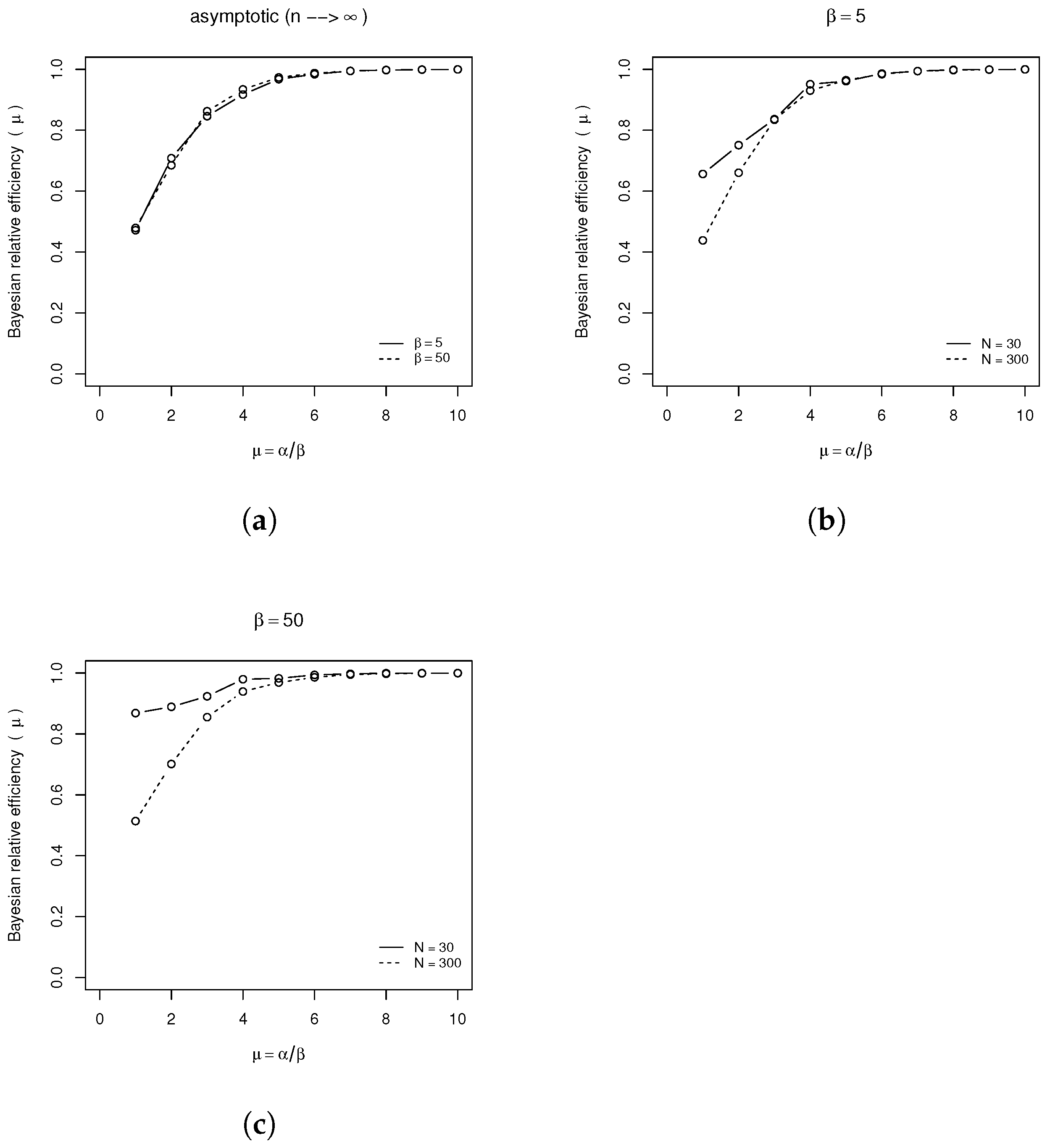3.1. Simulation Study
We present a simulation study to evaluate Bayesian estimation of the ZMPS class of models using the SGHMC algorithm. In this study, we considered samples
from the distributions ZMP (Poisson), ZMNB (negative binomial), and ZMGP (generalized Poisson) models, with sample size
, under the link functions
where
,
. We consider independent prior distributions, normal
with
and
for informative prior and
for vague prior, and a gamma distribution
for
. Let
.
