Feature Detection Based on Imaging and Genetic Data Using Multi-Kernel Support Vector Machine–Apriori Model
Abstract
1. Introduction
1.1. Background
1.2. Related Work
1.3. Proposed Framework
2. Materials and Methods
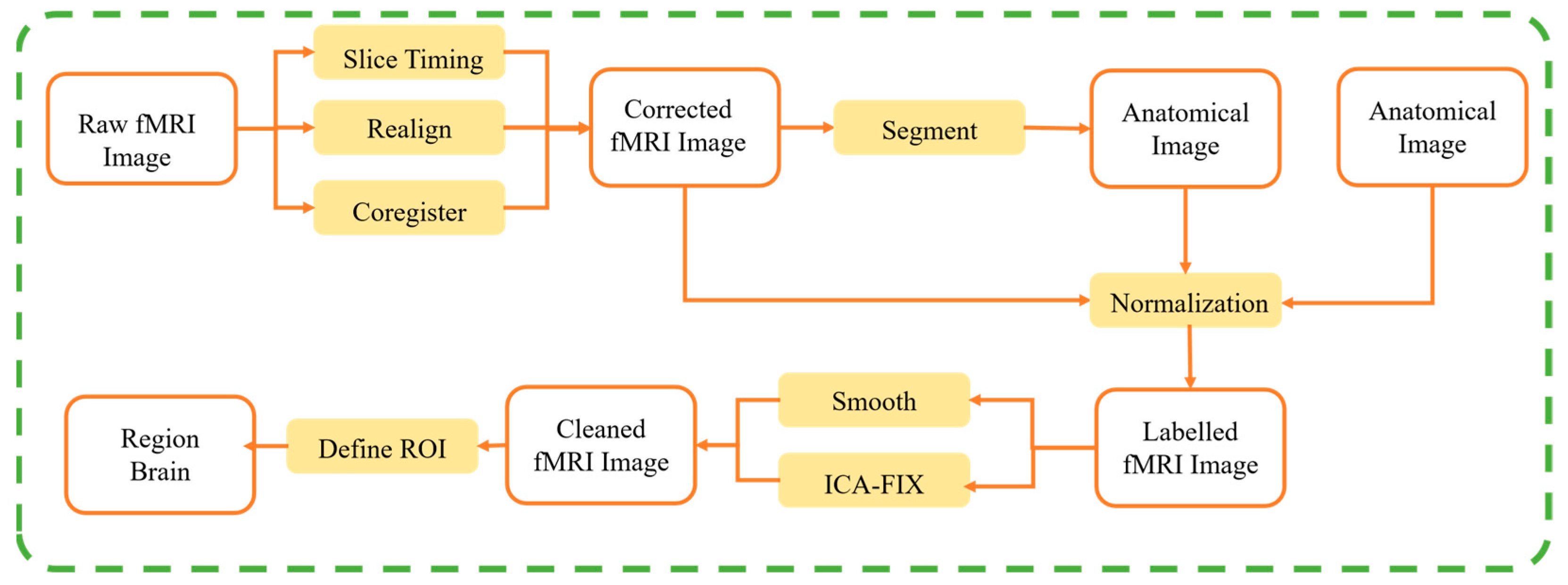
2.1. Data Pre-Processing
2.2. Features Fusion
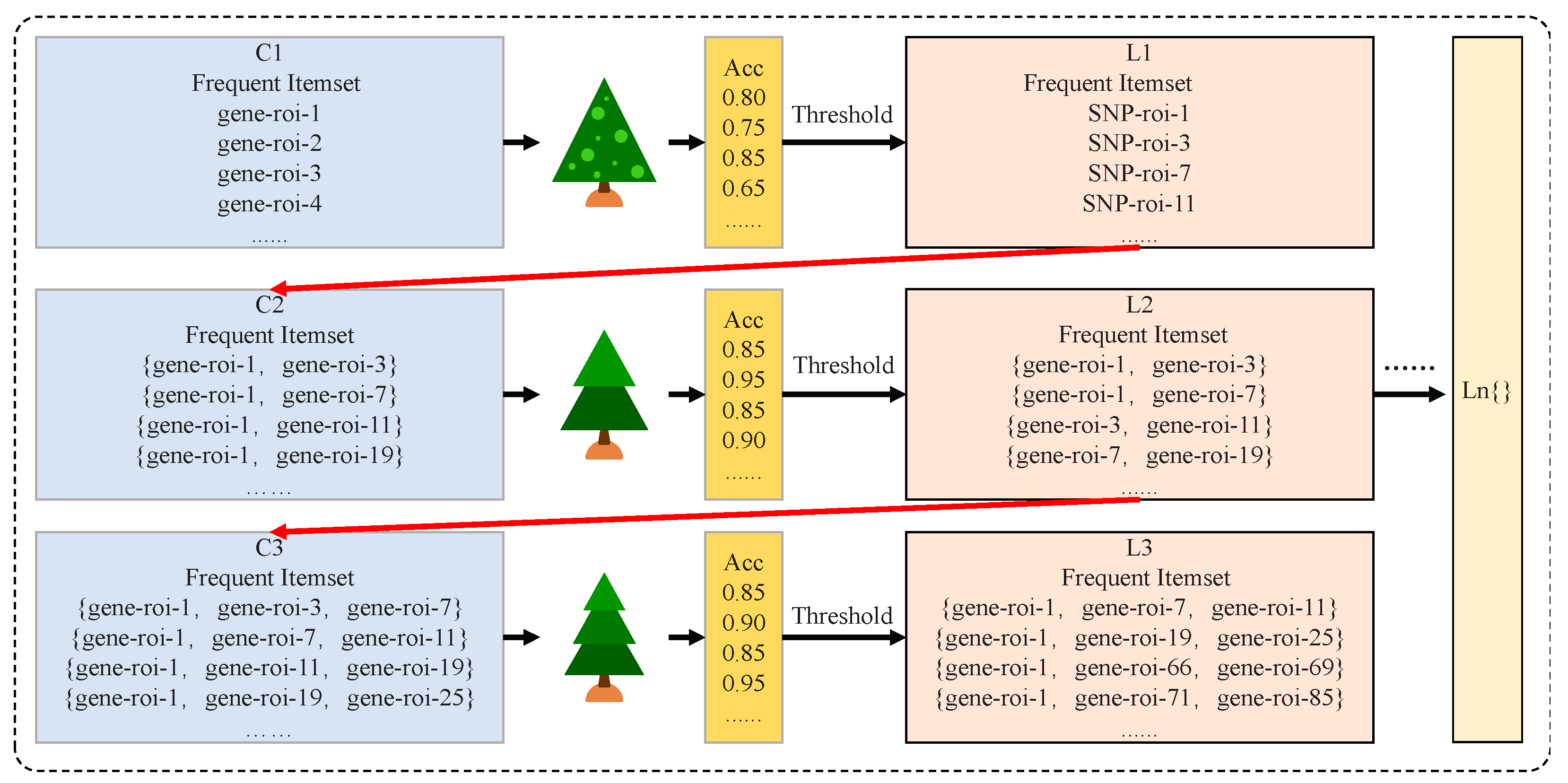
2.3. Multi-Kernel SVM-Apriori Model Construction
| Algorithm 1. The algorithm process of the Multi-Kernel SVM–Apriori Model |
| Input: experimental dataset {X,Y}; X is input, Y is the corresponding label (1 or −1) d, , Output: Multi-Kernel SVM–Apriori Model |
| 1: Initialize {X, Y}, w1, w2, w3, minsupport {X, Y} is experimental dataset, w1, w2, w3 are the weights of three kernels Equation (7), d is the degree of a polynomial (3), is the width of the Gaussian kernel (0.005), is the minimum support for generating frequent itemsets Equation (9). 2: Randomly select a subset of features as {Features}trak 3: Input {Features}trak according to 7:3 training, partitioning the {X, Y} into {X, Y}train, {X, Y}valid 4: Calculate predicted values: ypred = sign (w’ × x + b) Output: classifier: Acc = Σ (ypred == y)/length(y) 5: Input set {X, Y} to Multi-kernel SVM to obtain the Acc of all individual features and obtain the frequent itemset L1 which satisfies Acc > 6: For each k starting from k = 2: Repeat 7: Generate candidate feature set Ck by connecting frequent itemsets L (k − 1) – For each candidate feature set c in Ck: – For each transaction t in dataset S: – Check if c is a subset of t, and if so, increase the count of c – Calculate Acc for each candidate feature set based on Ck Update = mean(ΣACC L(k − 1)) Filter to obtain frequent feature set Lk which satisfies Acc > Until more frequent feature sets cannot be generated, return the frequent feature set column table L 8: Finally, perform Leave-One-Out Cross-Validation for each L (k − 1) and select the feature set with the highest ACC. |
2.4. Model Comparison
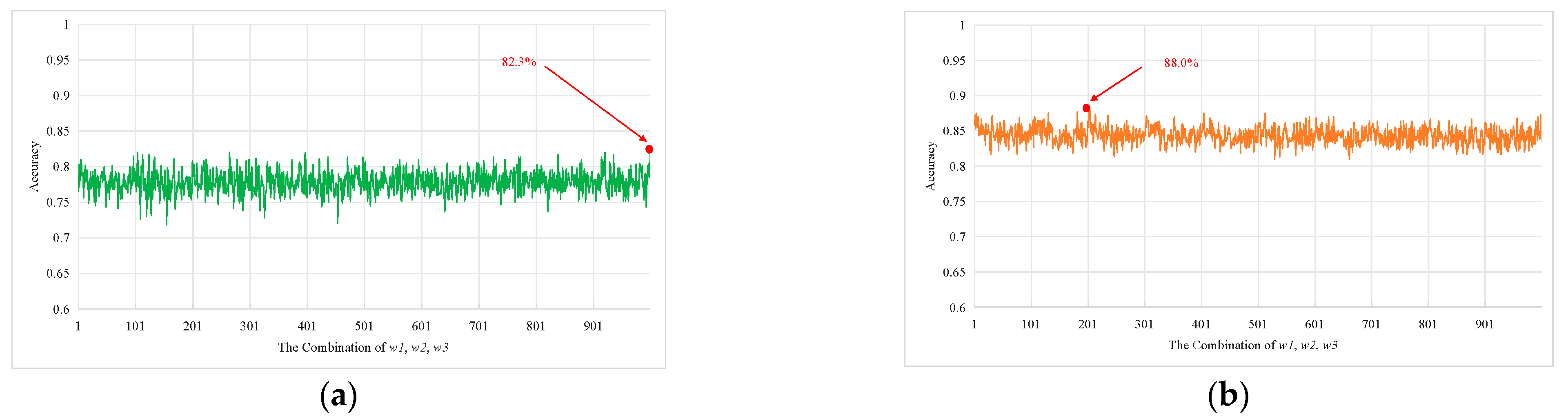
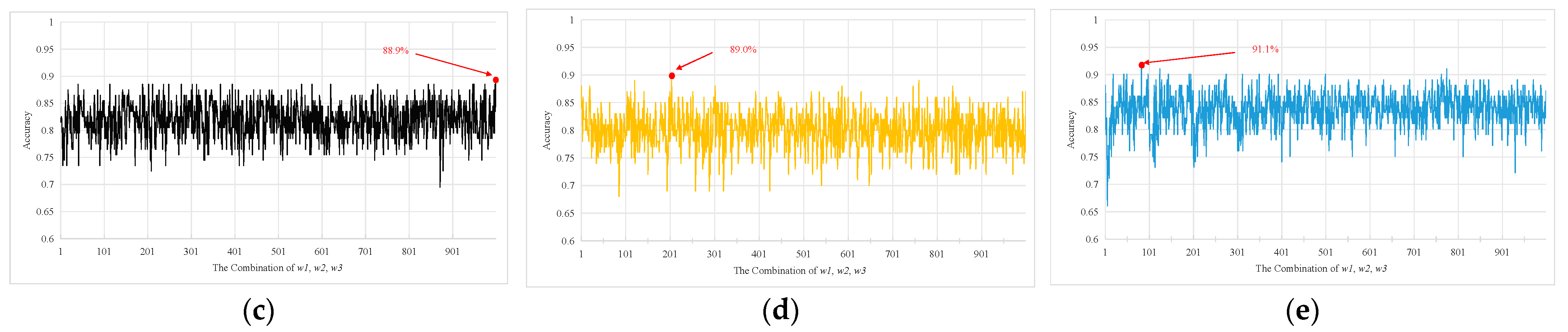
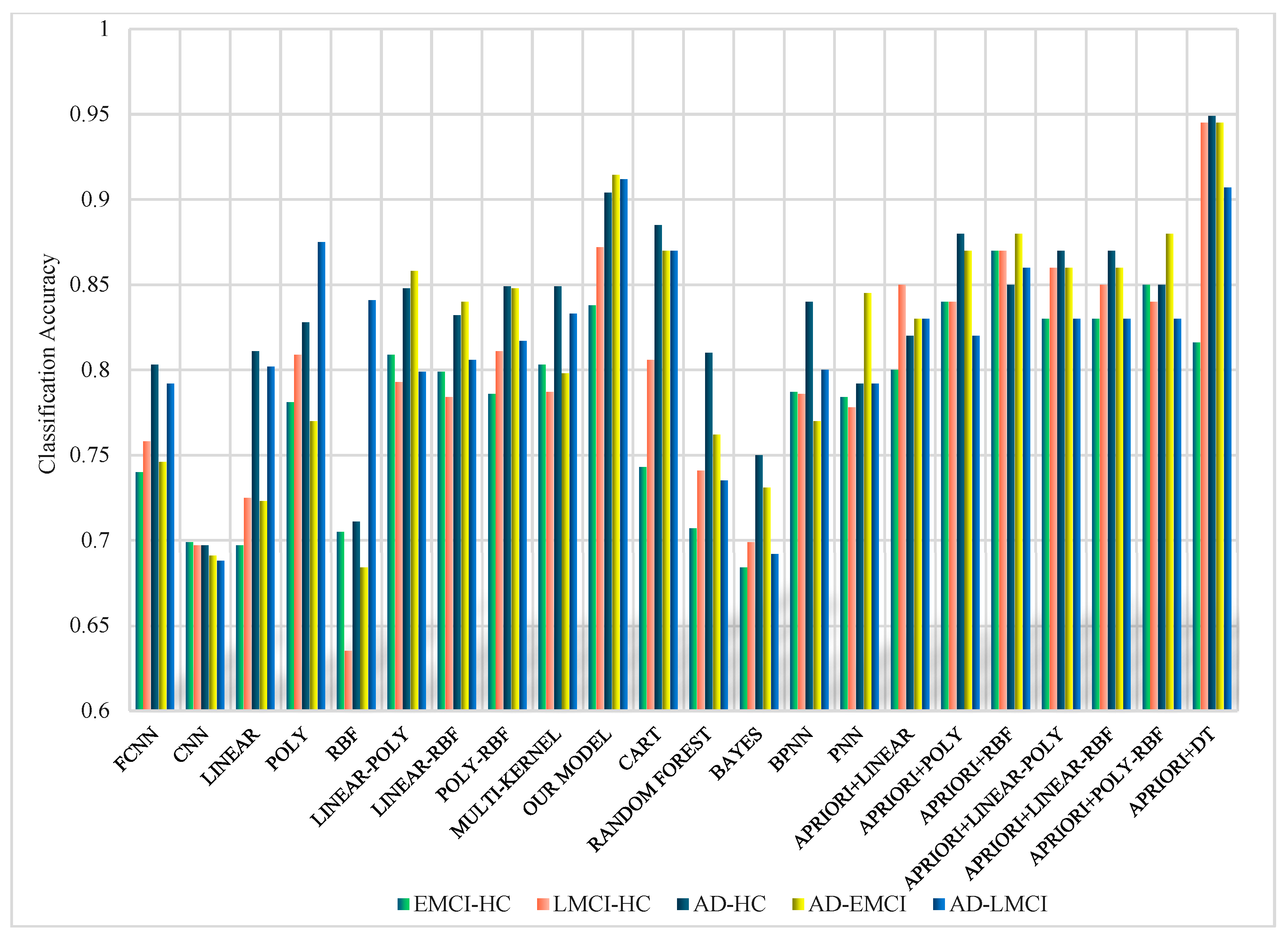
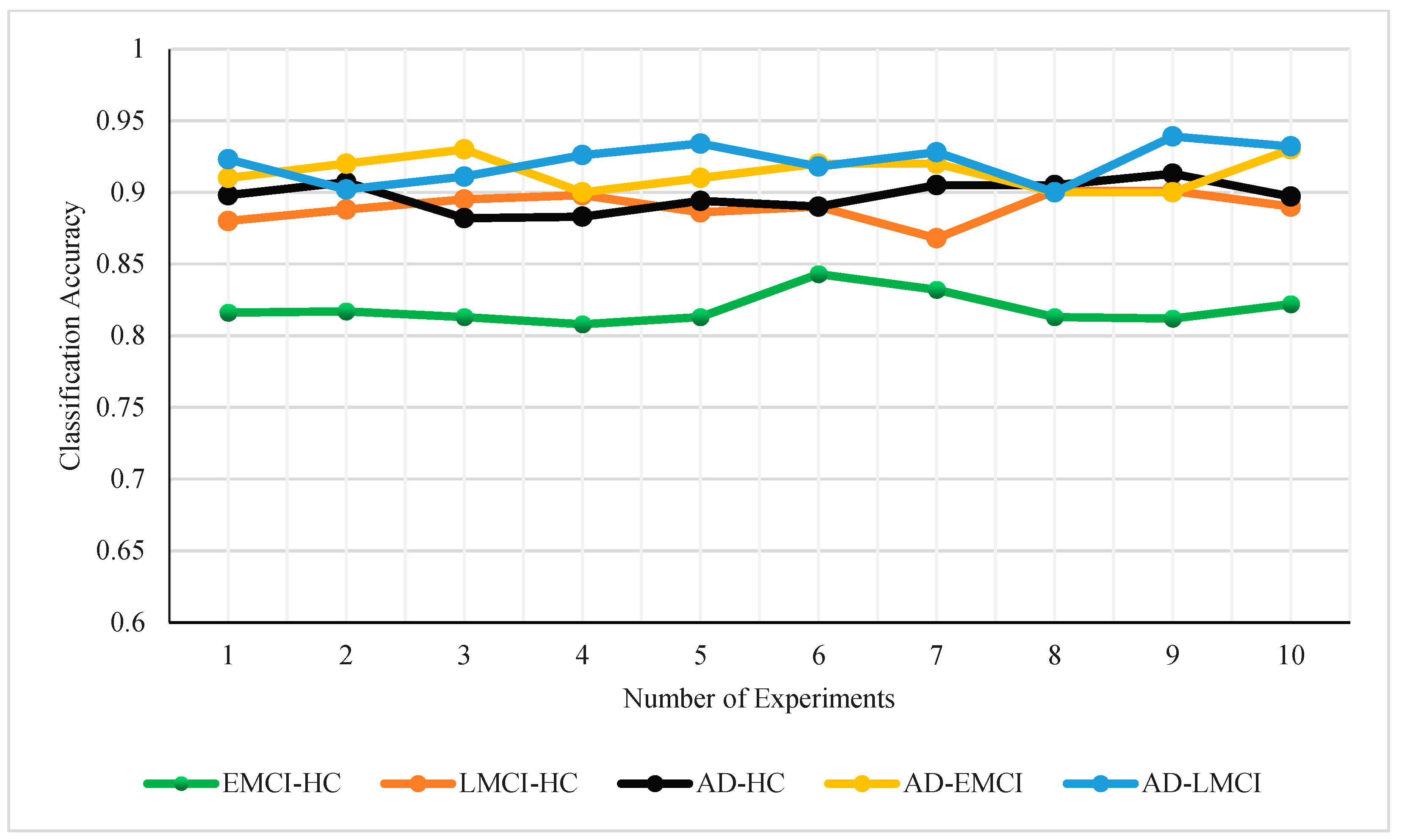
