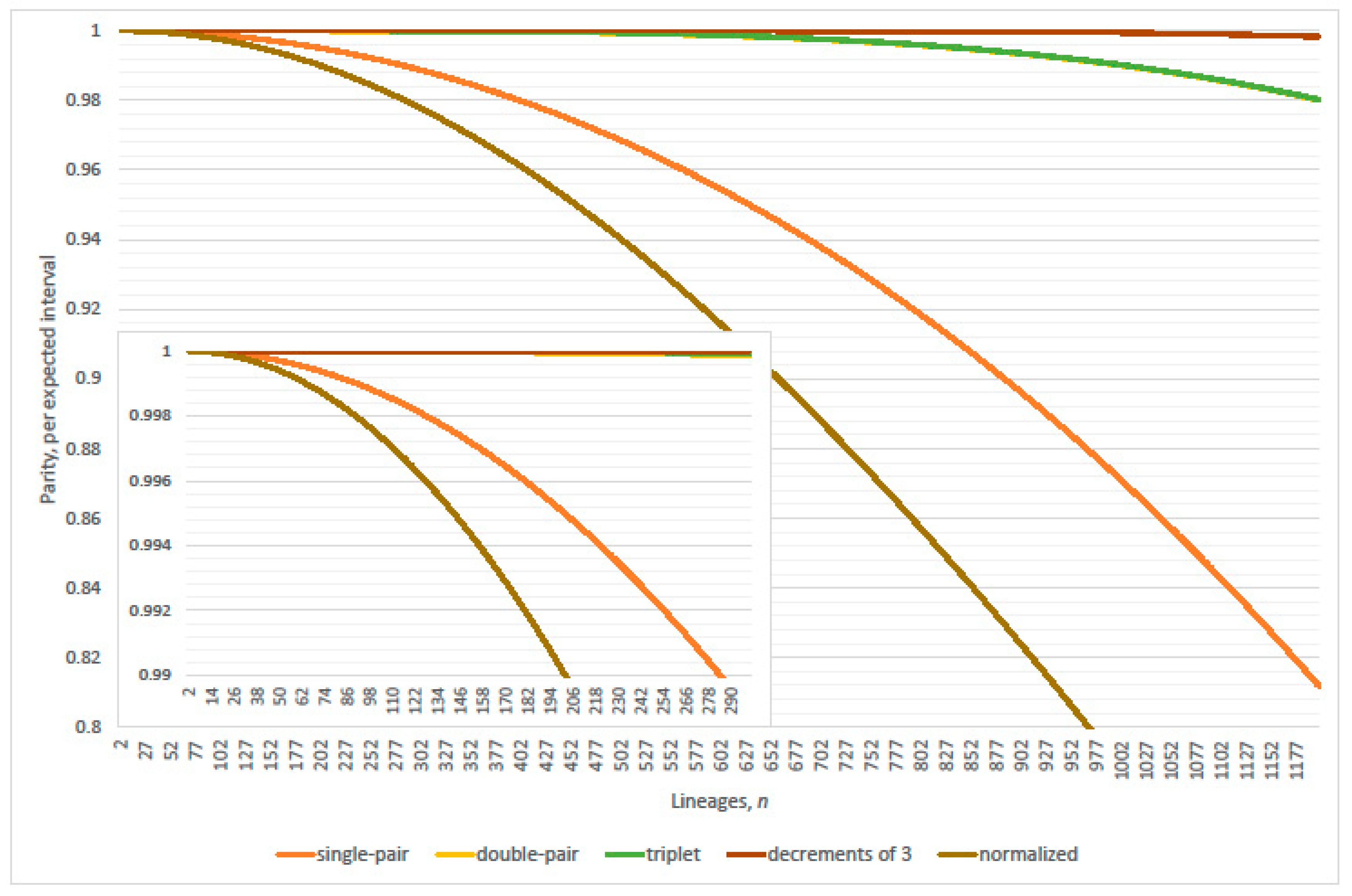
4.1. Linearization Errors
- parity, per expected interval, exceeds 99% where (approximately)(Table 2 verified this case where N = 2000; 20,000; 200,000; 2,000,000 and 20,000,000); and
- expected genealogical parity exceeds 99% where(precise, N = 20,000 and 200,000; plus one, N = 2,000,000 and 20,000,000; minus one, N = 2000).
- parity, per expected interval, exceeds 99% where(normalized parity criterion, per expected interval: n minus one N = 2000, 20,000; plus one 200,000; plus two 2,000,000; overestimates maximum lineages by 1.49% when 20,000,000); and
- expected genealogical parity exceeds 99% where(normalized expected genealogical parity criterion: precise, N = 2000, 20,000, 200,000 and 2,000,000; n plus one when 20,000,000).
