1. Introduction
Causal structure learning (CSL) is a crucial component of causal discovery and inference and aims to identify a directed acyclic graph (DAG) that represents causal relationships among variables. The accuracy of the DAG structure directly impacts the precision of subsequent causal inference. In previous studies, experts primarily constructed DAGs through domain knowledge and randomized controlled experiments, which are subjective and time-consuming. With the explosion of data volume, significant attention has been given to learning causal structures purely from observational data [
1,
2]. However, due to the high-dimensional feature space and substantial time costs, traditional CSL is computationally challenging, especially in high-dimensional and continuous scenarios. CSL finds widespread applications in real-world scenarios, including genetic analysis [
3], financial risk prediction [
4], biomedical image processing [
5], fault diagnosis [
6], telemarketing call filtering [
7], and more. However, data-driven CSL commonly encounters the following issues: (1) massive data volume leads to exponential computational complexity with increasing feature dimensions, rendering it an NP-hard problem [
8] and (2) inconsistent data quality, where sample noise significantly affects the effectiveness of CSL.
CSL helps reveal causal relationships within data, enhancing the interpretability and predictive capabilities of models. Understanding causal structures in complex systems is crucial for causal reasoning and decision-making. Existing CSL methods primarily fall into three categories: (1) Constraint-based (CB) methods, such as the IC (independent causal) [
9] and PC (Peter and Clark) algorithms [
10], mainly rely on conditional independence (CI) and fidelity constraints. These methods suffer from equivalence class issues in the output structure, preventing them from fully capturing complete causal information. (2) Score-based (SB) methods focus on designing scoring functions and optimizing searches. These methods formalize possible network structures and node parameters into linear descriptions, enhancing the intelligence of learning algorithms and problem-solving capabilities, but they reach certain limits in individual problem domains. (3) Gradient-based (GB) methods have also been formulated. Zheng et al. [