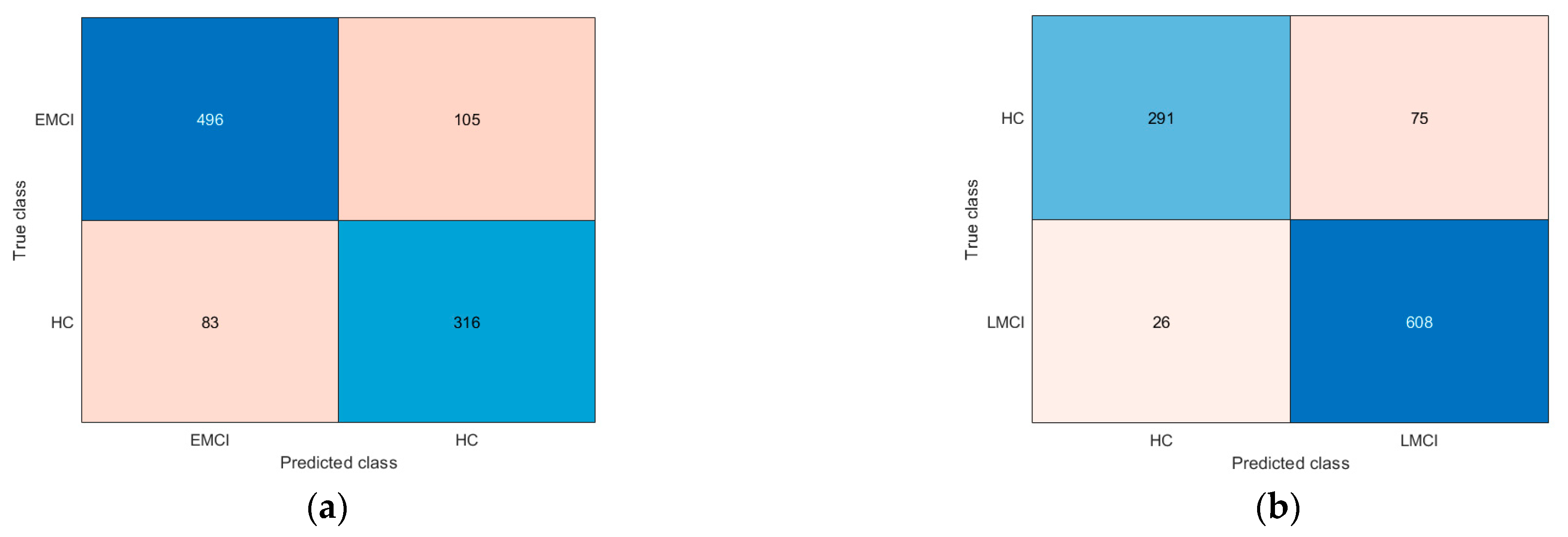
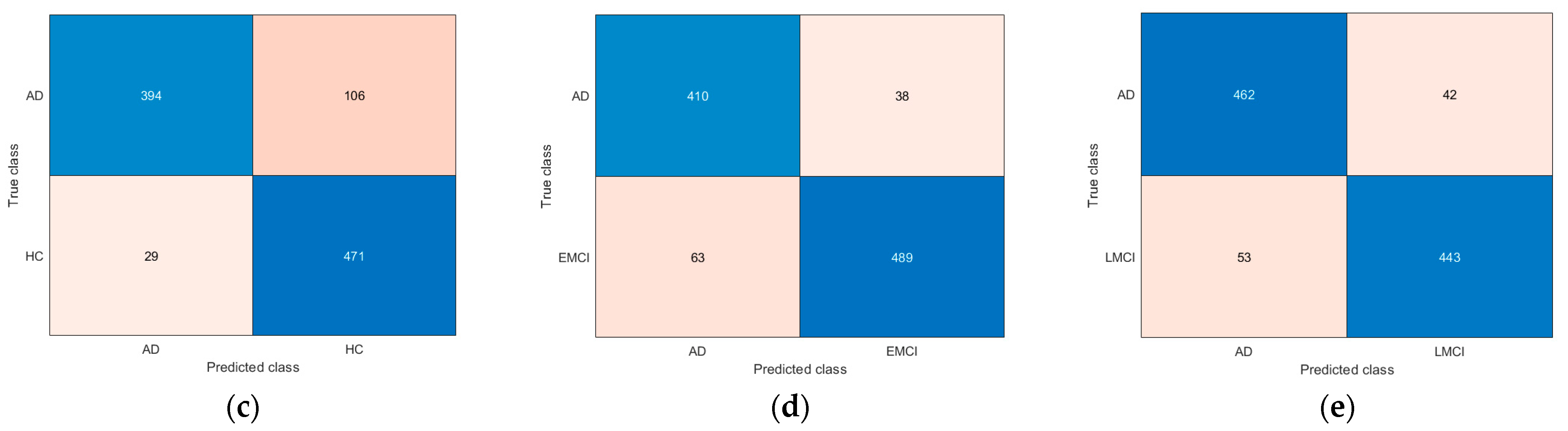
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Association, A.S. 2019 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2019, 15, 321–387. [Google Scholar] [CrossRef]
- Jiao, Z.; Ji, Y.; Jiao, T.; Wang, S. Extracting Sub-Networks from Brain Functional Network Using Graph Regularized Nonnegative Matrix Factorization. Comput. Model. Eng. Sci. 2020, 123, 845–871. [Google Scholar] [CrossRef]
- Agosta, F.; Galantucci, S.; Filippi, M. Advanced magnetic resonance imaging of neurodegenerative diseases. Neurol. Sci. 2017, 38, 41–51. [Google Scholar] [CrossRef]
- Moradi, E.; Pepe, A.; Gaser, C.; Huttunen, H.; Tohka, J. Machine learning framework for early MRI-based Alzheimer’s conversion prediction in MCI subjects. NeuroImage 2015, 104, 398–412. [Google Scholar] [CrossRef]
- Huang, Y.; Su, Y.; Byun, Y.; Lee, Y.; Kim, S. Analysis of multiple chronic disease characteristics in middle-aged and elderly South Koreans by exercise habits based on association rules mining algorithm. BMC Public Health 2023, 23, 1232. [Google Scholar] [CrossRef]
- Vougas, K.; Sakellaropoulos, T.; Kotsinas, A.; Foukas, G.-R.P.; Ntargaras, A.; Koinis, F.; Polyzos, A.; Myrianthopoulos, V.; Zhou, H.; Narang, S.; et al. Machine learning and data mining frameworks for predicting drug response in cancer: An overview and a novel in silico screening process based on association rule mining. Pharmacol. Ther. 2019, 203, 107395. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, L.; Miao, R.; Ren, H. A Data Mining Algorithm for Association Rules with Chronic Disease Constraints. Comput. Intell. Neurosci. 2022, 2022, 8526256. [Google Scholar] [CrossRef] [PubMed]
- Hadavi, S.M.S.; Oliaei, S.; Saidi, S.; Nadimi, E.; Kazemi-Galougahi, M.H. Using Data Mining and Association Rules for Early Diagnosis of Esophageal Cancer. Gulf J. Oncol. 2022, 1, 38–46. [Google Scholar]
- Zhang, D.; Wang, Y.; Zhou, L.; Yuan, H.; Shen, D. Multimodal classification of Alzheimer’s disease and mild cognitive impairment. NeuroImage 2011, 55, 856–867. [Google Scholar] [CrossRef] [PubMed]