In this simulation study, we conducted a sensitivity analysis to evaluate the effect of initial conditions on the convergence of the generated chains. We tested three different chains, each initialized with conditions based on the mean of the prior, (i) 1.4 times the mean (i.e., plus 40%), and (ii) 0.4 times the mean (i.e., minus 60%). After verifying the burn-in period and ensuring the method’s convergence, we selected the mean of the prior distribution as the reference point for subsequent analysis.
We employed the SGHMC algorithm to generate 50,000 samples of
from its full posterior distribution, discarding the first 50% as burn-in. This resulted in a chain consisting of 25,000 samples of
. The final sample was used to calculate the Monte Carlo estimates of each model parameter’s posterior mean and variance, as given by (
19) and (20). We repeated this procedure
times. With
M estimates of the model parameters, we evaluated their performance using metrics such as mean bias (B), the ratio of mean squared error (MSE) to variance (Var), and the average Bayesian efficiency (BE), calculated for each model parameter
:
where
is given by (
25) and
is given by (20) for each
,
. Additionally, we have also computed the coverage probability (
) of the Bayesian credible intervals expressed as percentages as
where
assumes 1 if the
kth Bayesian credible interval (BCI) contains the true value
and 0 otherwise.
A summary of the
simulations is shown in
Table 2. In this summary, we can see the effect of the prior information on the calculation of the standard deviation (s.d.) and the amplitude of the highest posterior density (HPD) credibility interval for each parameter.
As previously discussed, the performance of the estimators using the metrics in (
28)–(
31) is presented in
Table 3. The results show improved efficiency of the estimators when the prior distribution is more informative.
3.2. Application: Number of Femicide Cases
Violence against women encompasses a wide range of physical, psychological, sexual, and property-related attacks, often occurring on a continuum that can tragically culminate in murder, the most extreme manifestation of violence inflicted upon women.
Femicide refers to the murder of a woman specifically because she is a woman, typically driven by hatred, contempt, or a sense of lost control or ownership over women. Brazil’s Feminicide Law (Law 13.104 of 9 March 2015) classifies homicide as femicide when committed against a woman due to her gender. The law defines such crimes as those involving domestic or family violence, contempt, or discrimination against women and also includes femicide in the category of heinous crimes.
Of the five measures to combat gender-based violence recommended by the United Nations, Brazil has implemented only one: online services. This includes the “Red Light” campaign launched by the National Council of Justice and the Brazilian Association of Magistrates, which functions similarly to emergency alert systems. However, there has yet to be an assessment of the campaign’s impact on protecting women in violent situations. Factors contributing to the increase in gender-based violence include victims’ difficulty in reporting, a reduction in crime reporting at police stations, and a decline in the number of emergency protective measures granted to women.
This study aims to determine whether femicides increased during the COVID-19 pandemic. We analyze data from 642 municipalities in São Paulo for the years 2019 and 2020 [
46], fitting ZMPS regression models with the Municipal Human Development Index (MHDI) as an explanatory variable. The MHDI, a key measure of municipal development in Brazil, evaluates three dimensions of human development: longevity, education, and income. It ranges from 0 to 1, with higher values indicating greater development. By interpreting the mean
of the model and the parameter
(which accounts for zero inflation or deflation) as functions of the MHDI, we gain insights into femicide patterns. A summary of the sample is presented in
Table 4.
We apply the proposed Bayesian approach to fit ZMPS models to the femicide data, using the MHDI as a regressor. Specifically, we consider the zero-modified Poisson (ZMP), zero-modified negative binomial (ZMNB), zero-modified generalized Poisson (ZMGP), and zero-modified COM-Poisson (ZMCOMP) distributions. For the parameters
and
, we use normal priors
, while for
, we apply a gamma prior
with
and
. We employ three information criteria, LogCPO (
A8), EBIC (
A9), and WAIC (
A10), to identify the distribution that best fits the data, with higher LogCPO values and lower WAIC and EBIC values indicating better model performance. The values of these criteria for each fitted model are presented in
Table 5, with the selected model for each sample highlighted in bold.