11] developed a continuous optimization method for learning causal structures, overcoming the computational cost of non-cyclic constraints. This method models traditional combinatorial optimization problems as continuous optimization problems, improving the precision and efficiency of structure learning. It has been extended to handle intervention data, confounding factors, and time series. Using likelihood-based objectives, Ng et al. [
12] formulated the problem as an unconstrained optimization problem involving only soft constraints, while Yu et al. [
13] developed an equivalent representation of DAGs, allowing for continuous optimization in DAG space without the need for equality constraints. However, these methods are mainly based on structural equation modeling (SEM) equations of specific structures, and the accuracy of learned structures depends on the quality of the modeled data. As the number of sample nodes increases, algorithm performance quickly deteriorates due to the influence of data noise.
Motivated by this situation, this work explores how to mitigate the impact of sample data noise on algorithms and proposes a continuous optimization algorithm based on curriculum learning (CL): termed CL-NOTEARS. CL, introduced by Bengio in 2009 at ICML [
14], mimics human cognitive learning by gradually transitioning from simpler to more complex samples. The ultimate goal is to guide the model’s learning direction, reduce the influence of sample noise, and make machine learning models more efficient and accurate. Over the subsequent years, many researchers have developed CL strategies tailored to specific applications such as weakly supervised object localization, object detection, and neural machine translation [
15]. Despite its success across various domains, mainstream research on CL remains relatively limited.
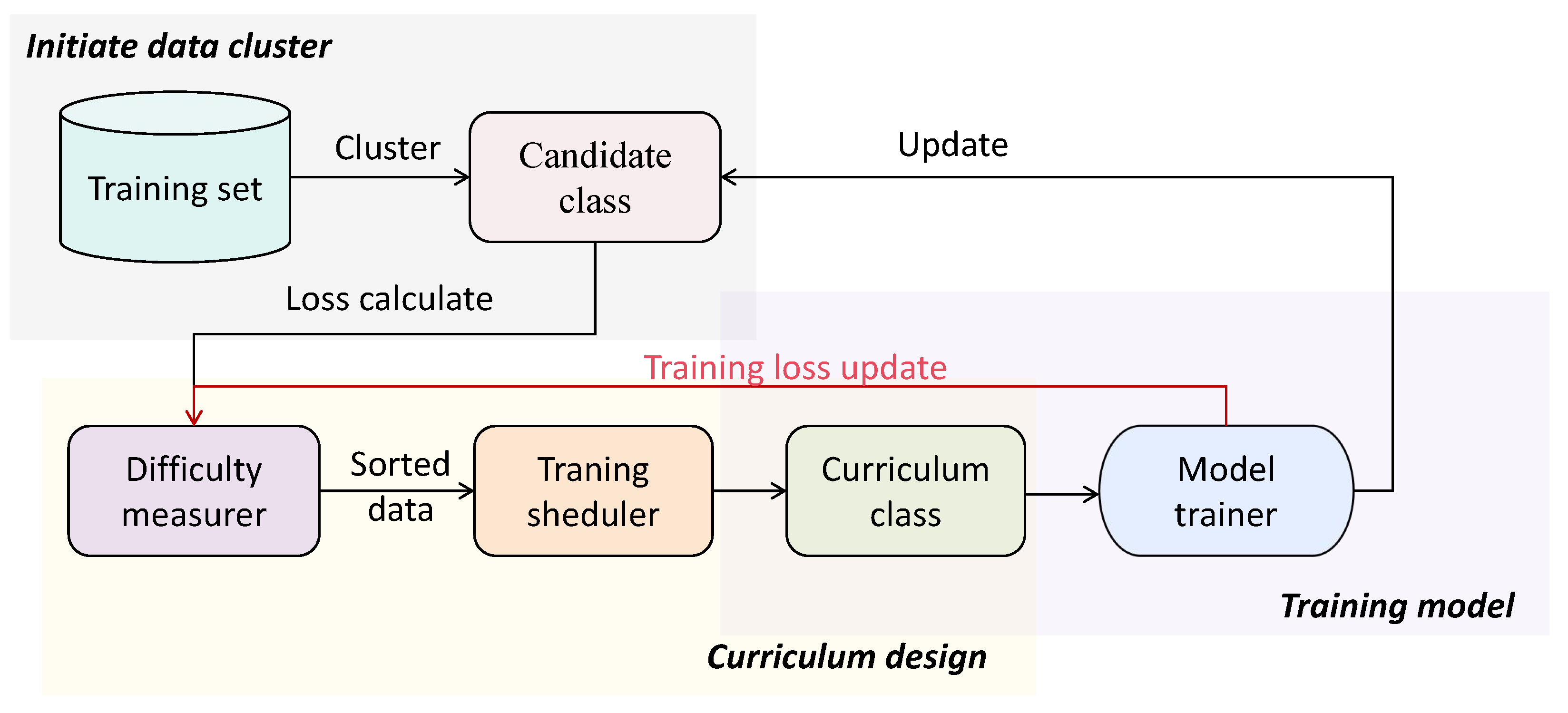
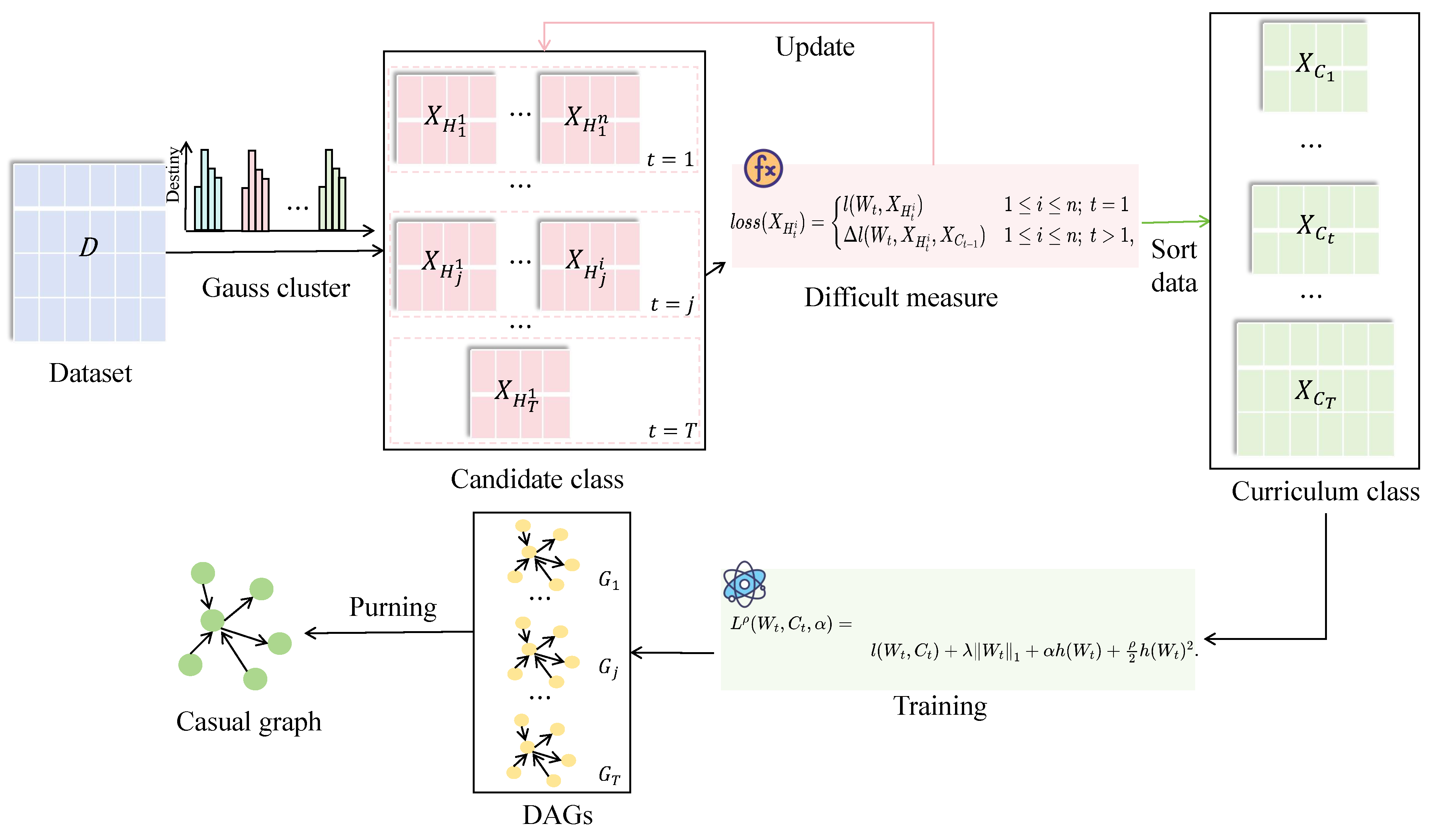
Based on the deep guidance and denoising mechanisms provided by CL, this work constructs a framework for curriculum sample learning aimed at mitigating the impact of sample noise on algorithms. By formulating a curriculum loss function, the framework adaptively adjusts the model’s sample learning sequence. The weights of the final causal diagram are dynamically updated based on the causal structure learned at different curriculum stages. The overall framework is illustrated in
Figure 1.
