Estimating the Relative Risks of Spatial Clusters Using a Predictor–Corrector Method
Abstract
1. Introduction
2. Retrospective Analysis of Spatial Clusters
2.1. Overview
2.2. U.S. COVID-19 Mortality Data
2.3. Spatial Statistical Scan
2.4. Significant Clusters Identified
3. Modeling Mortality Risk Prediction
3.1. The Markov Chain Model
3.2. Estimating the Parameters of
3.3. Predictors of the Transition Matrix
3.3.1. Derivation of (Exponential Smoothing)
3.3.2. Derivation of (Multiple Linear Regression)
4. Predicting the Relative Risks of Clusters
4.1. Exponential Smoothing
4.2. Multiple Linear Regression
5. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kulldorff, M. A Spatial Scan Statistic. Commun. Stat.-Theory Methods 1997, 26, 1481–1496. [Google Scholar] [CrossRef]
- Kulldorff, M.; Mostashari, F.; Duczmal, L.; Yih, K.; Kleinman, K.; Platt, R. Multivariate spatial scan statistics for disease surveillance. Stat. Med. 2007, 26, 1824–1833. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Huang, L.; Tiwari, R.; Zuo, J.; Kulldorff, M.; Feuer, E. Weighted normal spatial scan statistic for heterogeneous population data. J. Am. Stat. Assoc. 2009, 104, 886–898. [Google Scholar] [CrossRef]
- Kulldorff, M.; Huang, L.; Pickle, L.; Duczmal, L. An elliptic spatial scan statistic. Stat. Med. 2006, 25, 3929–3943. [Google Scholar] [CrossRef]
- Nordsborg, R.B.; Sloan, C.D.; Shahid, H.; Jacquez, G.M.; De Roos, A.J.; Cerhan, J.R.; Cozen, W.; Severson, R.; Ward, M.H.; Morton, L. Investigation of spatio-temporal cancer clusters using residential histories in a case–control study of non-Hodgkin lymphoma in the United States. Environ. Health 2015, 14, 48. [Google Scholar] [CrossRef] [PubMed]
- Amin, R.; Hall, T.; Church, J.; Schlierf, D.; Kulldorff, M. Geographical surveillance of COVID-19: Diagnosed cases and death in the United States. MedRxiv 2020. [Google Scholar] [CrossRef]
- Tran, T.; Bani-Yaghoub, M.; DeLisle, J.R. Non-emergency responses in the 311 system during the early stage of the COVID-19 pandemic: A case study of Kansas city. Disaster Prev. Resil. 2023, 2, 3. [Google Scholar]
- AlQadi, H.; Bani-Yaghoub, M. Incorporating global dynamics to improve the accuracy of disease models: Example of a COVID-19 SIR model. PLoS ONE 2022, 17, e0265815. [Google Scholar] [CrossRef] [PubMed]
- AlQadi, H.; Bani-Yaghoub, M.; Balakumar, S.; Wu, S.; Francisco, A. Assessment of retrospective COVID-19 spatial clusters with respect to demographic factors: Case study of Kansas City, Missouri, United States. Int. J. Environ. Res. Public Health 2021, 18, 11496. [Google Scholar] [CrossRef] [PubMed]
- AlQadi, H.; Bani-Yaghoub, M.; Wu, S.; Balakumar, S.; Francisco, A. Prospective spatial-temporal clusters of COVID-19 in local communities: Case study of Kansas City, Missouri, United States. Epidemiol. Infect. 2023, 151, e178. [Google Scholar] [CrossRef] [PubMed]
- Andrade, L.A.; Gomes, D.S.; Lima, S.V.M.A.; Duque, A.M.; Melo, M.S.; Góes, M.A.O.; Ribeiro, C.J.N.; Peixoto, M.V.S.; Souza, C.D.F.; Santos, A.D. COVID-19 Mortality in an area of northeast Brazil. Epidemiol. Infect. 2020, 148, e288. [Google Scholar] [CrossRef] [PubMed]
- Leveau, C.M. Variaciones espacio-temporales de la mortalidad por COVID-19 en barrios de la Ciudad Autónoma de Buenos Aires. Rev. Argent. Salud Pública 2021, 13, 9. [Google Scholar]
