Analysis of Caputo Fractional-Order Co-Infection COVID-19 and Influenza SEIR Epidemiology by Laplace Adomian Decomposition Method
Abstract
:1. Introduction
2. Preliminaries
2.1. Basic Definitions and Prepositions
2.2. Mathematical Model Formulation
2.2.1. Deterministic Epidemic Models—Compartment Approach
2.2.2. SEIR Model [16]
2.2.3. Fractional-Order SEIR (FO-SEIR) Model
2.2.4. Classical Co-Infection SEIR Model [4]
2.3. Model Analysis—Stability Analysis and Equilibria [16,29]
Reproduction Number [18]
3. Fractional-Order Co-Infection SEIR (Co-Infection FO-SEIR) Model
3.1. Fractional-Order Co-Infection SEIR Model (Co-Infection FO-SEIR)
3.2. Local Stability Analysis of COVID-19-Only Model
3.2.1. Disease (COVID-19)-Free Equilibrium (DFE)
3.2.2. Non-Negative Solution [15]
3.2.3. Reproduction Number (COVID-19-Only Model)
3.3. Local Stability Analysis of Influenza-Only Model
3.3.1. Influenza-Free Equilibrium
3.3.2. Non-Negative Solution [15]
3.3.3. Reproductive Number
3.4. The Co-Infection SEIR Model of COVID-19 and Influenza
3.4.1. Disease (Co-Infection)-Free Equilibrium
3.4.2. Non-Negative Solution
3.4.3. Reproductive Number for COVID-19 and Influenza [4]
4. Numerical Simulations
4.1. Using LADM [14]
4.2. Graphical Analysis Using LADM
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Azar, A.T.; Hassanien, A.E. Modeling, Control and Drug Development for COVID-19 Outbreak Prevention; Springer: Berlin/Heidelberg, Germany, 2021. [Google Scholar]
- Tchoumi, S.; Diagne, M.; Rwezaura, H.; Tchuenche, J. Malaria and COVID-19 co-dynamics: A mathematical model and optimal control. Appl. Math. Model. 2021, 99, 294–327. [Google Scholar] [CrossRef]
- Derouich, M.; Boutayeb, A.A. An avian influenza mathematical model. Appl. Math. Sci. 2008, 2, 1749–1760. [Google Scholar]
- Ojo, M.M.; Benson, T.O.; Peter, O.J.; Goufo, E.F.D. Nonlinear optimal control strategies for a mathematical model of COVID-19 and influenza co-infection. Phys. A Stat. Mech. Its Appl. 2022, 607, 128173. [Google Scholar] [CrossRef] [PubMed]
- Karim, S.; Razali, R. A Proposed Mathematical Model of Influenza A, H1N1 for Malaysia. J. Appl. Sci. 2011, 11, 1457–1460. [Google Scholar] [CrossRef]
- Jung, E.; Iwami, S.; Takeuchi, Y.; Jo, T.C. Optimal control strategy for prevention of avian influenza pandemic. J. Theor. Biol. 2009, 260, 220–229. [Google Scholar] [CrossRef]
- Lee, S.; Chowell, G.; Castillo-Chávez, C. Optimal control for pandemic influenza: The role of limited antiviral treatment and isolation. J. Theor. Biol. 2010, 265, 136–150. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado-Payán, E.; Montagud-Marrahi, E.; Torres-Elorza, M.; Bodro, M.; Blasco, M.; Poch, E.; Soriano, A.; Piñeiro, G.J. SARS-CoV-2 and influenza virus co-infection. Lancet 2020, 395, e84. [Google Scholar] [CrossRef]
- Ali, H.M.; Ameen, I.G. Stability and optimal control analysis for studying the transmission dynamics of a fractional-order MSV epidemic model. J. Comput. Appl. Math. 2023, 434, 115352. [Google Scholar] [CrossRef]
- Guo, P. The Adomian Decomposition Method for a Type of Fractional Differential Equations. J. Appl. Math. Phys. 2019, 7, 2459–2466. [Google Scholar] [CrossRef]
- Kimeu, J.M. Fractional Calculus: Definitions and Applications. Master’s Theses, Western Kentucky University, Bowling Green, KY, USA, 2009. Available online: https://digitalcommons.wku.edu/theses/115 (accessed on 1 April 2009).