Specifically, the contributions of this work are as follows:
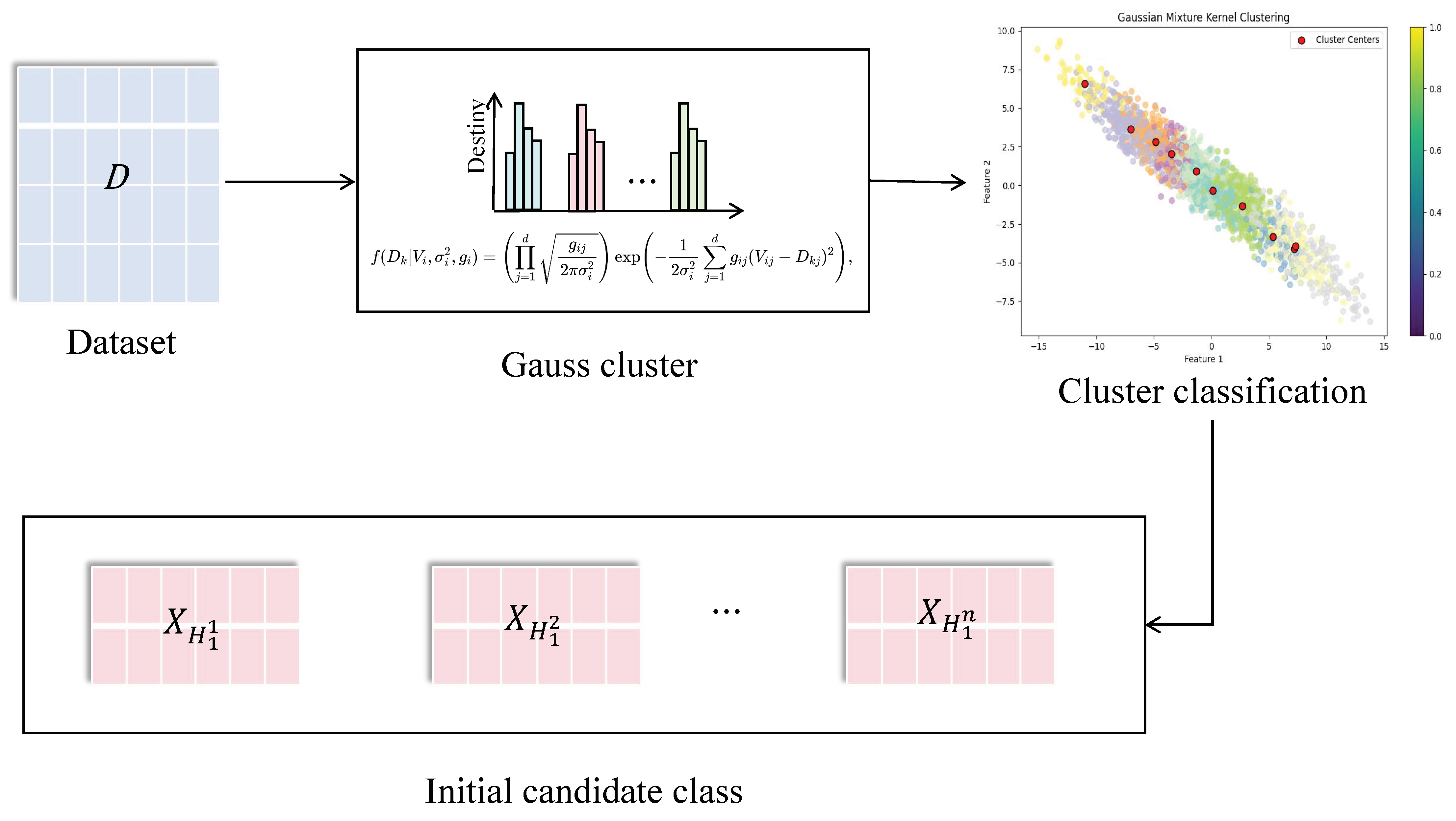
To evaluate the difficulty of learning data samples, this work proposes a similarity clustering method based on sample features. This method integrates similarity and entropy values among various sample features to classify samples into distinct types.