- Bonnet, E.; Bodson, O.; Le Marcis, F.; Faye, A.; Sambieni, N.E.; Fournet, F.; Boyer, F.; Coulibaly, A.; Kadio, K.; Diongue, F.B.; et al. The COVID-19 Pandemic in Francophone West Africa. Res. Sq. 2020, 21, 1490. [Google Scholar]
- Besag, J.; Clifford, P. Sequential Monte Carlo p-values. Biometrika 1991, 78, 301–330. [Google Scholar] [CrossRef]
- Silva, I.; Assunção, R.M.; Costa, M. Power of the sequential Monte Carlo test. Seq. Anal. 2009, 28, 163–174. [Google Scholar] [CrossRef]
- Tchole, A.I.M.; Li, Z.W.; Wei, J.T.; Ye, R.Z.; Wang, W.J.; Du, W.Y.; Wang, H.T.; Yin, C.N.; Ji, X.K.; Xue, F.Z.; et al. Epidemic and control of COVID-19 in Niger. J. Glob. Health 2020, 10, 020513. [Google Scholar] [CrossRef] [PubMed]
- Badaloni, C.; Asta, F.; Michelozzi, P.; Mataloni, F.; Di Rosa, E.; Scognamiglio, P.; Vairo, F.; Davoli, M.; Leone, M. Spatial analysis for detecting clusters of cases during the COVID-19 emergency in Rome and in the Lazio Region (Central Italy). Epidemiol. Prev. 2020, 44, 144–151. [Google Scholar] [PubMed]
- Zhang, Y.; Shen, Z.; Ma, C.; Jiang, C.; Feng, C.; Shanker, N.; Yang, P.; Sun, W.; Wang, Q. Avian Influenza A (H7N9) cluster analysis. Int. J. Environ. Res. Public Health 2015, 12, 816–828. [Google Scholar] [CrossRef] [PubMed]
- Souris, M.; Selenic, D.; Khaklang, S.; Ninphanomchai, S.; Minet, G.; Gonzalez, J.P.; Kittayapong, P. Poultry farm vulnerability and avian influenza risk. Int. J. Environ. Res. Public Health 2014, 11, 934–951. [Google Scholar] [CrossRef]
- Baygents, G.; Bani-Yaghoub, M. Cluster analysis of hemorrhagic disease in Missouri’s white-tailed deer population: 1980–2013. BMC Ecol. 2018, 18, 35. [Google Scholar] [CrossRef]
- Gautam, R.; Srinath, I.; Clavijo, A.; Szonyi, B.; Bani-Yaghoub, M.; Park, S.; Ivanek, R. Identifying areas of high risk of human exposure to coccidioidomycosis in Texas using serology data from dogs. Zoonoses Public Health 2013, 60, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.S.; Yokoe, D.S.; Stelling, J.; Placzek, H.; Kulldorff, M.; Kleinman, K.; O’Brien, T.F.; Calderwood, M.S.; Vostok, J.; Dunn, J.; et al. Automated outbreak detection in hospitals. PLoS Med. 2010, 7, e1000238. [Google Scholar] [CrossRef] [PubMed]
- Abboud, C.S.; Monteiro, J.; Franca, J.I.D.; Pignatari, A.C.; De Souza, E.E.; Camargo, E.C.G.; Monteiro, A.M.V.; Dos Santos, R.G.; Kiffer, C.R.V. A space-time model for carbapenemase-producing Klebsiella pneumoniae in hospitals. Epidemiol. Infect. 2015, 143, 2648–2652. [Google Scholar] [CrossRef] [PubMed]
- Natale, A.; Stelling, J.; Meledandri, M.; Messenger, L.A.; D’Ancona, F. Detection of antimicrobial resistance clusters. Eurosurveillance 2017, 22, 30484. [Google Scholar] [PubMed]
- Kleinman, K.P.; Abrams, A.M.; Kulldorff, M.; Platt, R. Space-time scan statistic in syndromic surveillance. Epidemiol. Infect. 2005, 133, 409–419. [Google Scholar] [CrossRef]
- Horst, M.A.; Coco, A.S. Spread of illnesses in a community using GIS. J. Am. Board Fam. Med. 2010, 23, 32–41. [Google Scholar] [CrossRef]
- Whittaker, J.A.; Rekab, K.; Thomason, M.G. A Markov chain model for predicting the reliability of multi-build software. Inf. Softw. Technol. 2000, 42, 889–894. [Google Scholar] [CrossRef]
- Coronavirus (COVID-19) Data in the United States, 2021. The New York Times. 21 June 2023. Available online: https://github.com/nytimes/covid-19-data (accessed on 12 May 2023).