4.2. Parity Paradox
5. Conclusions
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B
References
- Wakeley, J. Coalescent Theory: An Introduction, 1st ed.; Roberts and Company Publishers: Greenwood Village, CO, USA, 2009; ISBN 978-0-9747077-5-4. [Google Scholar]
- Hein, J.; Schierup, M.H.; Wiuf, C. Gene Genealogies, Variation and Evolution: A Primer in Coalescent Theory, 1st ed.; Oxford University Press: Oxford, UK, 2005; ISBN 0-19-852996-1. [Google Scholar]
- Tavaré, S. Ancestral inference in population genetics, Part 1. In Ecole d’Eté de Probabilités de Saint-Flour XXXI—2001, 1st ed.; Picard, J., Ed.; Lectures on Probability Theory and Statistics, 1837; Springer: Berlin/Heidelberg, Germany, 2004; pp. 1–188. ISBN 3-540-20832-1. [Google Scholar]
- Kingman, J.F.C. On the genealogy of large populations. J. Appl. Probab. 1982, 19, 27–43. [Google Scholar] [CrossRef]
- Kingman, J.F.C. The coalescent. Stoch. Proc. Appl. 1982, 13, 235–248. [Google Scholar] [CrossRef]
- Kingman, J.F.C. Exchangeability and the evolution of large populations, In Exchangeability in Probability and Statistics, 1st ed.; Koch, G., Spizzichino, F., Eds.; North-Holland: Amsterdam, The Netherlands, 1982; pp. 97–112. ISBN 04448644032. [Google Scholar]
- Kingman, J.F.C. Origins of the coalescent: 1974–1982. Genetics 2000, 156, 1461–1463. [Google Scholar] [PubMed]
- Yang, T.; Deng, H.W.; Niu, T. Critical assessment of coalescent simulators in modelling recombination hotspots in genomic sequences. BMC Bioinform. 2014, 15, 3. [Google Scholar] [CrossRef] [PubMed]
- Allman, E.S.; Degnan, J.H.; Rhodes, J.A. Identifying the rooted species tree from the distribution of unrooted gene trees under the coalescent. J. Math. Biol. 2011, 62, 833–862. [Google Scholar] [CrossRef] [PubMed]
- Steel, M. Phylogeny: Discrete and Random Processes in Evolution, 1st ed.; CMBS-NSF Regional Conference Series in Applied Mathematics 89; Society for Industrial and Applied Mathematics (SIAM): Philadelphia, PA, USA, 2016; ISBN 978-1-611974-47-8. [Google Scholar]
- Crane, H. The ubiquitous Ewens Sampling Formula. Stat. Sci. 2016, 31, 1–19. [Google Scholar] [CrossRef]
- Crane, H. Rejoinder: The ubiquitous Ewens Sampling Formula. Stat. Sci. 2016, 31, 37–39. [Google Scholar] [CrossRef]
- Kingman, J.F.C. The genealogy of the Wright-Fisher model, appendix II. In Mathematics of Genetic Diversity, 1st ed.; CMBS-NSF Regional Conference Series in Applied Mathematics 34; Society for Industrial and Applied Mathematics (SIAM): Philadelphia, PA, USA, 1980; pp. 63–66. ISBN 0-89871-166-5. [Google Scholar]
- Felsenstein, J. Trees of genes in populations, chapter 1. In Reconstructing Evolution: New Mathematical and Computational Advances, 1st ed.; Steel, M., Gascuel, O., Eds.; Oxford University Press: Oxford, UK, 2007; pp. 3–29. ISBN 978-0-19-920822-7. [Google Scholar]
- Wakeley, J.; Takahashi, T. Gene genealogies when the sample size exceeds the effective size of the population. Mol. Biol. Evol. 2003, 20, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.X. Exact coalescent for the Wright-Fisher model. Theor. Popul. Biol. 2006, 69, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Bhaskar, A.; Clark, A.G.; Song, Y.S. Distortion of genealogical properties when the sample is very large. Proc. Natl. Acad. Sci. USA 2014, 111, 2385–2390. [Google Scholar] [CrossRef] [PubMed]
- Wakeley, J. Coalescent theory has many new branches. Theor. Popul. Biol. 2013, 87, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Lessard, S. Recurrence equations for the probability distribution of sample configurations in exact population genetic models. J. Appl. Probab. 2010, 47, 732–751. [Google Scholar] [CrossRef][Green Version]
- Möhle, M. Robustness results for the coalescent. J. Appl. Probab. 1998, 35, 438–447. [Google Scholar] [CrossRef]
- Möhle, M. Ancestral processes in population genetics—The coalescent. J. Theor. Biol. 2000, 204, 629–638. [Google Scholar] [CrossRef] [PubMed]
- Möhle, M.; Sagitov, S. A classification of coalescent processes for haploid exchangeable population models. Ann. Probab. 2001, 29, 1547–1562. [Google Scholar] [CrossRef]
- Kingman, J.F.C. Random discrete distributions. J. R. Stat. Soc. B 1975, 37, 1–22. [Google Scholar]
- Kingman, J.F.C. Random partitions in population genetics. Proc. R. Soc. Lond. A 1978, 361, 1–20. [Google Scholar] [CrossRef]
- Kingman, J.F.C. The representation of partition structures. J. Lond. Math. Soc. 1978, 18, 374–380. [Google Scholar] [CrossRef]