To alleviate the interference of data noise, this work designs a loss function to measure the learning difficulty of samples for curriculum-level selection. In each curriculum stage, candidate samples are used as new learning samples for structural learning. The optimal candidate samples are then selected as new curriculum samples for the next stage based on the measurement of the loss function value during the learning process. The order of model sample learning is adaptively adjusted according to the loss function value, leading to the final optimal network structure.
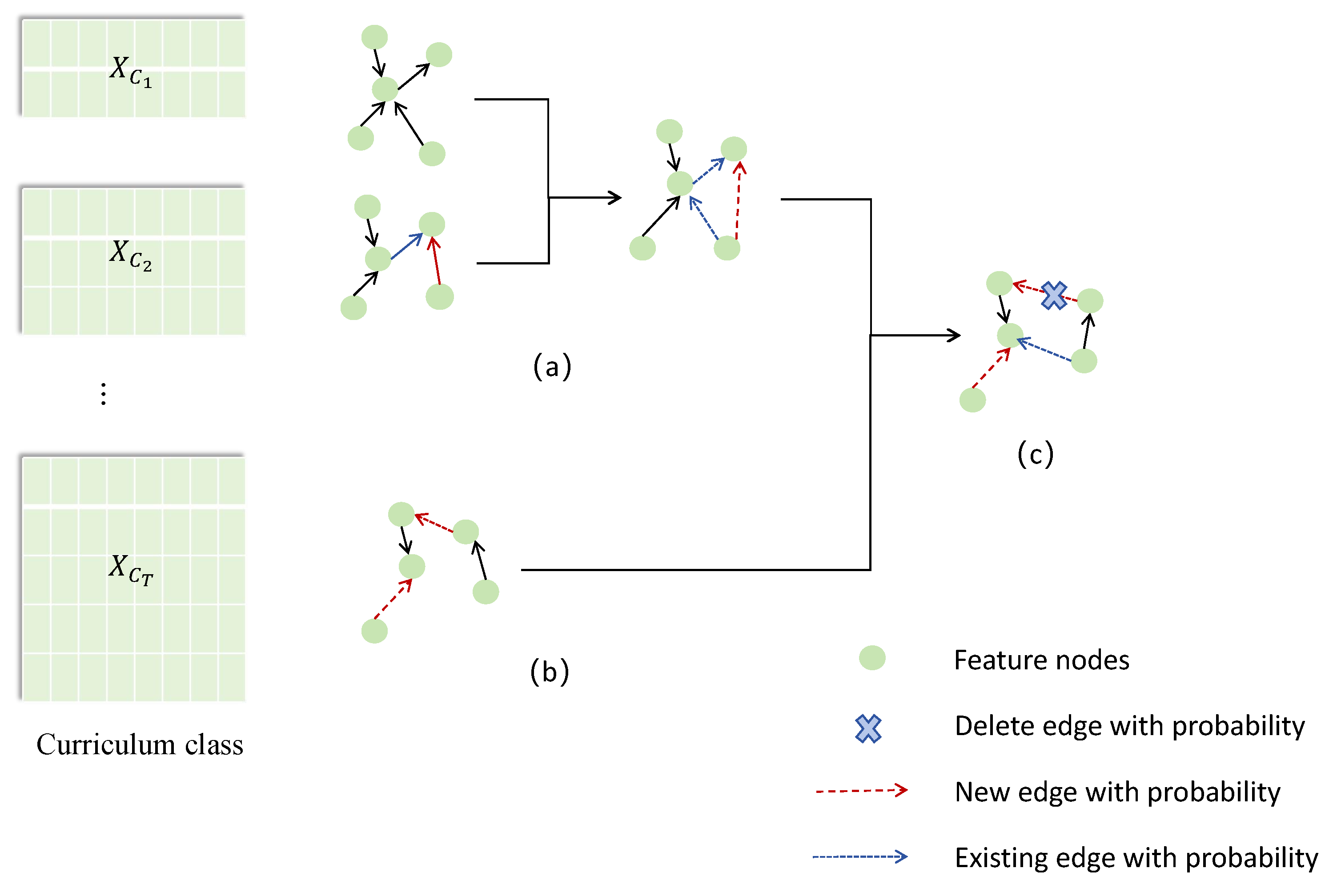
This work dynamically adjusts the weights of learned edges based on the results from different stages of the curriculum, filtering to obtain the final causal structure.
Experiments conducted on multiple synthetic datasets with varying scales and on a real dataset demonstrate that CL-NOTEARS exhibits good generalization ability and higher accuracy across networks with different scales and complexities.
Table 1 summarizes the notations and descriptions.