- An, L.; Adeli, E.; Liu, M.; Zhang, J.; Shen, D. Semi-supervised Hierarchical Multimodal Feature and Sample Selection for Alzheimer’s Disease Diagnosis. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016, Athens, Greece, 17–21 October 2016; pp. 79–87. [Google Scholar]
- Wu, A.T.H.; Lawal, B.; Wei, L.; Wen, Y.-T.; Tzeng, D.T.W.; Lo, W.-C. Multiomics Identification of Potential Targets for Alzheimer Disease and Antrocin as a Therapeutic Candidate. Pharmaceutics 2021, 13, 1555. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Lin, X.; Lin, C.; Lin, X.; Chen, Z.; Zhang, L. Identification of endoplasmic reticulum stress-associated genes and subtypes for prediction of Alzheimer’s disease based on interpretable machine learning. Front. Pharmacol. 2022, 13, 975774. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Lu, Y.; Cao, Y.; Dang, C.; Wang, N.; Tian, K.; Luo, Q.; Guo, E.; Luo, S.; Wang, L.; et al. Identification of diagnostic signatures associated with immune infiltration in Alzheimer’s disease by integrating bioinformatic analysis and machine-learning strategies. Front. Aging Neurosci. 2022, 14, 919614. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Bao, W.; Hong, S. Alzheimer-Compound Identification Based on Data Fusion and forgeNet_SVM. Front. Aging Neurosci. 2022, 14, 931729. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Petersen, M.; Johnson, L.; Hall, J.; O’Bryant, S.E. Recursive Support Vector Machine Biomarker Selection for Alzheimer’s Disease. J. Alzheimer’s Dis. 2021, 79, 1691–1700. [Google Scholar] [CrossRef] [PubMed]
- Olatunji, S.O.; Alansari, A.; Alkhorasani, H.; Alsubaii, M.; Sakloua, R.; Alzahrani, R.; Alsaleem, Y.; Alassaf, R.; Farooqui, M.; Basheer Ahmed, M.I.; et al. Preemptive Diagnosis of Alzheimer’s Disease in the Eastern Province of Saudi Arabia Using Computational Intelligence Techniques. Comput. Intell. Neurosci. 2022, 2022, 5476714. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Z.; Chen, S.; Shi, H.; Xu, J. Multi-Modal Feature Selection with Feature Correlation and Feature Structure Fusion for MCI and AD Classification. Brain Sci. 2022, 12, 80. [Google Scholar] [CrossRef] [PubMed]