- Sagitov, S. The general coalescent with asynchronous mergers of ancestral lines. J. Appl. Probab. 1999, 36, 1116–1125. [Google Scholar] [CrossRef]
- Pitman, J. Coalescents with multiple collisions. Ann. Probab. 1999, 27, 1870–1902. [Google Scholar] [CrossRef]
- Sagitov, S. Convergence to the coalescent with simultaneous multiple mergers. J. Appl. Probab. 2003, 40, 839–854. [Google Scholar] [CrossRef]
- Sargsyan, O.; Wakeley, J. A coalescent process with simultaneous multiple mergers for approximating the gene genealogies of many marine organisms. Theor. Popul. Biol. 2008, 74, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, P.; Kurtz, T. Particle representations for measure-valued population models. Ann. Probab. 1999, 27, 166–205. [Google Scholar] [CrossRef]
- Birkner, M.; Blath, J.; Capaldo, M.; Etheridge, A.; Möhle, M.; Schweinsberg, J.; Wakolbinger, A. α-stable branching and β-coalescents. Electron. J. Probab. 2005, 10, 303–325. [Google Scholar] [CrossRef]
- Steinrücken, M.; Birkner, M.; Blath, J. Analysis of DNA sequence variation within marine species using β-coalescents. Theor. Popul. Biol. 2013, 87, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Heuer, B.; Sturm, A. On spatial coalescents with multiple mergers in two dimensions. Theor. Popul. Biol. 2013, 87, 90–104. [Google Scholar] [CrossRef] [PubMed]
- Huillet, T.; Möhle, M. On the extended Moran model and its relation to coalescents with multiple collisions. Theor. Popul. Biol. 2013, 87, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Dong, R.; Gnedin, A.; Pitman, J. Exchangeable partitions derived from Markovian coalescents. Ann. Appl. Probab. 2007, 17, 1172–1201. [Google Scholar] [CrossRef]
- Freund, F.; Möhle, M. On the number of allelic types for samples taken from exchangeable coalescents with mutation. Adv. Appl. Probab. 2009, 41, 1082–1101. [Google Scholar] [CrossRef][Green Version]
- Bertoin, J. The structure of the allelic partition of the total population for Galton-Watson processes with neutral mutations. Ann. Probab. 2009, 37, 1502–1523. [Google Scholar] [CrossRef]
- Burden, C.J.; Simon, H. Genetic drift in populations governed by a Galton-Watson branching process. Theor. Popul. Biol. 2016, 109, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L. fsc26 Manual, online documentation for Fastsimcoal Version 2.6, Swiss Institute of Bioinformatics, Lausanne, Switzerland. 2016. Available online: http://cmpg.unibe.ch/software/fastsimcoal2 (accessed on 23 November 2017).
- Excoffier, L.; Dupanloup, I.; Huerta-Sánchez, E.; Foll, M. Robust demographic inference from genomic and SNP data. PLoS Genet. 2013, 9, e1003905. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Foll, M. Fastsimcoal: A continuous-time coalescent simulator of genomic diversity under arbitrarily complex evolutionary scenarios. Bioinformatics 2011, 27, 1332–1334. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Novembre, J.; Schneider, S. SIMCOAL: A general coalescent program for the simulation of molecular data in interconnected populations with arbitrary demography. J. Hereditary 2000, 91, 506–509. [Google Scholar] [CrossRef]
- Anderson, C.N.K.; Ramakrishnan, U.; Chan, Y.L.; Hadley, E.A. Serial SimCoal: A population genetics model for data from multiple populations and points in time. Bioinformatics 2005, 21, 1733–1734. [Google Scholar] [CrossRef] [PubMed]
- Rudman, S.A.; Barbour, M.A.; Csillérry, K.; Gienapp, P.; Guillaume, F.; Hairston, N.G., Jr.; Hendry, A.P.; Lasky, J.R.; Rafajlović, M.; Räsänen, K.; et al. What genomic data can reveal about eco-evolutionary dynamics. Nature Ecol. Evol. 2018, 2, 9–15. [Google Scholar] [CrossRef] [PubMed]




| Population Size, N | Single-Pair | Double-Pair | Triplet | Decrements of Three |
|---|---|---|---|---|
| 2000 | 10 | 29 | 34 | 52 |
| 20,000 | 30 | 98 | 102 | 164 |
| 200,000 | 91 | 316 | 319 | 516 |
| 2,000,000 | 284 | 1002 | 1004 | 1633 |
| 20,000,000 | 895 | 3174 | 3178 | 5159 |
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Slade, P.F. Linearization of the Kingman Coalescent. Mathematics 2018, 6, 82. https://doi.org/10.3390/math6050082
Slade PF. Linearization of the Kingman Coalescent. Mathematics. 2018; 6(5):82. https://doi.org/10.3390/math6050082
Chicago/Turabian StyleSlade, Paul F. 2018. "Linearization of the Kingman Coalescent" Mathematics 6, no. 5: 82. https://doi.org/10.3390/math6050082
APA StyleSlade, P. F. (2018). Linearization of the Kingman Coalescent. Mathematics, 6(5), 82. https://doi.org/10.3390/math6050082