2. Related Work
Mining and inferring causal relationships are crucial for causal reasoning and decision-making in complex systems, especially in biomedical research, disease diagnosis, and public policy evaluation. The learned causal structure is represented as a DAG and is denoted as , where represents the set of nodes (features), and is the set of edges, representing causal relationships between different variables. The primary objective of causal discovery is to recover the underlying causal structure and the associated conditional probability distributions. Methods for learning causal structures from observational data mainly fall into three categories:
CB methods: CB methods are represented as constraint satisfaction problems and primarily explore causal skeletons by testing conditional independencies among different variables. They then orient edges from the skeleton to Markov equivalence classes. Some noteworthy algorithms in this category include the IC [
9], PC [
10], and IAMB algorithms [
16]. The IC algorithm searches for separation sets in the node set for each pair of variables; if there are no separation sets, the variables remain associated. The PC algorithm laid the foundation for CB methods and was preceded by the SGS algorithm [
17]. The PC algorithm relies mainly on efficient CI tests, assuming that causal edges between non-conditionally independent nodes should be retained. It has been effective for discovering causal relationship structures with high-dimensional sparse connections. However, its output depends on the variable processing order, which becomes more pronounced in high-dimensional environments, leading to highly variable results. Consequently, many researchers have studied and improved the PC algorithm. Li et al. [
18] alleviated the problem of high-order CI tests by introducing the FEPC algorithm. Additionally, some scholars addressed the instability caused by the PC algorithm’s dependency on node order and proposed the PC-stable algorithm [
19] and the PC-parallel algorithm [
20]. CB methods can handle a wider range of data types and distributions, and they boast high computational efficiency, making them highly interpretable. However, the accuracy of the learning process depends on the number of CI tests performed and the size of the constraint set. CB methods are sensitive to CI testing and data noise, and higher-order dependencies are unreliable for large-scale networks and complex data. Therefore, some research has introduced enhanced CI tests, including kernel-based causal learning methods such as the kernel-based Hilbert-Schmidt norms [
21] and the kernel-based conditional independence test (KCI) [
22]. However, these approaches require the assumption that random variables in different statistical tests are independent of each other, and research on such methods is still in its early stages. CB methods are based on the Markov assumption and require certain assumptions to infer CI relationships between variables for causal analysis, but they cannot provide the direction of edges in Markov equivalence classes.
SB methods: SB methods primarily assess the fit between the network structure and data using scoring functions and employ search algorithms to find the optimal structure. Common scoring functions include BIC [
23], BDeu [
24], MDL [
25], and AIC [
26]. SB methods mainly consist of two parts: scoring functions and search algorithms. The scoring function measures the fit between the structure and sample data; the better the fit, the higher the score obtained. The search algorithm is then used to find the network structure with the highest score, essentially seeking a network structure
that satisfies:
Many studies have proposed using greedy searches and heuristic searches to solve combinatorial problems and improve algorithm performance, with examples including genetic algorithms [
27], particle swarm optimization [
28], ant colony optimization [
29], and bee algorithms [
30]. However, the heuristic nature of these algorithms often leads to local optima. In addition to these heuristic search algorithms, some methods transform combinatorial optimization problems into continuous optimization problems.
GB methods: From the perspective of the data distribution characteristics of the causal mechanism, some scholars model data generation methods based on the structural equation model and transform the combinatorial optimization problem into a continuous optimization problem using gradient-based approaches through the smooth score function and the smooth representation of acyclicity, as represented by the NOTEARS algorithm [
11]. The NOTEARS algorithm utilizes the smoothing feature of the weighted adjacency matrix to model the data: aiming to find the optimal structure while satisfying the aperiodic constraints. Subsequently, DAG-GNN [
13] and GAE [
31] extended the NOTEARS idea to nonlinear scenarios, assuming that all variables undergo a common nonlinear transformation. Gran-DAG [
32] models variable distributions using multilayer perceptrons (MLPs) and constructs equivalent weighted adjacency matrices. Zhu et al. [
33] utilized reinforcement learning algorithms to search for the best-scoring DAG, resulting in training times that far exceed those of other algorithms due to the exploratory nature of reinforcement learning.
During the training of learning algorithms based on observational data, a common phenomenon emerges: there are significant disparities in performance as the sample size increases and with variations in sampling capabilities. It is evident that both the quality and scale of the data have a considerable impact on algorithm performance. The objective is to explore a hierarchical learning framework that involves designing sample prioritization evaluation metrics to partition samples and to develop curriculum loss functions for the model training process. These steps aim to enable adaptive adjustments to the sequence of training data samples throughout the model training process.
3. Problem Formulation
Let the DAG be represented as
, where
represents the set of nodes, with
d being the number of nodes. The set of edges
E is defined as
, indicating edges from node
to node
. Each node
is associated with a random variable
. Probability models associated with
G assume data independence such that, given the parent nodes,
is independent of its non-descendant nodes. The joint probability distribution of the data can be decomposed into the product of the conditional probabilities of each individual node, given its parent nodes, following the probability chain rule:
where
represents the conditional probability of
given its parent nodes. The parent nodes of
are represented as
.