- Block, R. Software review: Scanning for clusters in space and time: A tutorial review of SatScan. Soc. Sci. Comput. Rev. 2007, 25, 272–278. [Google Scholar] [CrossRef]
- Ghaderpour, E.; Pagiatakis, S.D.; Hassan, Q.K. A survey on change detection and time series analysis with applications. Appl. Sci. 2021, 11, 6141. [Google Scholar] [CrossRef]
- Sen-Crowe, B.; McKenney, M.; Elkbuli, A. Social distancing during the COVID-19 pandemic: Staying home save lives. Am. J. Emerg. Med. 2020, 38, 1519. [Google Scholar] [CrossRef]
- Gostin, L.O.; Wiley, L.F. Governmental public health powers during the COVID-19 pandemic: Stay-at-home orders, business closures, and travel restrictions. JAMA 2020, 323, 2137–2138. [Google Scholar] [CrossRef]
- Gostin, L.O.; Cohen, I.G.; Koplan, J.P. Universal masking in the United States: The role of mandates, health education, and the CDC. JAMA 2020, 324, 837–838. [Google Scholar] [CrossRef]
- Benjamin-Chung, J.; Reingold, A. Measuring the success of the US COVID-19 vaccine campaign—it’s time to invest in and strengthen immunization information systems. Am. J. Public Health 2021, 111, 1078–1080. [Google Scholar] [CrossRef] [PubMed]
- Covid, C.D.C.; Team, V.B.C.I.; Birhane, M.; Bressler, S.; Chang, G.; Clark, T.; Dorough, L.; Fischer, M.; Watkins, L.F.; Goldstein, J.M.; et al. COVID-19 vaccine breakthrough infections reported to CDC—United States, January 1–April 30, 2021. Morb. Mortal. Wkly. Rep. 2021, 70, 792. [Google Scholar]