- Syaifullah, A.H.; Shiino, A.; Kitahara, H.; Ito, R.; Ishida, M.; Tanigaki, K. Machine Learning for Diagnosis of AD and Prediction of MCI Progression From Brain MRI Using Brain Anatomical Analysis Using Diffeomorphic Deformation. Front. Neurol. 2021, 11, 576029. [Google Scholar] [CrossRef]
- Houria, L.; Belkhamsa, N.; Cherfa, A.; Cherfa, Y. Multi-modality MRI for Alzheimer’s disease detection using deep learning. Phys. Eng. Sci. Med. 2022, 45, 1043–1053. [Google Scholar] [CrossRef]
- Yan, C.-G.; Wang, X.-D.; Zuo, X.-N.; Zang, Y.-F. DPABI: Data Processing & Analysis for (Resting-State) Brain Imaging. Neuroinformatics 2016, 14, 339–351. [Google Scholar] [CrossRef]
- Jenkinson, M.; Beckmann, C.F.; Behrens, T.E.; Woolrich, M.W.; Smith, S.M.J.N. Fsl. Neuroimage 2012, 62, 782–790. [Google Scholar] [CrossRef]
- Tzourio-Mazoyer, N.; Landeau, B.; Papathanassiou, D.; Crivello, F.; Etard, O.; Delcroix, N.; Mazoyer, B.; Joliot, M. Automated Anatomical Labeling of Activations in SPM Using a Macroscopic Anatomical Parcellation of the MNI MRI Single-Subject Brain. NeuroImage 2002, 15, 273–289. [Google Scholar] [CrossRef]
- Saykin, A.J.; Shen, L.; Foroud, T.M.; Potkin, S.G.; Swaminathan, S.; Kim, S.; Risacher, S.L.; Nho, K.; Huentelman, M.J.; Craig, D.W.; et al. Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimer’s Dement. 2010, 6, 265–273. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Zhang, X.; Xie, Y.; Tang, J.; Qin, W.; Liu, F.; Ding, H.; Ji, Y.; Yang, B.; Zhang, P.; Li, W.; et al. Dissect Relationships Between Gene Co-expression and Functional Connectivity in Human Brain. Front. Neurosci. 2021, 15, 797849. [Google Scholar] [CrossRef]
- Jeong, B.; Lee, J.; Kim, H.; Gwak, S.; Kim, Y.K.; Yoo, S.Y.; Lee, D.; Choi, J.-S. Multiple-Kernel Support Vector Machine for Predicting Internet Gaming Disorder Using Multimodal Fusion of PET, EEG, and Clinical Features. Front. Neurosci. 2022, 16, 856510. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Wu, Y.; Liu, W.; Wang, Y.; Xu, Z.; Jiao, Z. Research on Voxel-Based Features Detection and Analysis of Alzheimer’s Disease Using Random Survey Support Vector Machine. Front. Neuroinform. 2022, 16, 856295. [Google Scholar] [CrossRef] [PubMed]