Considering the limitations of traditional SEM, where some variables can only be observed and measured empirically, and inspired by recent advancements in continuous optimization, we model X via a SEM defined by the weighted adjacency matrix . Consequently, we operate not in the discrete space but in , which is the continuous space of real matrices.
Let be the binary matrix such that and is zero otherwise. Then defines the adjacency matrix of a directed graph . In addition to the graph , defines the linear SEM through , where is a random vector, and is an arbitrary noise vector. Assuming z is mutually independent with no unobserved confounding factors, the linear SEM is thus fully defined.
Zheng et al. [
12] proved that the matrix
is a DAG if and only if
holds. Here, ∘ denotes the Hadamard product, and
is the matrix exponential of
W. The resulting continuous constrained optimization problem is:
where
represents the
norm, which promotes sparsity in the matrix. Specifically,
, resulting in the regularized score function, with
as the regularization coefficient. Here,
is the least squares objective, where
denotes the Frobenius norm. The Frobenius norm measures the square root of the sum of the absolute squares of the matrix elements.
5. Experiments
To encompass more general and challenging cases in the CSL scenario, this work addresses the following three research questions:
: In scenarios involving linear Gaussian (LG) noise and linear non-Gaussian (LN) noise, is the CL-NOTEARS algorithm superior to other models?
: Are the curriculum mechanism and continual update strategy effective for CSL?
: In real-world scenarios, can the CL-NOTEARS algorithm still reduce noise in the data and provide a greater advantage?
5.1. General Settings
All the experiments were performed on a computer equipped with an Intel(R) Core(TM) i7-1165G7 CPU at 2.80 GHz and 16 GB of memory, and the compiler environment is PyCharm 2021.1.2.
Datasets Following previous research, we generated six types of synthetic datasets to answer RQ1. For RQ2, we chose four types of LN noise datasets. For RQ3, we utilized real BN data from Sachs.
Simulated data: The work employed two graph sampling schemes—Erdős–Rényi (ER) and scale-free (SF) [
43]—varying across four aspects: the data generation process, number of nodes, edge sparsity, and graph type. The data generation process included LG/LN ANM. For the LG autoregressive model, data were sampled as
, with independent noise
. For the LN autoregressive model, the work compared two noise distributions: additive noise
from either an exponential or uniform distribution with uniformly sampled variances. Each dataset type was randomly sampled according to ER or SF schemes, and graphs with nodes
and edges
were considered.
Real BN data: The work adopts a real BN dataset—specifically, the Sachs dataset. Sachs is a multivariate proteomic dataset that is widely used in causal discovery. It contains measurements of various proteins and phospholipids found in human immune system cells, including 11 phosphorylated proteins and phospholipids (PKC, PKA, P38, Jnk, Raf, Mek, Erk, Akt, Plcg, PIP2, and PIP3).
Evaluation index: The work evaluates the quality of learned network structures using the proposed CL-NOTEARS approach through the following measurements:
Structural Hamming distance (SHD): The SHD is a standard distance used to compare graphs by using their adjacency matrices. It involves calculating the differences between two (binary) adjacency matrices: each missing or non-existent edge in the target graph is considered an error. The smaller the SHD, the fewer incorrect edges are learned, and the better the learning effect.
True positive rate (TPR): In binary classification, the TPR, also known as sensitivity or recall, measures the proportion of actual positive cases that are correctly identified by a classification model. It is calculated as the number of true positive predictions divided by the sum of true positive and false negative predictions.
False discovery rate (FDR): The FDR is the ratio of all discoveries that are incorrect or reversed in direction. The FDR helps determine potential false discoveries. The smaller the FDR, the lower the error rate of the learned network structure, and the better the algorithm’s performance.
Using the SHD, TPR, and FDR to measure the benefits of DAG provides a comprehensive evaluation, assessing the network structure, classification accuracy, and performance of feature selection from different perspectives.
Baseline: On the dataset, the work compared CL-NOTEARS against several classical and state-of-the-art methods, including PC [
10], NOTEARS [
11], DAG-GNN [
13], GraN-DAG [
32], and DirectLiNGAM [
44]. The codes for these baseline methods are available in the pyagrum, case-learn2, and gCastle3 packages. Since PC may learn undirected edges, MCSL [
45] was used to treat undirected edges as true edges if the true graph has a directed edge instead.
5.2. Comparison of Linear Models with Gaussian Noise
This section evaluates the proposed methods using LG models with equal-variance Gaussian noise and LiNGAM [
46] data models, where the true DAGs are known to be identifiable [
47]. The parameters
and
are used to generate the observed data.
In this section, we first present a comparison between CL-NOTEARS and state-of-the-art methods using synthetic datasets generated by linear SEMs.
Table 3 and
Table 4 show the variations in FDR and SHD for the CL-NOTEARS model on ER- and SF-sampled datasets of LG ANM, respectively.