- Christie, A.; Mbaeyi, S.A.; Walensky, R.P. CDC interim recommendations for fully vaccinated people: An important first step. JAMA 2021, 325, 1501–1502. [Google Scholar] [CrossRef] [PubMed]
- Murthy, N. Advisory committee on immunization practices recommended immunization schedule for adults aged 19 years or older—United States, 2022. MMWR Morb. Mortal. Wkly. Rep. 2022, 71, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Shekhar, R.; Garg, I.; Pal, S.; Kottewar, S.; Sheikh, A.B. COVID-19 vaccine booster: To boost or not to boost. Infect. Dis. Rep. 2021, 13, 924–929. [Google Scholar] [CrossRef] [PubMed]
- Anwar, A.; Malik, M.; Raees, V.; Anwar, A. Role of mass media and public health communications in the COVID-19 pandemic. Cureus 2020, 12, e10453. [Google Scholar] [CrossRef] [PubMed]
- Raamkumar, A.S.; Tan, S.G.; Wee, H.L. Measuring the outreach efforts of public health authorities and the public response on Facebook during the COVID-19 pandemic in early 2020: Cross-country comparison. J. Med. Internet Res. 2020, 22, e19334. [Google Scholar] [CrossRef]
- Kemeny, J.; Snell, L. Finite Markov Chains; Springer: New York, NY, USA, 1976. [Google Scholar]
- Whittaker, J.; Thomason, M. Markov chain model for statistical software testing. IEEE Trans. Softw. Eng. 1994, 20, 812–824. [Google Scholar] [CrossRef]
- Rohatgi, V. An Introduction to Probability Theory and Mathematical Statistics; Wiley: New York, NY, USA, 1976. [Google Scholar]