- Murray, A.N.; Chandler, H.L.; Lancaster, T.M. Multimodal hippocampal and amygdala subfield volumetry in polygenic risk for Alzheimer’s disease. Neurobiol. Aging 2021, 98, 33–41. [Google Scholar] [CrossRef] [PubMed]
- An, N.; Fu, Y.; Shi, J.; Guo, H.-N.; Yang, Z.-W.; Li, Y.-C.; Li, S.; Wang, Y.; Yao, Z.-J.; Hu, B.; et al. Synergistic Effects of APOE and CLU May Increase the Risk of Alzheimer’s Disease: Acceleration of Atrophy in the Volumes and Shapes of the Hippocampus and Amygdala. J. Alzheimer’s Dis. 2021, 80, 1311–1327. [Google Scholar] [CrossRef]
- Caesar, M.H.; Nateka, L.J.; Abbi, R.H.; Lori, L.M. Impairments in Fear Extinction Memory and Basolateral Amygdala Plasticity in the TgF344-AD Rat Model of Alzheimer’s Disease Are Distinct from Nonpathological Aging. eNeuro 2022, 9, ENEURO.0181-0122.2022. [Google Scholar] [CrossRef]
- Feng, Q.; Niu, J.; Wang, L.; Pang, P.; Wang, M.; Liao, Z.; Song, Q.; Jiang, H.; Ding, Z. Comprehensive classification models based on amygdala radiomic features for Alzheimer’s disease and mild cognitive impairment. Brain Imaging Behav. 2021, 15, 2377–2386. [Google Scholar] [CrossRef]
- Hu, X.; Pickering, E.; Liu, Y.C.; Hall, S.; Fournier, H.; Katz, E.; Dechairo, B.; John, S.; Van Eerdewegh, P.; Soares, H.; et al. Meta-Analysis for Genome-Wide Association Study Identifies Multiple Variants at the BIN1 Locus Associated with Late-Onset Alzheimer’s Disease. PLoS ONE 2011, 6, e16616. [Google Scholar] [CrossRef]
- Barral, S.; Bird, T.; Goate, A.; Farlow, M.R.; Diaz-Arrastia, R.; Bennett, D.A.; Graff-Radford, N.; Boeve, B.F.; Sweet, R.A.; Stern, Y.; et al. Genotype patterns at PICALM, CR1, BIN1, CLU, and APOE genes are associated with episodic memory. Neurology 2012, 78, 1464. [Google Scholar] [CrossRef] [PubMed]