- Acay, B.; Bas, E.; Abdeljawad, T. Non-local fractional calculus from different viewpoint generated by truncated M-derivative. J. Comput. Appl. Math. 2020, 366, 112410. [Google Scholar] [CrossRef]
- Adomian, G. A review of the decomposition method and some recent results for nonlinear equations. Comput. Math. Appl. 1991, 21, 101–127. [Google Scholar] [CrossRef]
- Zeng, Y. A Laplace Adomian Decomposition Method for Fractional Order Infection Model. J. Appl. Math. Comput. 2023, 6, 529–534. [Google Scholar] [CrossRef]
- Tamilalagan, P.; Karthiga, S.; Manivannan, P. Dynamics of fractional order HIV infection model with antibody and cytotoxic T-lymphocyte immune responses. J. Comput. Appl. Math. 2020, 382, 113064. [Google Scholar] [CrossRef]
- Okundalaye, O.; Othman, W.; Oke, A. Toward an efficient approximate analytical solution for 4-compartment COVID-19 fractional mathematical model. J. Comput. Appl. Math. 2022, 416, 114506. [Google Scholar] [CrossRef]
- Diekmann, O.; Heesterbeek, J.A.P.; Roberts, M.G. The construction of next-generation matrices for compartmental epidemic models. J. R. Soc. Interface 2009, 57, 873–885. [Google Scholar] [CrossRef]
- Selvam, G.M.A.; Britto, J.S.D.; Vianny, A. Analysis of Fractional Order SIR Model. Int. J. Eng. Res. Technol. 2017, 5. Available online: https://www.ijert.org/research/analysis-of-fractional-order-sir-model-IJERTCONV5IS04012.pdf (accessed on 1 June 2024).
- Gumel, A.B.; Iboi, E.A.; Ngonghala, C.N.; Ngwa, G.A. Mathematical assessment of the roles of vaccination and non-pharmaceutical interventions on COVID-19 dynamics: A multigroup modeling approach. MedRxiv 2021. [Google Scholar] [CrossRef]
- Jin, Z.; Zhang, J.; Song, L.P.; Sun, G.Q.; Kan, J.; Zhu, H. Modelling and analysis of influenza A (H1N1) on networks. BMC Public Health 2011, 11, S9. [Google Scholar] [CrossRef]
- Kanyiri, C.W.; Mark, K.; Luboobi, L. Mathematical Analysis of Influenza A Dynamics in the Emergence of Drug Resistance. Comput. Math. Methods Med. 2018, 2434560. [Google Scholar] [CrossRef]
- Wessel, L.; Hua, Y.; Wu, J.; Moghadas, S.M. Public health interventions for epidemics: Implications for multiple infection waves. BMC Public Health 2011, 11, S2. [Google Scholar] [CrossRef]