- Ji, D.; Dunn, E.; Frahm, J. Spatio-temporally consistent correspondence for dense dynamic scene modeling. In Proceedings of the Computer Vision–ECCV 2016: 14th European Conference, Amsterdam, The Netherlands, 11–14 October 2016; Proceedings, Part VI 14. Springer International Publishing: Berlin/Heidelberg, Germany, 2016; pp. 3–18. [Google Scholar]
- Bani-Yaghoub, M.; Elhomani, A.; Catley, D. Effectiveness of motivational interviewing, health education and brief advice in a population of smokers who are not ready to quit. BMC Med. Res. Methodol. 2018, 18, 52. [Google Scholar] [CrossRef] [PubMed]
- Xiang, R.; Chen, L.; Han, X. Hybrid Markov chain and neural network models for spatial prediction. IEEE Trans. Neural Netw. Learn. Syst. 2022, 33, 3623–3635. [Google Scholar]
- Dietterich, T.G. Ensemble methods in machine learning. In Handbook of Learning and Approximation; Springer: Berlin/Heidelberg, Germany, 2000; Volume 1, pp. 1–16. [Google Scholar]
- Yang, H.; Zou, Y.; Wang, Z.; Wu, B. A hybrid method for short-term freeway travel time prediction based on wavelet neural network and Markov chain. Can. J. Civ. Eng. 2018, 45, 77–86. [Google Scholar] [CrossRef]
- Caprarelli, G.; Fletcher, S. A brief review of spatial analysis concepts and tools used for mapping, containment and risk modelling of infectious diseases and other illnesses. Parasitology 2014, 141, 581–601. [Google Scholar] [CrossRef] [PubMed]