- Carrasquillo, M.M.; Belbin, O.; Hunter, T.A.; Ma, L.; Bisceglio, G.D.; Zou, F.; Crook, J.E.; Pankratz, V.S.; Sando, S.B.; Aasly, J.O.; et al. Replication of BIN1 Association with Alzheimer’s Disease and Evaluation of Genetic Interactions. J. Alzheimer’s Dis. 2011, 24, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Cheng, R.; Barral, S.; Reitz, C.; Medrano, M.; Lantigua, R.; Jiménez-Velazquez, I.Z.; Rogaeva, E.; St. George-Hyslop, P.H.; Mayeux, R. Identification of Novel Loci for Alzheimer Disease and Replication of CLU, PICALM, and BIN1 in Caribbean Hispanic Individuals. Arch. Neurol. 2011, 68, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Tezuka, K.; Handa, T.; Sato, R.; Takeuchi, H.; Takao, M.; Tano, M.; Uchida, Y. Upregulation of ribosome complexes at the blood-brain barrier in Alzheimer’s disease patients. J. Cereb. Blood Flow Metab. 2022, 42, 2134–2150. [Google Scholar] [CrossRef]
- Kelleher, D.J.; Gilmore, R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 2005, 16, 47R–62R. [Google Scholar] [CrossRef]
- Honma, K.; Iwao-Koizumi, K.; Takeshita, F.; Yamamoto, Y.; Yoshida, T.; Nishio, K.; Nagahara, S.; Kato, K.; Ochiya, T. RPN2 gene confers docetaxel resistance in breast cancer. Nat. Med. 2008, 14, 939–948. [Google Scholar] [CrossRef]
- Wilson, C.M.; High, S. Ribophorin I acts as a substrate-specific facilitator of N-glycosylation. J. Cell Sci. 2007, 120, 648–657. [Google Scholar] [CrossRef]
- Rentzos, M.; Zoga, M.; Paraskevas, G.P.; Kapaki, E.; Rombos, A.; Nikolaou, C.; Tsoutsou, A.; Vassilopoulos, D. IL-15 Is Elevated in Cerebrospinal Fluid of Patients With Alzheimer’s Disease and Frontotemporal Dementia. J. Geriatr. Psychiatry Neurol. 2006, 19, 114–117. [Google Scholar] [CrossRef]
- Asby, D.; Boche, D.; Allan, S.; Love, S.; Miners, J.S. Systemic infection exacerbates cerebrovascular dysfunction in Alzheimer’s disease. Brain 2021, 144, 1869–1883. [Google Scholar] [CrossRef]
- Shorena, J.; Niklas, M.; Erik, S.; Olof, L.; Sebastian, P.; Henrik, Z.; Kaj, B.; Oskar, H. CSF biomarkers of neuroinflammation and cerebrovascular dysfunction in early Alzheimer disease. Neurology 2018, 91, e867. [Google Scholar] [CrossRef]

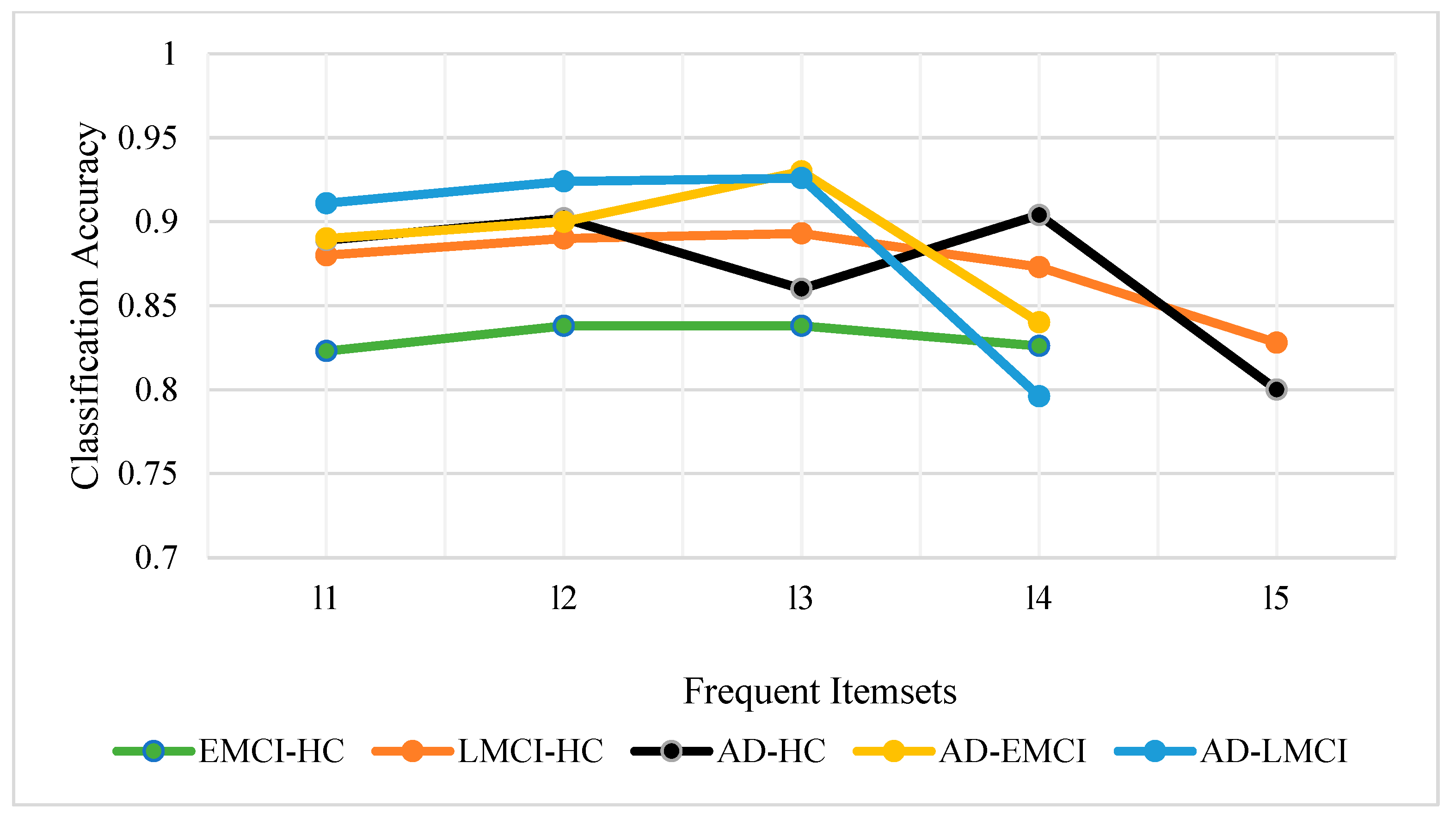
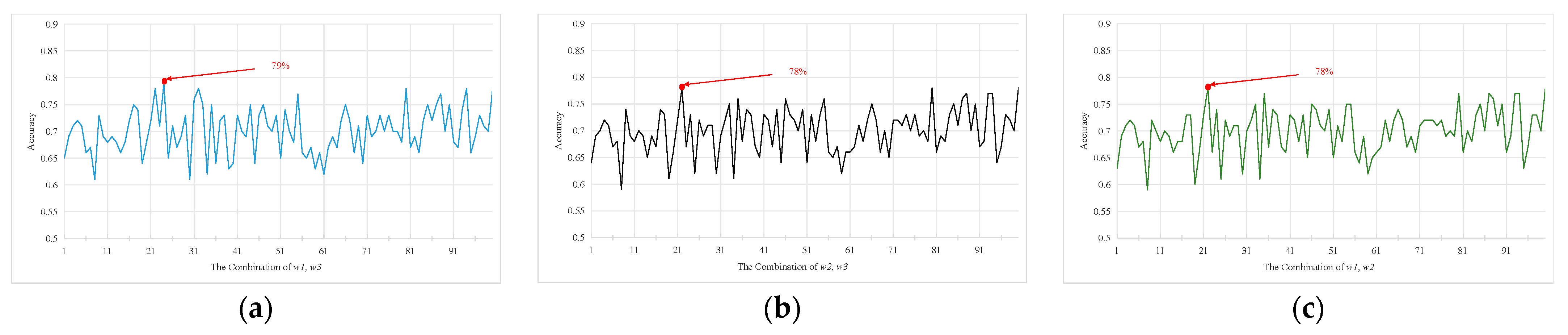
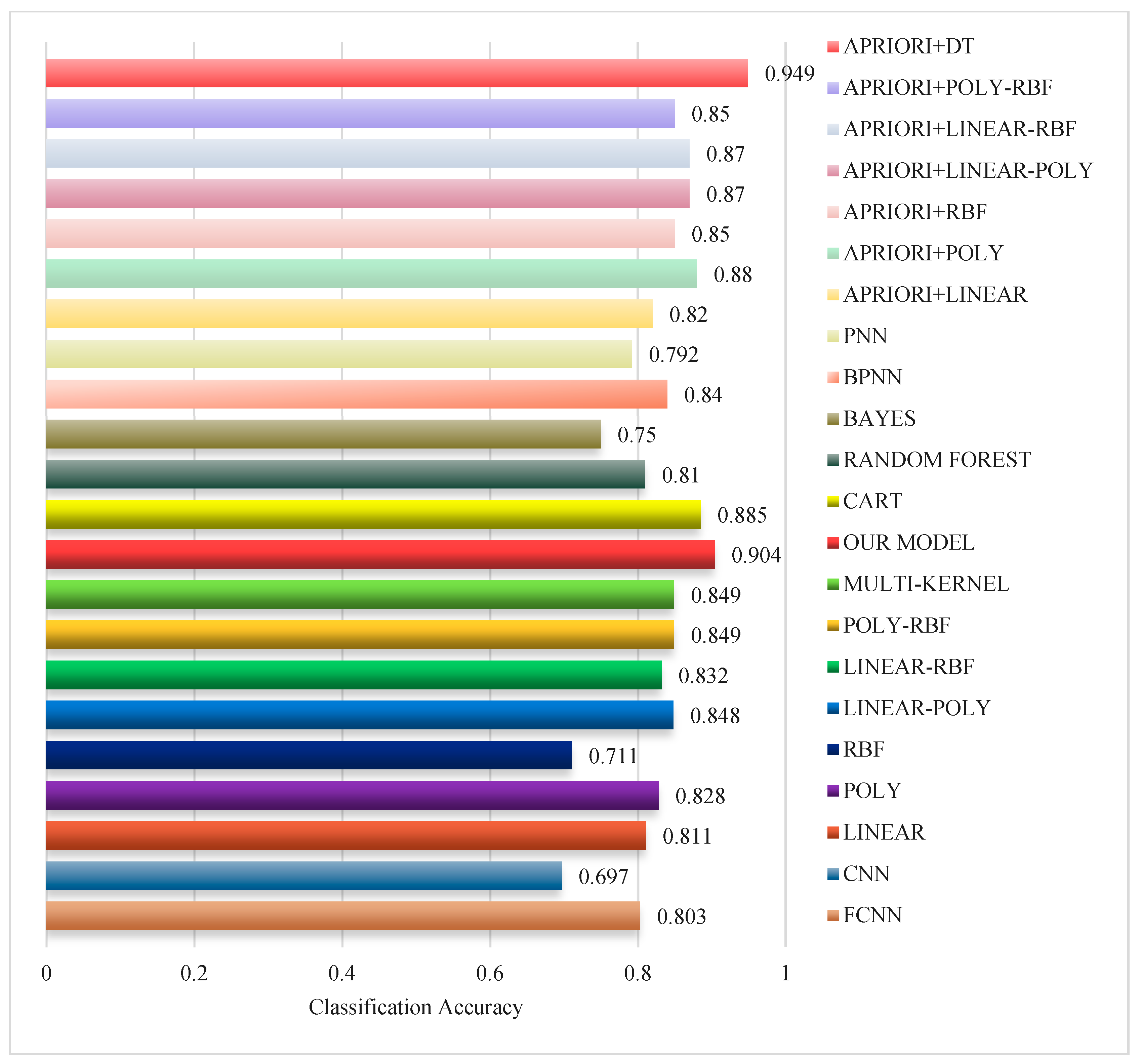
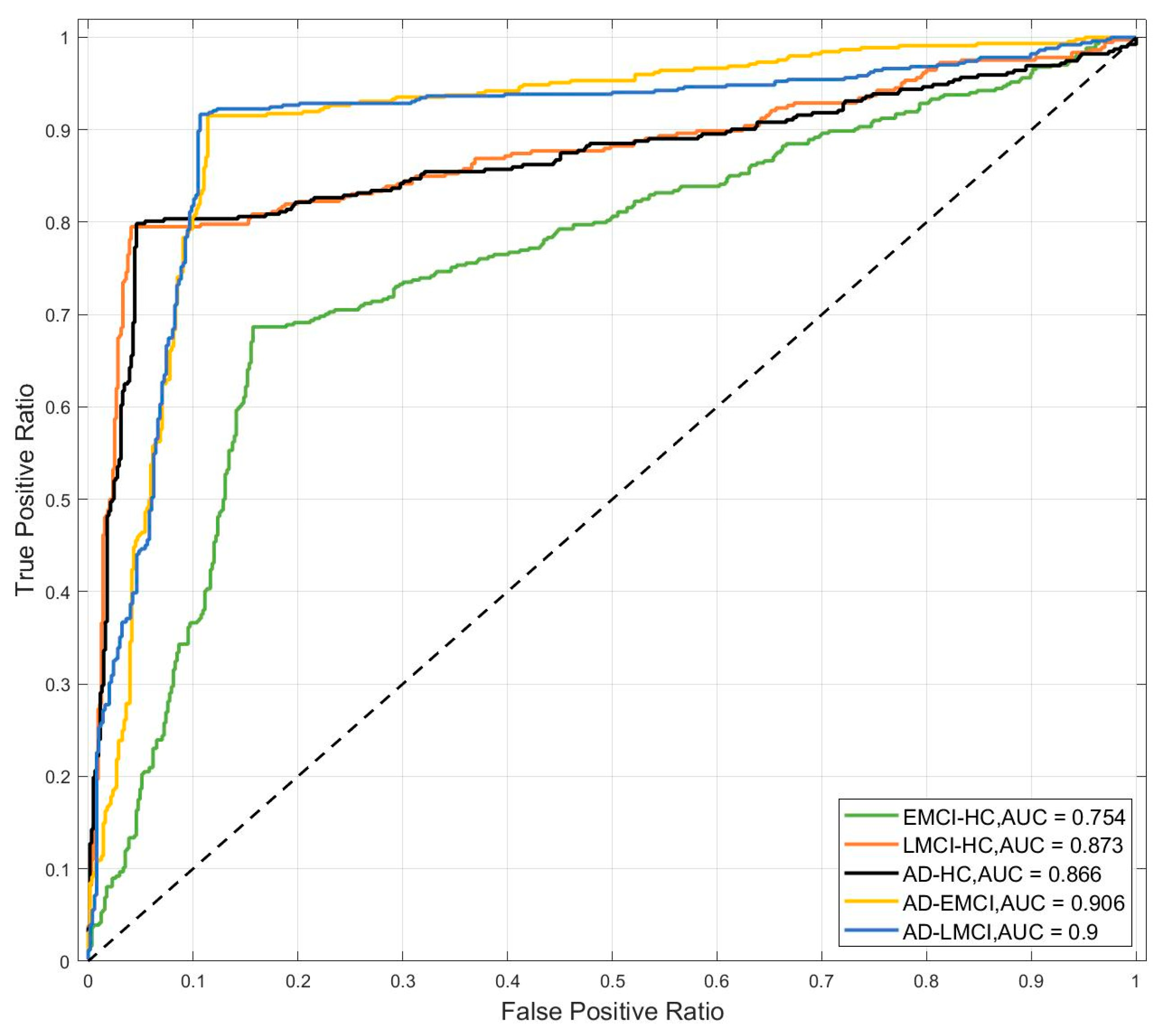
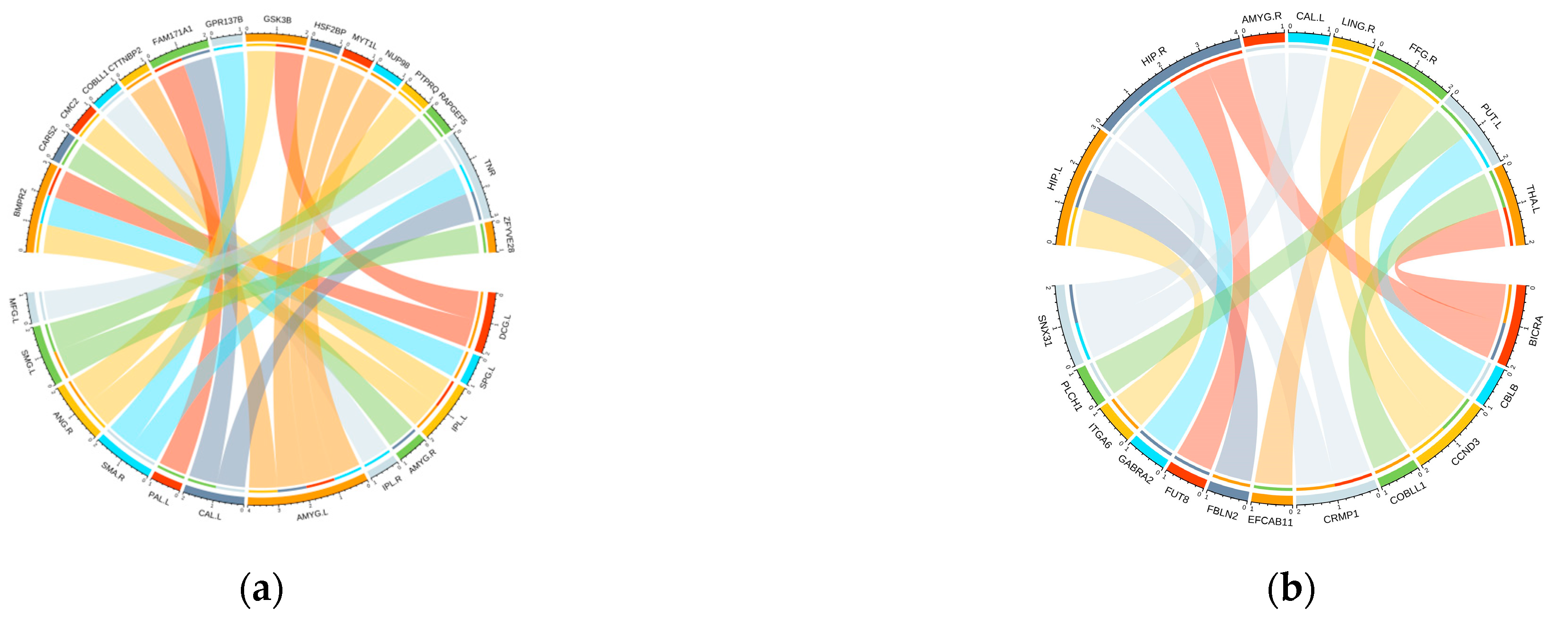
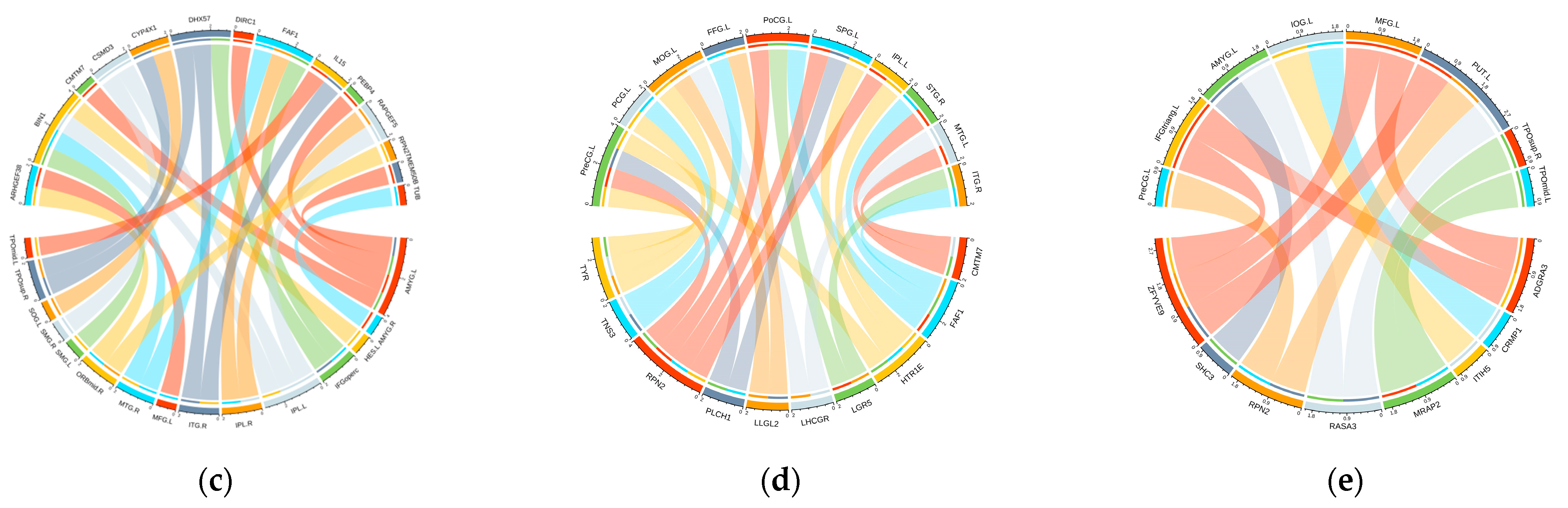