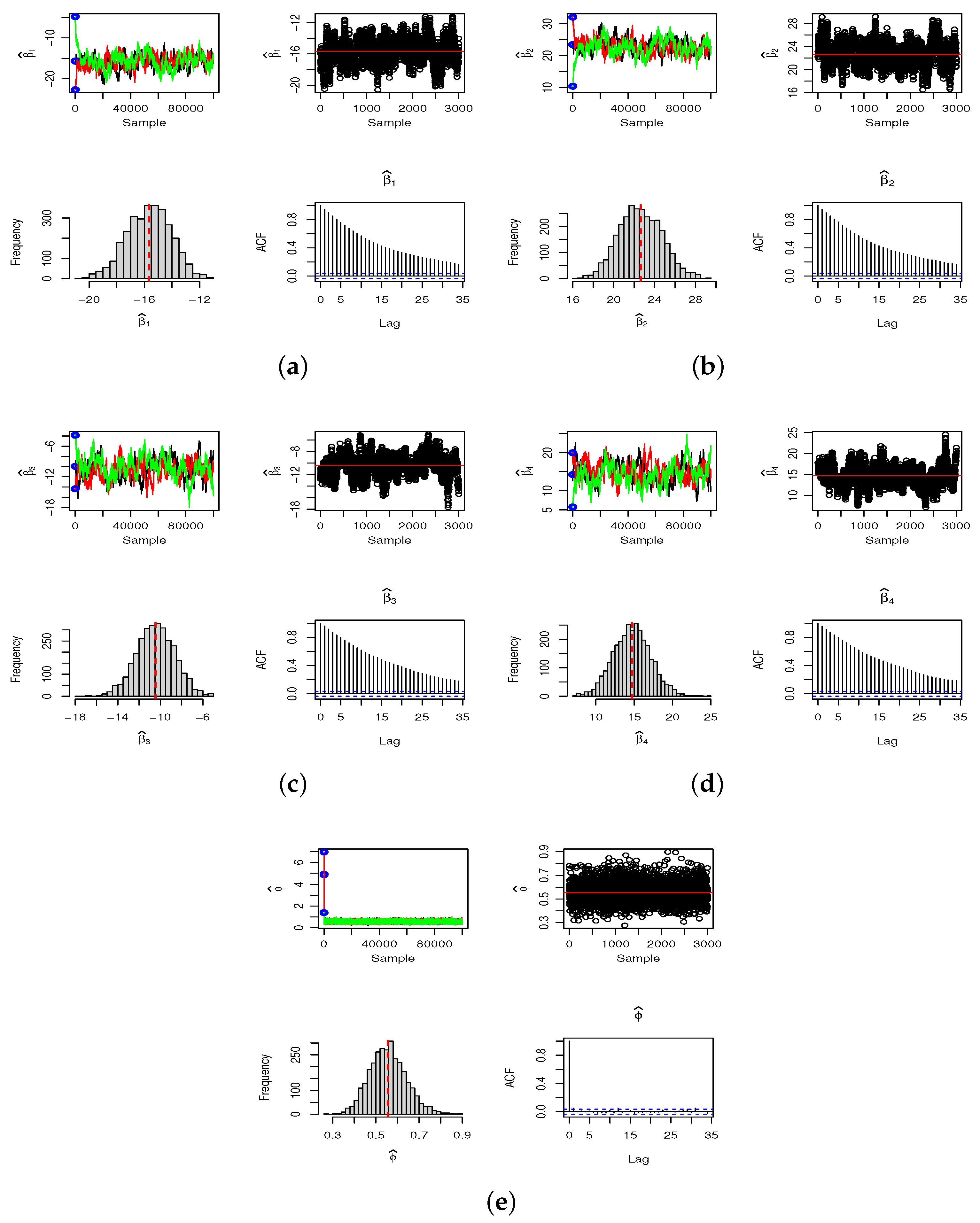
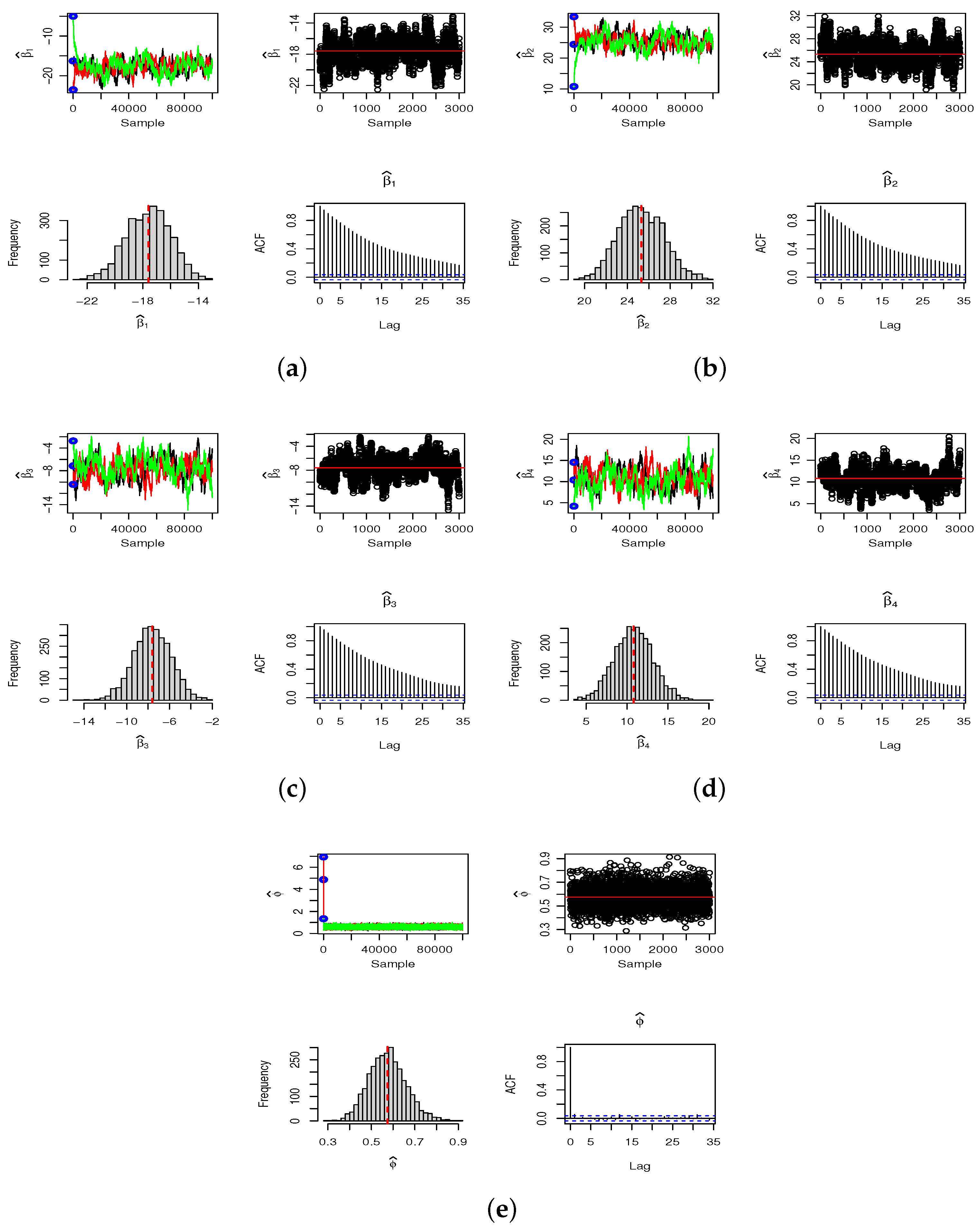
The three criteria identify the ZMNB model as the best fit for both 2019 and 2020. Posterior summaries and credible intervals for the parameters
,
, and
are presented in
Table 6. The results are derived using three Markov chains, each containing 100,000 samples, initialized with distinct starting conditions. To minimize the effect of the burn-in period, we discard 25% of each chain and select every 75th value from the remaining 75% (thinning), which helps reduce autocorrelation within the chains. We use the resulting final sample of 3000 observations to compute the Bayesian Monte Carlo estimates. The convergence details of the SGHMC algorithm are provided in
Appendix B.
Figure 1 displays the estimated mean
and the parameter
for the ZMNB models based on data from 2019 and 2020.
In
Figure 1a, we examine the mean number of reported femicides in relation to the MHDI. The graphs demonstrate that the mean number of notifications increases with the MHDI and is slightly higher during the COVID-19 pandemic. This trend indicates that municipalities with higher levels of development tend to report a greater mean number of femicides. This pattern is anticipated as more developed municipalities are likely to offer better protective measures for women and more effective crime reporting systems. The increase in reported cases in 2020, compared with the averages in 2019, can be attributed to the impacts of the pandemic.
The
parameters are depicted as a function of the MHDI in
Figure 1b. These parameters help interpret the frequency of zero observations within the sample. Typically, municipalities with lower mean notification numbers,
, are expected to exhibit zero inflation, meaning
. However,
Figure 1b shows that for municipalities with an MHDI below 0.7 (i.e., those with low average
), the estimates of
exceed 1. This indicates a deflation of zero observations at this level of the covariate, suggesting that municipalities with a low MHDI experience less zero inflation than expected. In fact, they report more cases than the standard model would predict (
). Furthermore, the increase in registrations during 2020, marked by a rise in deflation from zero, can be attributed to the impact of the COVID-19 pandemic, as seen in the comparison of
estimates between 2019 and 2020.