Figure 5 illustrates the TPR performance of CL-NOTEARS across these six classes of datasets. The following observations can be made: (1) PC performs poorly due to the dense graphs in the generated data. (2) Both NOTEARS and DAG-GNN exhibit high performance in causal discovery. (3) GraN-DAG performs worse because it uses a two-layer feedforward neural network to model causal relationships, which limits its ability to effectively learn the ideal causal structures. (4) When
, the CL-NOTEARS algorithm does not show significant superiority over DAG-GNN, whereas when
, CL-NOTEARS demonstrates strong advantages. Moreover, with the same number of nodes, CL-NOTEARS exhibits increasing superiority as the number of edges grows. This is because with the increase in both nodes and edges, the network becomes more complex, introducing relatively more noise into the sampled datasets. CL-NOTEARS leverages the curriculum-grained framework for noise reduction, allowing it to learn relatively accurate causal structures even in higher-noise datasets, thus demonstrating significant advantages. (5) For each node-size dataset, the FDR value of the CL-NOTEARS algorithm ranges between 0 and 0.01, confirming that the CL framework effectively mitigates the impact of data noise and reduces the occurrence of erroneous edges.
5.3. Comparison of Linear Models with Non-Gaussian Noise
LN data models were evaluated with noise types set to exponential and uniform distributions. For both cases, and , generating observational data with 2000 instances per dataset. Four causal learning algorithms were compared: the traditional CB method PC algorithm, the GB methods DAG-GNN and GraN-DAG, and the NOTEARS algorithm.
For ER and SF graphs of SEM with LN noise, the experimental results are presented in
Table 5 and
Table 6. The results show that DAG-GNN significantly outperforms the other algorithms. This is due to its ability to capture complex interactions and nonlinear transformations among features in linear models under non-Gaussian noise. In contrast, the CL-NOTEARS model performs less effectively with non-Gaussian noise compared to Gaussian noise. This is because the initial partitioning of the CL-NOTEARS algorithm relies on Gaussian mixture models, making it sensitive to the clustering effects of non-Gaussian noise.
5.4. The Effect of Different Curriculum Difficulty Levels on the Learning Process
This work revisited Bengio’s proposed CL prototype model, as highlighted in the literature discussion in
Section 5. It explored various challenges in the curriculum mechanism by considering three cases:
Curriculum mechanism: a basic setup that progresses from easy to difficult, where learning begins with simple sample examples and gradually advances to more complex ones.
Anti-curriculum mechanism: under anti-curriculum conditions, two mechanisms based on the original CL-NOTEARS model architecture were designed: (1) initially learning from difficult sample tasks and then transitioning from simple to complex samples in subsequent learning phases and (2) learning progresses from complex samples to simple samples throughout the entire process.
Random curriculum mechanism: learning from samples of varying difficulties selected randomly.
The specific design is illustrated in
Figure 6.
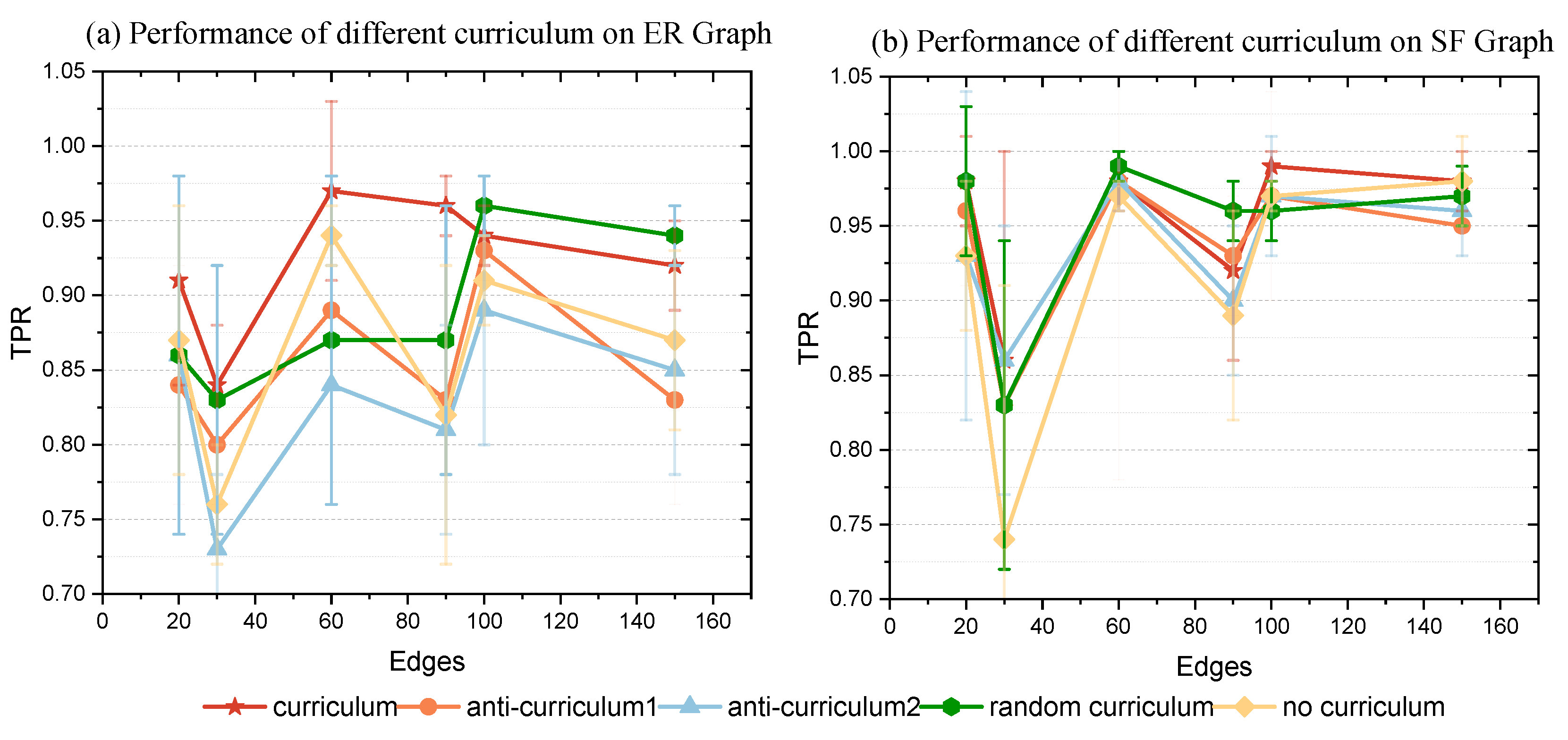
Based on these three curriculum mechanisms, the CL-NOTEARS algorithm was integrated and compared with the NOTEARS algorithm, which lacks a curriculum mechanism. Observational data were generated with and , with each dataset containing 2000 instances.
Table 7 shows the FDR and SHD performances for different curriculum mechanisms on the datasets, while