- Bani-Yaghoub, M.; Gautam, R.; Shuai, Z.; Van Den Driessche, P.; Ivanek, R. Reproduction numbers for infections with free-living pathogens growing in the environment. J. Biol. Dyn. 2012, 6, 923–940. [Google Scholar] [CrossRef]

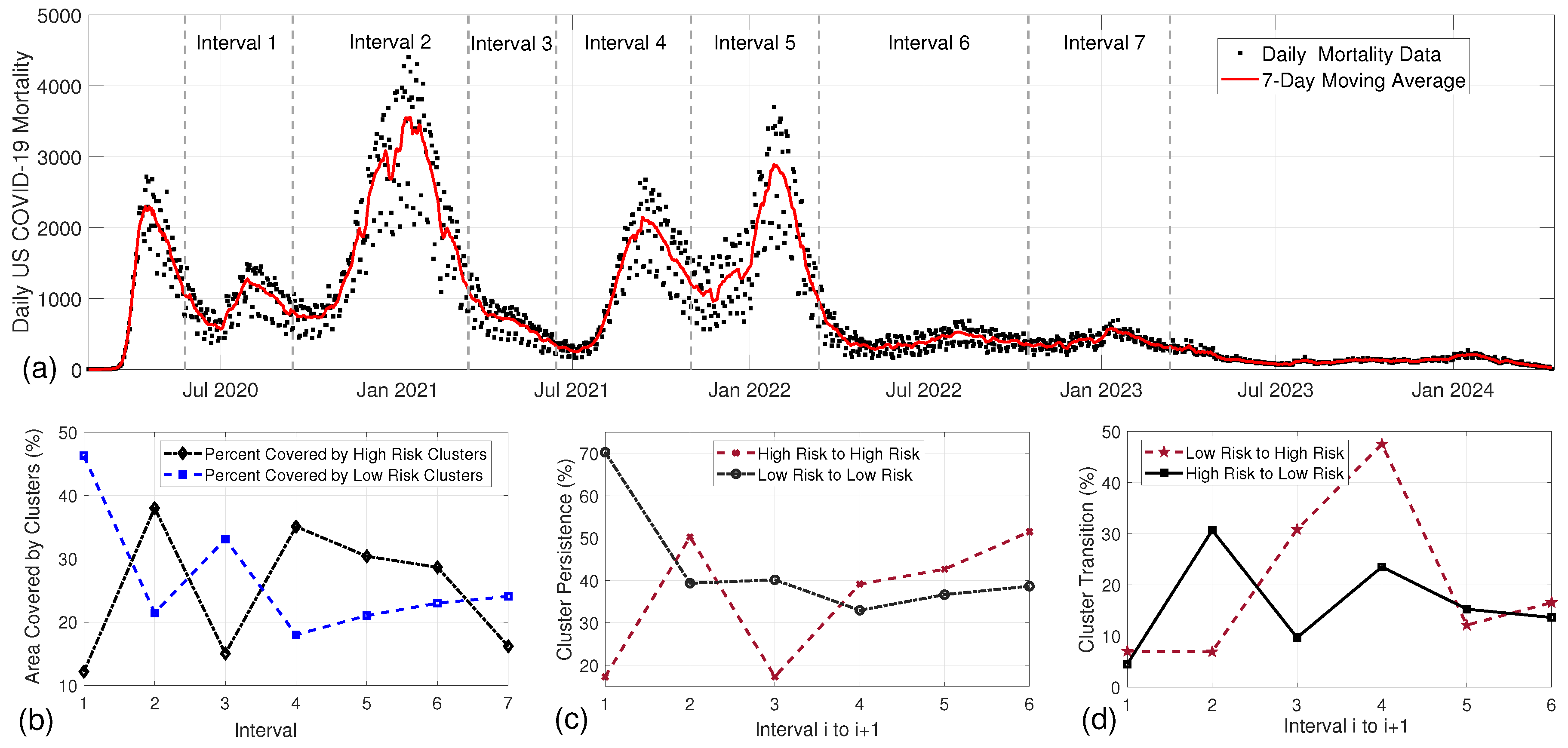
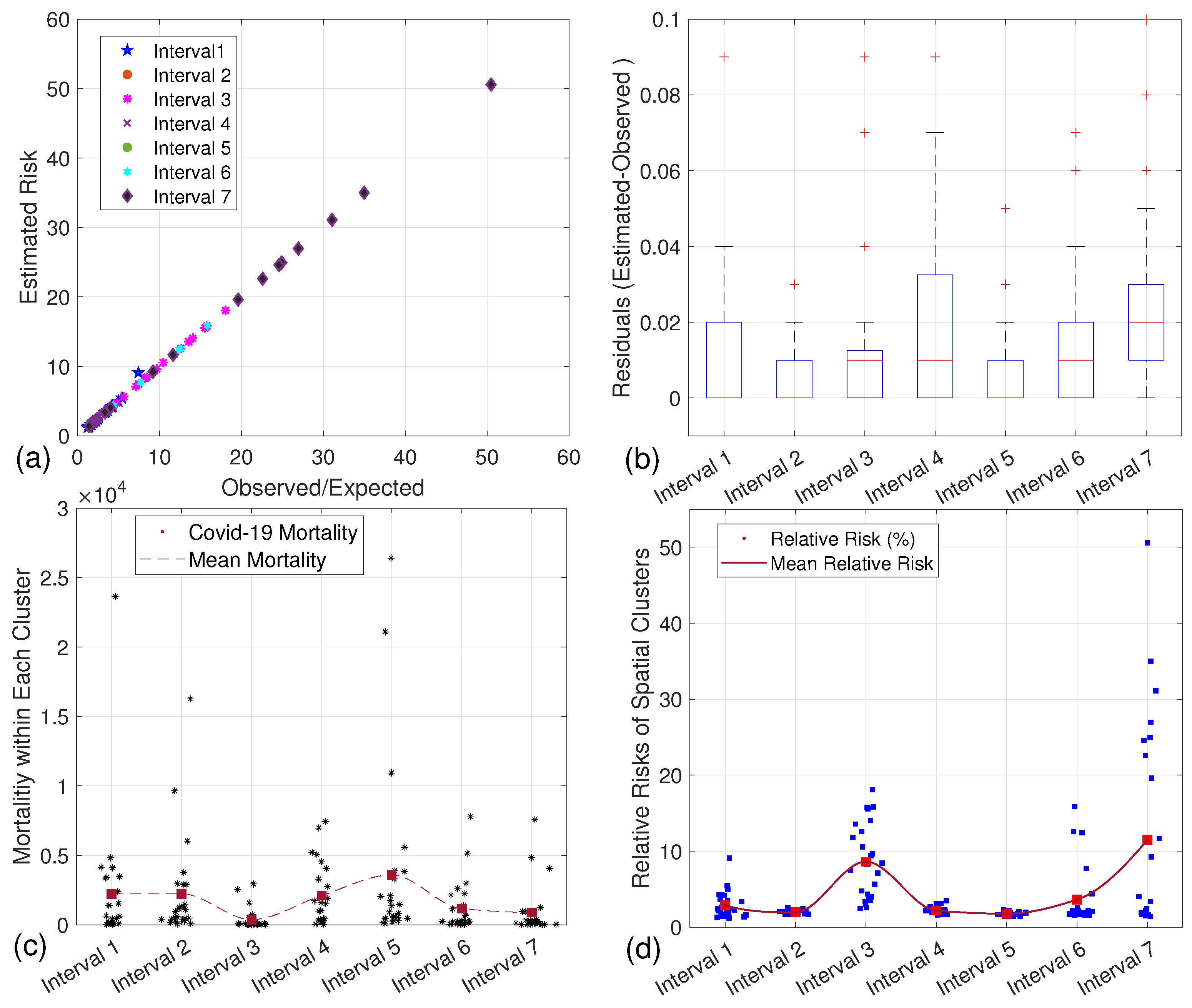
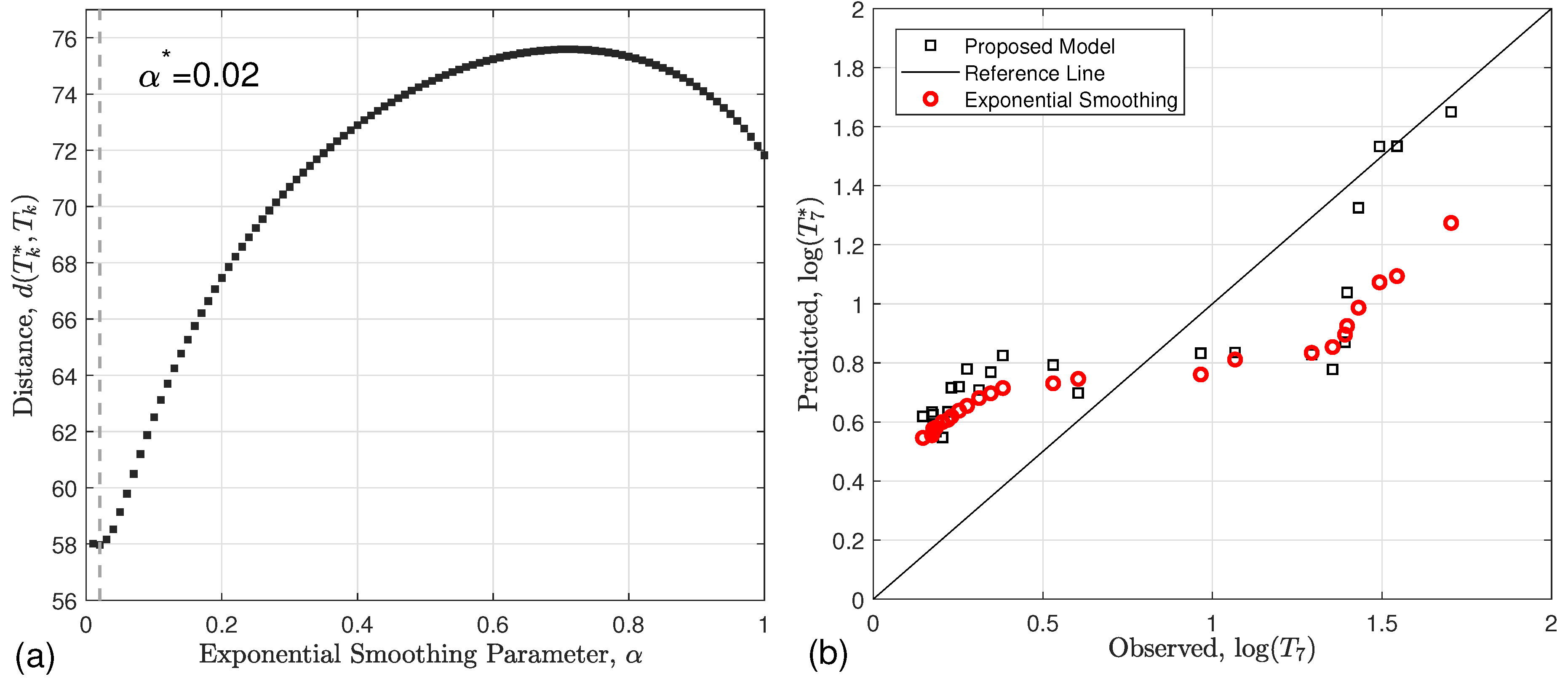
| Interval | Variants | Control and Preventive Measures | Refs. |
|---|---|---|---|
| 1. 5/24/2020–9/13/2020 | Original strain (Wuhan) | Social distancing measures, use of face masks, limitations on gatherings, travel restrictions | [31,32] |
| 2. 9/13/2020–3/14/2021 | D614G variant | Expanded mask mandates, introduction of COVID-19 vaccines (Pfizer, Moderna), enhanced testing and contact tracing | [33,34] |
| 3. 3/14/2021–6/13/202 | Alpha variant (B.1.1.7) | Vaccination campaigns ramped up, continued mask-wearing in high-transmission areas | [35,36] |
| 4. 6/13/2021–10/31/2021 | Alpha variant predominant | CDC recommendations for vaccinated individuals, return to in-person learning in colleges and schools, ongoing vaccination efforts | [36,37] |
| 5. 10/31/2021–3/13/2022 | Delta variant (B.1.617.2) | Booster shots recommended, reinstated mask mandates in certain areas, increased indoor venue capacity restrictions | [37,38] |
| 6. 3/13/2022–10/16/2022 | Omicron variant (B.1.1.529) | Vaccination updates including boosters, recommendations for masks in crowded indoor settings, continued public health surveillance | [31,37] |