| Subjects | HC | EMCI | LMCI | AD | P |
|---|---|---|---|---|---|
| Number | 42 | 31 | 24 | 24 | - |
| Gender (M/F) | 19/23 | 12/19 | 14/10 | 8/16 | <0.001 |
| Age (Mean ± sd) | 74.2 ± 6.1 | 72.8 ± 6.4 | 70.9 ± 8.3 | 72.0 ± 7.6 | <0.001 |
| EDU (Mean ± sd) | 16.5 ± 2.7 | 15.8 ± 2.7 | 16.8 ± 2.6 | 15.5 ± 2.9 | <0.001 |
| L1 | L2 | L3 | L4 | … | Ln | |
|---|---|---|---|---|---|---|
| Generate From | S | L1 | L2 | L3 | … | Ln−1 |
| Accuracy Threshold | 0.8 | mean(ACCL1) | mean(ACCL2) | mean(ACCL3) | … | mean(ACCLn−1) |
| Model | Accuracy | F1 Score | Recall | Precision |
|---|---|---|---|---|
| LINEAR | 76.25% ± 2.64% | 74.8% ± 4.64% | 79% ± 6.46% | 71.8% ± 6.94% |
| POLY | 76.88% ± 3.02% | 74.8% ± 3.91% | 79.6% ± 5.52% | 71.6% ± 7.52% |
| RBF | 78.13% ± 3.29% | 77.6% ± 4.4% | 81.8% ± 6.43% | 74.8% ± 6.96% |
| LINEAR-POLY | 76.25% ± 2.64% | 75.6% ± 4.45% | 81.9% ± 6.82% | 71% ± 6.8% |
| LINEAR-RBF | 76.88% ± 3.02% | 74.9% ± 4.15% | 77.3% ± 5.76% | 73% ± 4.03% |
| POLY-RBF | 76.88% ± 3.02% | 76.5% ± 3.37% | 76.8% ± 4.61% | 76.4% ± 3.75% |
| MULTI-KERNEL | 76.88% ± 4.22% | 76.5% ± 4.53% | 79.9% ± 6.12% | 74.2% ± 5.07% |
| OUR MODEL | 90.20% ± 1.54% | 93.20% ± 1.08% | 93.30% ± 1.19% | 93.50% ± 1.02% |
| CART | 84.44% ± 2.34% | 82.2% ± 3.46% | 82% ± 5.03% | 83% ± 5.08% |
| RANDOM FOREST | 84.44% ± 2.34% | 81.8% ± 3.79% | 83.3% ± 4.45% | 80.8% ± 4.92% |
| BAYES | 85% ± 2.68% | 84.9% ± 2.92% | 85.2% ± 2.82% | 84.9% ± 3.07% |
| BPNN | 72.94% ± 4.11% | 76.5% ± 3.72% | 82.2% ± 3.71% | 71.4% ± 4.2% |
| PNN | 82.94% ± 1.86% | 84.1% ± 1.91% | 86.6% ± 2.27% | 81.8% ± 1.87% |
| APRIORI+LINEAR | 86.6% ± 3.47% | 90.6% ± 2.41% | 91.5% ± 2.17% | 90.4% ± 2.46% |
| APRIORI+POLY | 83.4% ± 3.69% | 88.7% ± 2.45% | 88.9% ± 2.64% | 89% ± 2.49% |
| APRIORI+RBF | 87.8% ± 3.19% | 91.3% ± 2.16% | 92.1% ± 2.08% | 91.3% ± 2.16% |
| APRIORI+LINEAR-POLY | 82.1% ± 4.79% | 85.5% ± 4.33% | 85.5% ± 4.14% | 85.7% ± 4.3% |
| APRIORI+LINEAR-RBF | 88.6% ± 2.84% | 89.1% ± 2.56% | 89.9% ± 2.51% | 88.9% ± 2.81% |
| APRIORI+POLY-RBF | 82% ± 3.74% | 87.7% ± 2.71% | 88.2% ± 2.57% | 87.6% ± 2.59% |
| APRIORI+DT | 94.57% ± 0.78% | 95.6% ± 0.52% | 95.6% ± 0.52% | 96.5% ± 0.53% |