- Abbott, S. The Laplace transform: Theory and applications, by Joel L. Schiff., Pp. 233. £27.50. 2000. ISBN 0 387 98698 7 (Springer-Verlag). Math. Gaz. 2001, 85, 178–179. [Google Scholar] [CrossRef]
- Caputo, M.; Fabrizio, M. A new definition of fractional derivative without singular kernel. Prog. Fract. Differ. Appl. 2015, 1, 73–85. [Google Scholar]
- Caputo, M. Elasticita e Dissipazione; Zanichelli: Modena, Italy, 1965. [Google Scholar]
- Meena, B. An Overview of Definitions of Riemann-Liouville’s Fractional Derivative and Caputo’s Fractional Derivative. Int. J. Sci. Res. IJSR 2019, 10, 2319–7064. [Google Scholar]
- Paul, S.; Mahata, A.; Mukherjee, S.; Roy, B.; Salimi, M.; Ahmadian, A. Study of Fractional Order SEIR Epidemic Model and Effect of Vaccination on the Spread of COVID-19. Int. J. Appl. Comput. Math. 2022, 8, 237. [Google Scholar] [CrossRef] [PubMed]
- Li, M.Y. An Introduction to Mathematical Modeling of Infectious Diseases; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Farman, M.; Saleem, M.U.; Ahmad, A.; Ahmad, M. Analysis and numerical solution of SEIR epidemic model of measles with non-integer time fractional derivatives by using Laplace Adomian Decomposition Method. Ain Shams Eng. J. 2018, 9, 3391–3397. [Google Scholar] [CrossRef]
- Solís-Pérez, J.; Gómez-Aguilar, J.; Atangana, A. Novel numerical method for solving variable-order fractional differential equations with power, exponential and Mittag-Leffler laws. Chaos Solitons Fractals 2018, 114, 175–185. [Google Scholar] [CrossRef]
- Khuri, S.A. A Laplace decomposition algorithm applied to a class of nonlinear differential equations. J. Appl. Math. 2001, 1, 141–155. [Google Scholar] [CrossRef]
- Abbaoui, K.; Cherruault, Y. New ideas for proving convergence of decomposition methods. Comput. Math. Appl. 1995, 29, 103–108. [Google Scholar] [CrossRef]
- Cherruault, Y.; Adomian, G.; Abbaoui, K.; Rach, R. Further remarks on convergence of decomposition method. Int. J. Bio-Med. Comput. 1995, 38, 89–93. [Google Scholar] [CrossRef]
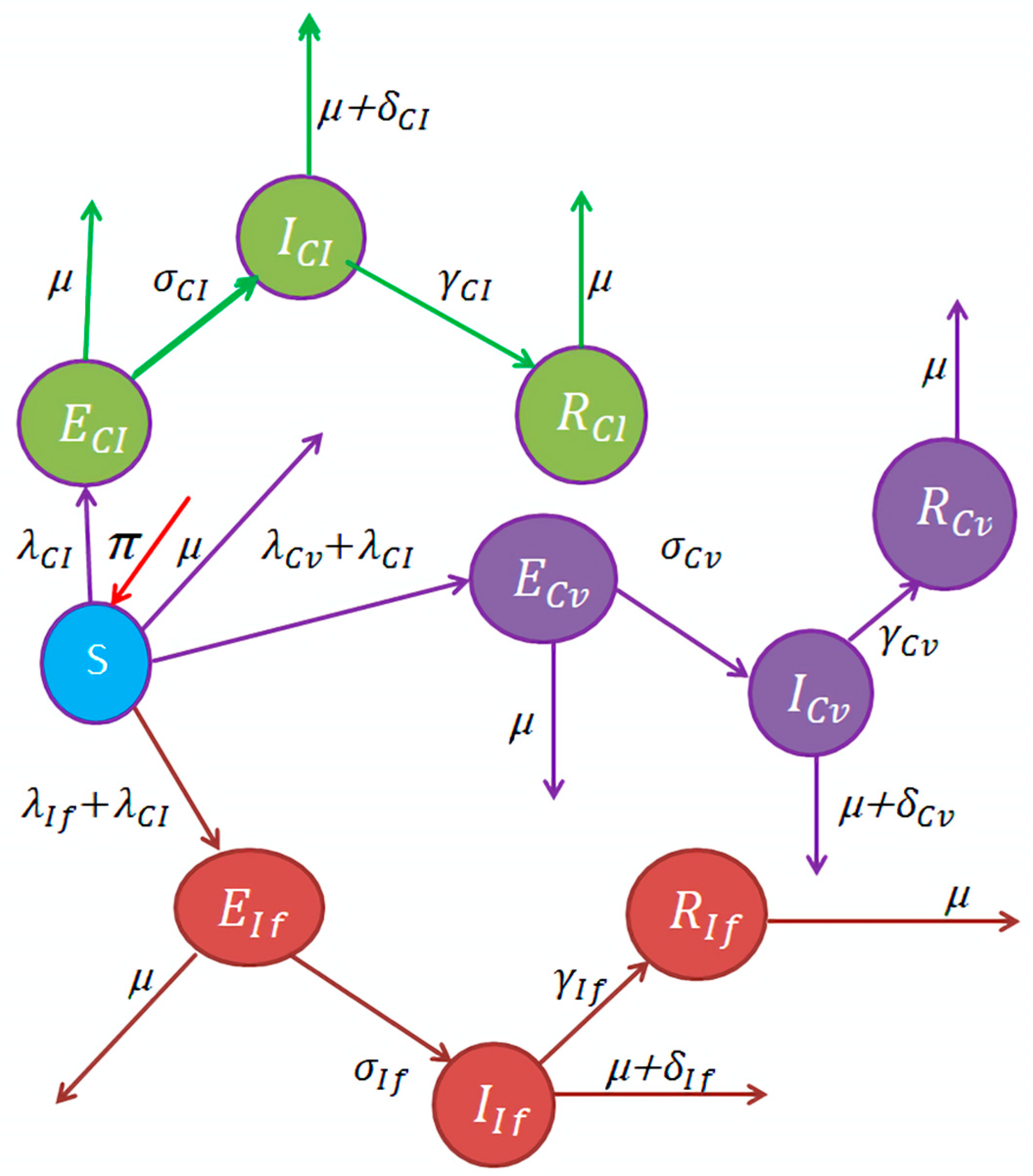




| Variable | Explanation |
|---|---|
| S(t) | Vulnerable human population. |
| (t) | Human population exposed to COVID-19 only. |
| (t) | Individuals exposed to Influenza only. |
| (t) | Individuals exposed to COVID-19 and Influenza [4]. |
| Individuals infected with COVID-19 [4]. | |
