3.2. Method Based on the Historical Data Under the Normal Distribution
First, consider the normal distribution case. To establish an upper bound for the count of daily new COVID-19 cases, a high content upper tolerance bound can be considered, such as an upper 0.99-content and 0.95 level tolerance bound. Since in this study, the past 7- or 14-day data are used to construct upper tolerance bounds, the sample size is 7 or 14. The values of
in (2) corresponding to different sample sizes can be calculated using (3). The corresponding
values calculated using (3) of the 0.99-content and 0.95 level tolerance bounds for sample sizes 7 and 14 are provided in
Table 1.
Let , denote the daily data of the region that need to be predicted. Since the first 7 or 14 data points need to be used as historical data to construct the upper tolerance bound, only the data , with m = 8 or m = 15 are used for the prediction period in the 7-day or 14-day prediction case, respectively. Let , be the constructed upper tolerance bounds for the data. For the 7-day case, the first 7 days of data are used to construct the upper bound for the next 7 days of data , and then the 7 days of data are used to construct the upper bound for the next 7 days of data . Therefore, the values of are the same for every 7 days.
Take the USA data as an example. When considering using 7-day data to construct a tolerance bound, the first 7 USA data points (3–9 January 2020) in
Table S1 were used to construct the upper tolerance bound for the next 7 days (10–16 January 2020). Next, the 7 USA data points (10–16 January 2020) were used to construct the upper tolerance bound for the dates of 17–23 January 2020. Since the first case occurred on 20 January 2020, in the USA data, the data before 20 January are all zero. Since the mean and standard deviation of the data from 3 January to 9 January are both zero, using (2), the upper tolerance bound for 10 January to 16 January is 0. The upper tolerance bound is zero until using the 7-day data from 17 January to 23 January to construct the tolerance bound for 24 January to 30 January. The mean and standard deviation of data from 17 January to 23 January are 0.1429 and 0.3780. Consequently, using (2) and the
value in
Table 1, we have
This is the upper tolerance bound for 24 January to 30 January. Similarly, the upper tolerance bounds for other days can be obtained.
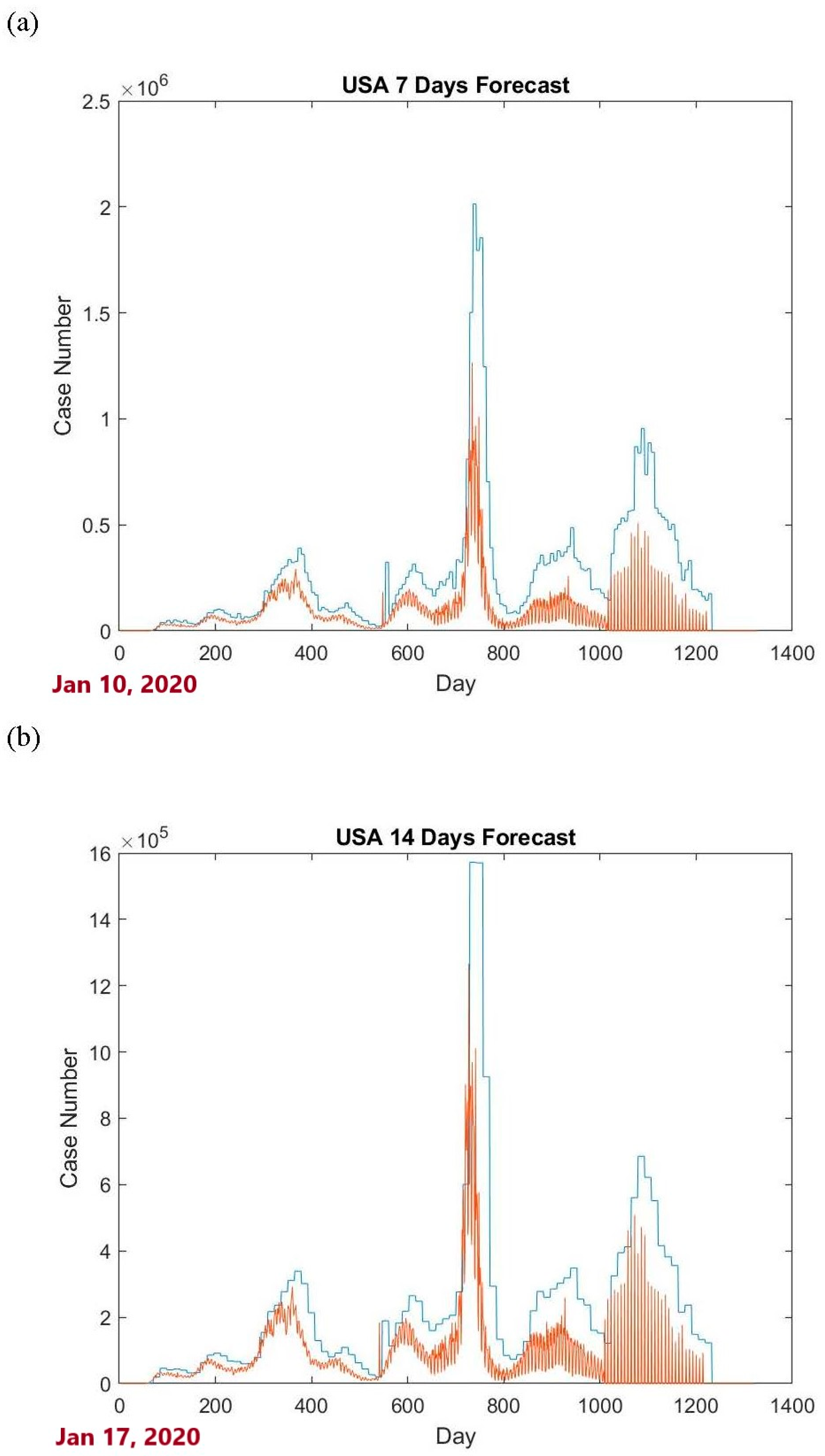
Figure 1 shows the upper tolerance bounds for the USA data case.