| 7. 10/16/2022–3/12/2023 | Omicron subvariants (BA.1, BA.2) | Focus on public health awareness campaigns, emphasis on personal responsibility regarding health measures | [39,40] |
| Interval | High-Risk | Low-Risk | Area of High Risk | Area of Low Risk | Overlap Area | Total Area |
|---|---|---|---|---|---|---|
| 1 | 25 | 38 | 1,170,531.55 (12.16%) | 4,457,499.54 (46.29%) | 4830.30 (0.05%) | 5,623,200.79 (58.40%) |
| 2 | 38 | 34 | 3,661,450.98 (38.02%) | 2,065,006.01 (21.45%) | 7781.26 (0.08%) | 5,718,675.73 (59.39%) |
| 3 | 40 | 39 | 1,447,800.92 (15.04%) | 3,189,789.31 (33.13%) | 5915.07 (0.06%) | 4,631,675.16 (48.10%) |
| 4 | 30 | 25 | 3,379,836.76 (35.10%) | 1,733,777.69 (18.01%) | 24,583.89 (0.26%) | 5,089,030.56 (52.85%) |
| 5 | 29 | 36 | 2,928,563.46 (30.41%) | 2,024,862.64 (21.03%) | 14,785.95 (0.15%) | 4,938,640.15 (51.29%) |
| 6 | 29 | 25 | 2,760,890.45 (28.67%) | 2,214,642.66 (23.00%) | 3259.16 (0.03%) | 4,972,273.95 (51.64%) |
| 7 | 28 | 30 | 1,556,809.44 (16.17%) | 2,316,998.49 (24.06%) | 5328.31 (0.06%) | 3,868,479.62 (40.17%) |
| Intervals | High-Risk Overlap | Low-Risk Overlap | High–Low Transition | Low–High Transition |
|---|---|---|---|---|
| 1 → 2 | 630,606 (53.87%, 17.22%) | 1,451,535 (32.56%, 70.29%) | 93,533 (7.99%, 4.53%) | 1,311,032 (29.41%, 35.81%) |
| 2 → 3 | 727,798 (19.88%, 50.27%) | 1,255,175 (60.78%, 39.35%) | 979,045 (26.74%, 30.69%) | 100,420 (4.86%, 6.94%) |