Figure 7 visually demonstrates the causal relationships captured by the different curriculum mechanisms. Observations indicate: (1) curriculum ≥ anti-curriculum 1 ≥ anti-curriculum 2; the first anti-curriculum mechanism performs better than the second one, as it only changes the difficulty of the initial learning task, resulting in less disturbance from noise compared to the second mechanism. (2) The random curriculum mechanism exhibits more variable performance compared to the curriculum mechanism.
Figure 7 shows that the errors associated with the random curriculum mechanism are significantly larger and essentially encompass the range of errors found with the curriculum mechanism. This variability is due to the random sampling approach, which introduces randomness into the learning process, occasionally leading to better outcomes than those achieved with the curriculum mechanism.
5.5. The Impact of the Number of Curriculum Stages on Algorithms
The number of curriculum stages in the algorithm influences the size of the learning sample set at different stages, which in turn affects the accuracy of separating the sample set at various noise levels. To investigate the effect of the curriculum stage size on the algorithm, four different sizes were considered:
. Observation data were generated with
and
. The dataset was randomly sampled using the ER scheme to create a real DAG consisting of 2000 sample instances. Evaluation metrics included TPR and SHD. Each experimental result was averaged over five sets of data, and the results are summarized in
Table 8.
It can be seen from
Table 8 that the best performance occurs when the number of curriculum stages is 10. Based on the experimental data, we speculate that if the number of curriculum stages is too large, sample division becomes too fine, making it difficult for the algorithm to differentiate between noise and signals in small samples, which results in poor learning performance. Conversely, if the number of curriculum stages is too small, the sample division is too coarse, and the sample set learned in each stage becomes noisy, and the model fails to balance noise differences effectively. Therefore, when dividing curriculum stages, the size of each stage should be adjusted based on the size of the sample set for that stage.
5.6. Real Data
The Sachs dataset [
48], which contains 11 nodes and 17 edges, is widely used in the study of graph models. The expression levels of proteins and phospholipids in this dataset can be used to uncover the underlying protein signaling network. This dataset is a common benchmark in graph modeling, and its experimental annotations are widely accepted by the biological community and encompass both observational and interventional aspects. The observational dataset includes
samples and is used to discover causal structures. The work also uses Gaussian process regression to simulate causality and to calculate the score. Since the real network structure is sparse, the SHD of an empty graph can be as low as 17. Comparing the best available algorithms, detailed results of the estimated graphs constructed by the different algorithms are shown in
Table 9. This includes the total number of edges, the number of correct edges, and the SHD. The PC algorithm, which outputs many undirected edges, is not included in this comparison.
As shown in
Table 9, the CL-NOTEARS algorithm demonstrates a significant advantage over other methods, achieving an optimal value of 13. Although the CL-NOTEARS algorithm identifies the same number of correct edges as the NOTEARS algorithm, it exhibits a substantial reduction in the SHD and a marked decrease in the number of erroneous edges. This improvement is attributed to the CL framework, which mitigates the impact of data noise on the algorithm, thereby significantly reducing the number of erroneous edges detected.