To evaluate the performance of the constructed upper tolerance bounds, the proportion of the number of COVID-19 cases that is lower than or equal to the upper tolerance bound is calculated. The proportion is defined as
where
denotes the indicator function.
Figure 1 shows that a large proportion of the data are below the calculated upper bounds. Since the upper bounds are the 0.99 content, 0.95 level upper tolerance bounds, it is expected that 99% of the data are below the constructed upper bounds. Using the criterion (13), for the 7-day and 14-day cases, the proportions are
and
, respectively. This indicates a good outcome. However, these bounds have not yet achieved the original goal that 99% of daily new COVID-19 confirmed cases fall below them. To improve the outcomes, the confirmed case numbers from another country can be used as auxiliary information to aid in prediction.
3.3. Method Based on the Historical Data and Auxiliary Data Under the Normal Distribution
In addition to constructing the upper tolerance bound using historical data, auxiliary data from other regions can be utilized to enhance predictions. As we know, a new strain of COVID-19 may transmit from one country to another. It is likely that the data of one country are correlated with the past data from another country with a time lag. Therefore, if other regions have already experienced an outbreak, then the data may be useful in predicting the outbreak in another country. In this study, data from one of the two investigated countries, either the USA or the UK, are utilized as auxiliary data to aid in predicting the number of COVID-19 cases in the other country.
Different time lag cases can be considered when using the auxiliary data. Let , denote the daily data from another region. The time lag is denoted as An appropriate time lag can be selected such that the absolute value of is the largest based on historical data because the data is expected to provide earlier outbreak information for . Then, the modified upper tolerance bound based on the historical data and auxiliary data using (6) can be calculated. The steps of calculating the upper tolerance bound using a 7-day timeframe are provided in Procedure 1.
Procedure 1
Step 1. Find a such that the absolute value of the is large, say .
Step 2. Consider a 7-day timeframe case using the data Calculate the estimators and for each based on these data.
Step 3. Let and the mean of these 7 values is denoted as And according to Theorem 1, calculate Then calculate using formula (8) by replacing with .
Step 4. Use and to calculate Then take the maximum value of and , which is the proposed upper tolerance bound .
In Step 1 of Procedure 1, when not enough historical data are available, such as only the 7 or 14 days of data used, it is not easy to find the best
value. In this case, we can consider the simple case by using
when performing a 7-day timeframe scenario and using
when performing a 14-day timeframe scenario. For the data used in this study, the entire dataset was used to find suitable
values by calculating the correlation between the USA and UK data for different time lags, which shows that time lags of 7 and 14 are appropriate (
Table 2).