| 3 → 4 | 583,839 (40.33%, 17.27%) | 696,469 (21.83%, 40.17%) | 168,040 (11.61%, 9.69%) | 1,042,772 (32.69%, 30.85%) |
| 4 → 5 | 1,146,392 (33.92%, 39.15%) | 667,377 (38.49%, 32.96%) | 476,609 (14.10%, 23.54%) | 1,391,524 (21.28%, 12.60%) |
| 5 → 6 | 1,178,061 (40.23%, 42.67%) | 811,950 (40.09%, 36.66%) | 337,960 (11.54%, 15.26%) | 335,357 (16.56%, 12.15%) |
| 6 → 7 | 802,151 (29.05%, 51.53%) | 895,447 (40.43%, 38.65%) | 315,871 (11.44%, 13.63%) | 257,680 (11.64%, 16.55%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bani-Yaghoub, M.; Rekab, K.; Pluta, J.; Tabharit, S. Estimating the Relative Risks of Spatial Clusters Using a Predictor–Corrector Method. Mathematics 2025, 13, 180. https://doi.org/10.3390/math13020180
Bani-Yaghoub M, Rekab K, Pluta J, Tabharit S. Estimating the Relative Risks of Spatial Clusters Using a Predictor–Corrector Method. Mathematics. 2025; 13(2):180. https://doi.org/10.3390/math13020180
Chicago/Turabian StyleBani-Yaghoub, Majid, Kamel Rekab, Julia Pluta, and Said Tabharit. 2025. "Estimating the Relative Risks of Spatial Clusters Using a Predictor–Corrector Method" Mathematics 13, no. 2: 180. https://doi.org/10.3390/math13020180
APA StyleBani-Yaghoub, M., Rekab, K., Pluta, J., & Tabharit, S. (2025). Estimating the Relative Risks of Spatial Clusters Using a Predictor–Corrector Method. Mathematics, 13(2), 180. https://doi.org/10.3390/math13020180