| Infectious individuals affected by Influenza [4]. | |
| Infectious individuals affected by COVID-19 and Influenza [4]. | |
| Individuals recovered from COVID-19 [4]. | |
| (t) | Individuals recovered from Influenza [4]. |
| (t) | Individuals recovered from both COVID-19 and Influenza [4]. |
| Parameters | Explanations | Specifications |
|---|---|---|
| Π | Susceptible individual’s entering rate | 1.2 × [4,19] |
| COVID-19’s transmission rate | 0.224334/day [19] | |
| Influenza’s transmission rate | 0.203 [20] | |
| The progression rate of COVID-19 from the exposed to others [4] | 0.40 [19] | |
| The progression rate of Influenza from the exposed to others [4] | 0.40 [22] | |
| The progression rate of infections caused by both (co-infection) from the exposed to others [4] | 0.40 Assumed | |
| μ | Natural mortality rate | 0.0003516 [19] |
| Death due to COVID-19 | 0.000573 per day [19] | |
| Death due to Influenza | 0.021 [21] | |
| Death due to co-infection | 0.021–0.026 assumed | |
| Rate of recovery from COVID-19 | 0.125 [19] | |
| Influenza infectious individuals’ recovery rate | 0.1998 [21] | |
| Co-infected infectious individuals’ recovery rate | 0.125–0.1998 assumed |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meenakshi, A.; Renuga, E.; Čep, R.; Karthik, K. Analysis of Caputo Fractional-Order Co-Infection COVID-19 and Influenza SEIR Epidemiology by Laplace Adomian Decomposition Method. Mathematics 2024, 12, 1876. https://doi.org/10.3390/math12121876
Meenakshi A, Renuga E, Čep R, Karthik K. Analysis of Caputo Fractional-Order Co-Infection COVID-19 and Influenza SEIR Epidemiology by Laplace Adomian Decomposition Method. Mathematics. 2024; 12(12):1876. https://doi.org/10.3390/math12121876
Chicago/Turabian StyleMeenakshi, Annamalai, Elango Renuga, Robert Čep, and Krishnasamy Karthik. 2024. "Analysis of Caputo Fractional-Order Co-Infection COVID-19 and Influenza SEIR Epidemiology by Laplace Adomian Decomposition Method" Mathematics 12, no. 12: 1876. https://doi.org/10.3390/math12121876
APA StyleMeenakshi, A., Renuga, E., Čep, R., & Karthik, K. (2024). Analysis of Caputo Fractional-Order Co-Infection COVID-19 and Influenza SEIR Epidemiology by Laplace Adomian Decomposition Method. Mathematics, 12(12), 1876. https://doi.org/10.3390/math12121876