Consider the case with a time lag of 7 days, where UK historical data are used as auxiliary information to construct upper bounds for USA data. For a forecast period of 7 days, the past 7 days of USA data are used to calculate
and
, while the past 14–8 days of UK data are used to calculate
and
. The past 7 days of UK data are then used to calculate the mean, which is
in Step 3 of Procedure 1. For example, the 7-day USA data (7–13 February 2020) and the 7-day UK data (31 January to 6 February 2020) were used to calculate
and
for providing the USA 7-day (14–20 February 2020) upper tolerance bound. Through calculation, we have
,
and
. Note that the period of these UK data is 7 days before the period of these USA data. Then, the 7 UK data points (7–13 February 2020) are used to calculate
in Step 3 of Procedure 1. As a result, we have
and
. Then
and
. The maximum value of
and
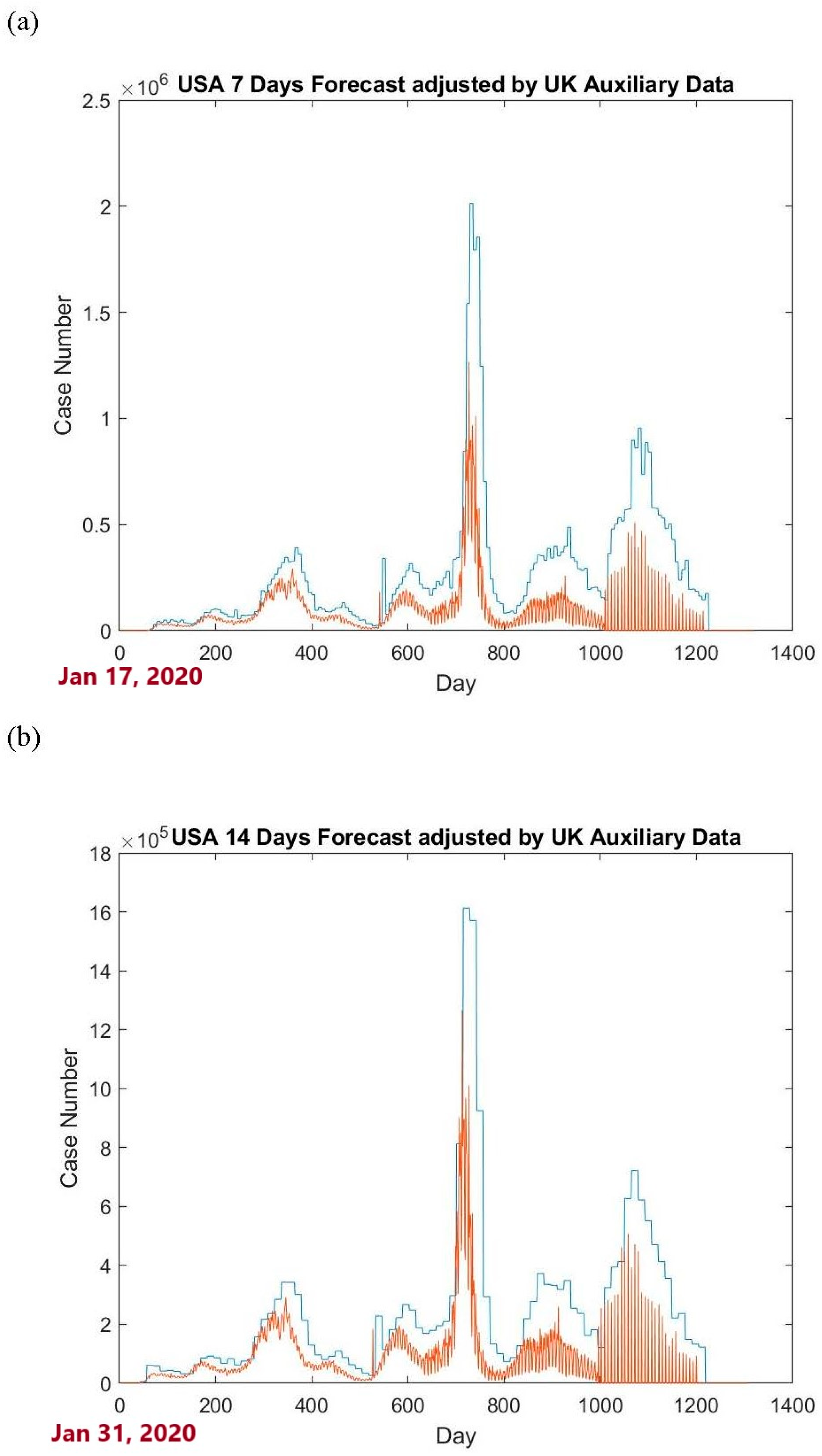
is 1.8796, which is the proposed upper tolerance bound for predicting the USA COVID-19 case numbers for the period (14–20 February 2020). The USA 7-day and 14-day forecasts incorporating auxiliary data from the UK are shown in
Figure 2.
Table 3 and
Table 4 present the proportions of the case numbers below the calculated upper bounds for the USA and UK forecasts, respectively. For the USA case, as mentioned in
Section 3.2, the proportions for the 7-day and 14-day forecasts based on the conventional upper tolerance bound are 0.9744 and 0.9281, respectively. When incorporating auxiliary data from the UK, the proportions for the 7-day and 14-day forecasts increase to 0.9803 and 0.9526, respectively.
Next, USA historical data are used as auxiliary information to incorporate historical data from the UK to construct upper bounds for predicting UK data. For the 7-day and 14-day forecasts, the proportions of the case numbers below the calculated upper bounds based on the conventional method are 0.9624 and 0.8850, respectively. When incorporating auxiliary data from the USA, the proportions for the 7-day and 14-day forecasts increase to 0.9720 and 0.9052, respectively.
Regarding the results presented in
Table 3 and
Table 4, while the proportions of the data below the upper bounds for these two methods do not reach 0.99, most of them are at least greater than 0.9. This means that these bounds can provide useful information for preparing sources for COVID-19 management.