| CNN(Overfitting) | 68.03% ± 1.33% | 26% ± 0% | 18% ± 0% | 50% ± 0% |
| FCNN(Overfitting) | 73.09% ± 1.15% | 31% ± 0% | 22% ± 0% | 50% ± 0% |
| Group | Accuracy | F1 Score | Recall | Precision |
|---|---|---|---|---|
| EMCI-HC | 81.88% ± 1.02% | 83.30% ± 0.78% | 83.60% ± 0.80% | 83.30% ± 0.78% |
| LMCI-HC | 88.97% ± 0.97% | 91.60% ± 0.80% | 92.80% ± 0.87% | 90.80% ± 0.60% |
| AD-HC | 89.73% ± 0.98% | 92.20% ± 0.75% | 94.00% ± 0.63% | 91.10% ± 0.83% |
| AD-EMCI | 90.20% ± 1.54% | 93.20% ± 1.08% | 93.30% ± 1.19% | 93.50% ± 1.02% |
| AD-LMCI | 92.13% ± 1.27% | 94.70% ± 1.00% | 94.70% ± 1.00% | 94.70% ± 1.00% |
| Brain Region | AD-EMCI | AD-HC |
|---|---|---|
| Left hippocampus | 0.178128395 | 0.001062739 |
| Right hippocampus | 0.13044139 | 0.055262965 |
| Left parahippocampal gyrus | 0.080443747 | 0.034482065 |
| Right parahippocampal gyrus | 0.045045533 | 0.008222377 |
| Left amydala | 0.11497812 | 0.044599828 |
| Right amydala | 0.089006046 | 0.117985895 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Z.; Tang, C.; Liang, Y.; Chang, S.; Ni, X.; Xiao, S.; Meng, X.; He, B.; Liu, W. Feature Detection Based on Imaging and Genetic Data Using Multi-Kernel Support Vector Machine–Apriori Model. Mathematics 2024, 12, 684. https://doi.org/10.3390/math12050684
Hu Z, Tang C, Liang Y, Chang S, Ni X, Xiao S, Meng X, He B, Liu W. Feature Detection Based on Imaging and Genetic Data Using Multi-Kernel Support Vector Machine–Apriori Model. Mathematics. 2024; 12(5):684. https://doi.org/10.3390/math12050684
Chicago/Turabian StyleHu, Zhixi, Congye Tang, Yingxia Liang, Senhao Chang, Xinyue Ni, Shasha Xiao, Xianglian Meng, Bing He, and Wenjie Liu. 2024. "Feature Detection Based on Imaging and Genetic Data Using Multi-Kernel Support Vector Machine–Apriori Model" Mathematics 12, no. 5: 684. https://doi.org/10.3390/math12050684
APA StyleHu, Z., Tang, C., Liang, Y., Chang, S., Ni, X., Xiao, S., Meng, X., He, B., & Liu, W. (2024). Feature Detection Based on Imaging and Genetic Data Using Multi-Kernel Support Vector Machine–Apriori Model. Mathematics, 12(5), 684. https://doi.org/10.3390/math12050684







