1. Introduction
The construction of several bivariate distributions has attracted considerable attention. Bivariate data find applications in a wide range of fields, including engineering, industry, finance, economics, technology, sports, drought, weather, medical sciences, and many more. In recent years, various methods have been explored to generate new bivariate distributions based on different approaches. These methods include the marginal transformation method [
1], copula method [
2], conditional specification method [
3], frailty approach [
4], and the Marshall–Olkin methodology [
5]. For more in-depth information on these methods and their application in establishing bivariate distributions, one can refer to the works of Ashkar and Aucoin [
1], Nelsen [
2], Vincent Raja and Gopalakrishnan [
4], and Pathak and Vellaisamy [
6].
In recent decades, copula theories have witnessed substantial growth in response to the increasing importance placed on modeling and describing diverse relationships among random variables. The emergence of new copula families has been motivated by the need to explore the structure of dependence in various fields, including the insurance industry [
7], actuarial science [
8,
9], finance [
10], hydrology [
11], and public health and medicine [
12], among others. This highlights the recognition of the significance of investigating the patterns of dependence between random variables in these domains.
In the field of statistical literature, copula structures have been widely employed by numerous authors as a method for establishing and modeling bivariate distributions. Flores [
13] presented various bivariate Weibull models based on different copula functions, such as the Farlie–Gumbel–Morgenstern (FGM), Clayton, Ali–Mikhail–Haq (AMH), Gumbel–Hougaard, Gumbel–Barnett, and Nelsen Ten copulas. Abd Elaal et al. [
14] introduced a bivariate generalized exponential distribution based on the FGM and Plackett copulas. Baharith et al. [
15] proposed two bivariate Pareto Type II distributions; one is derived from the Gaussian copula (BPIIG) and the other is based on the mixture and Gaussian copula (BPIImG). El-sherpieny et al. [
16] introduced the bivariate generalized Rayleigh distribution, denoted as Clayton-BGR, which is based on the Clayton copula. The study also derived the likelihood function for a progressive Type II censoring scheme with random removal and applied it to the Clayton-BGR distribution. Usman and Aliyu [
17] proposed a new bivariate Nadarajah–Haghighi distribution using two copula functions: the Gumbel–Barnett and Clayton copula functions. Qura et al. [
18] introduced a bivariate power Lomax distribution, named BFGMPLx, which is based on Farlie–Gumbel–Morgenstern (FGM) copulas. Fayomi et al. [
19] presented a novel family of bivariate continuous Lomax generators called the BFGMLG family. This family is constructed by utilizing univariate Lomax generator (LG) families and the Farlie–Gumbel–Morgenstern (FGM) copula. Fayomi et al. [
20] introduced two innovative continuous bivariate families, the FGM bivariate Kavya–Manoharan transformation and the Clayton bivariate Kavya–Manoharan transformation families, which are derived from the univariate Kavya–Manoharan transformation family and incorporate FGM and Clayton copulas, respectively.
In numerous life-testing experiments, it is typical for some test units’ lifetimes to remain unobserved. Censored observations arise when a sample is drawn from a complete population, but either the initial or final observations remain unknown. Type I and Type II censoring are the two predominant methods employed. Consider a scenario where
n items are simultaneously subjected to a life test, planned to end upon observing the
r-th failure (where
r is predetermined). Consequently, only the initial
r failures are recorded. This kind of data collected from a restricted life test is denoted as a Type II censored sample. In the context of bivariate censoring samples, Balakrishnan and Kim [
21] introduced the likelihood function for a bivariate distribution based on Type II censoring. Kim et al. [
22] have explored both maximum likelihood and Bayesian estimation methods for determining the unknown parameters in the bivariate generalized exponential distribution under a Type II censoring scheme. Almetwally [
23] has explored the estimation of parameters in bivariate models within a specific censoring framework. Bai et al. [
24] introduced the Type I progressive interval censoring schemes for the Marshall–Olkin bivariate Weibull (MOBW) distribution and discussed how confidence intervals depend on the percentile bootstrap of the unknown parameters. El-Morshedy et al. [
25] have examined both maximum likelihood and Bayesian methods for estimating the parameters of the bivariate Burr X-G family, considering various distributions and utilizing Type II censored data. Lin et al. [
26] used the Pareto type I distribution for the joint frailty–copula model, a well-known heavy-tailed distribution. They devised statistical inference techniques encompassing bivariate random censoring, semi-competing risks, and competing risks, alongside maximum likelihood estimation procedures. Haj Ahmed et al. [
27] introduced a novel bivariate model based on the FGM copula and the univariate modified extended exponential distribution, named the bivariate modified extended exponential distribution. They estimated the unknown parameters using both maximum likelihood and Bayesian estimation methods within a Type II censored sampling scheme.
The bivariate model, based on a censored sample, holds significant importance in various fields of data science. It facilitates the analysis of the relationship between two variables through copula functions, effectively reducing data collection costs while maintaining overall efficiency comparable to that of using complete samples. In recent times, the analysis of data has become increasingly challenging due to the prevalence of large and diverse datasets. A notable characteristic of these extensive datasets is their frequent redundancy. For further information on data science, refer to the works of Gramaje et al. [
28] and Tien [
29].
In this study, three novel bivariate distributions were developed using a special mixture of exponential and Lindley distributions, as detailed in Chouia and Zeghdoudi [
30]. The specific mixture of exponential and Lindley distributions mentioned is referred to as the XLindley (XL) distribution. This new model features a single parameter, making it easy to use, granting significant flexibility to its density function and suitability for modeling different data patterns. Moreover, it demonstrates an increasing hazard function, making it a valuable tool in various statistical analyses and modeling, as discussed in Chouia and Zeghdoudi [
30]. Furthermore, other papers have utilized the XLindley distribution to create new extended distributions, as seen in Ahsan-ul-Haq et al. [
31], Etaga et al. [
32], Musekwa and Makubate [
33], and Gemeay et al. [
34]. In the latter, Alotaibi et al. [
35] employed an adaptive Type II progressive hybrid censoring plan to tackle estimation challenges related to the XL distribution using both classical and Bayesian methods. Their study illustrated that datasets from chemical engineering could be effectively modeled using the XL distribution instead of traditional distributions such as gamma and Weibull distributions. Additionally, Alotaibi et al. [
36] delved into the estimation of constant-stress accelerated life tests (CSALT) for XL distribution based on progressive.
Type II censoring, applying it to model specific engineering domains. Based on previous studies investigating this distribution, it was proven that it is more efficient than many famous competing distributions, such as the Lindley, Xgamma, exponential, Zeghdoudi, log-normal, and Pareto distributions. Therefore, this was a strong motivation to use this distribution to create the best bivariate model.
Considering the above, the main objectives of this paper are as follows:
Introducing three innovative models derived from the XLindley distribution and copula functions:
The Frank Bivariate XL (FBXL) Distribution;
The Gumbel Bivariate XL (GBXL) Distribution;
The Clayton Bivariate XL (CBXL) Distribution.
These novel models offer enhanced flexibility and suitability for application across diverse fields, including the medical and industrial domains.
Sub-objectives of this paper include:
Examining multiple scenarios based on an XLindley distribution with a single parameter, enabling a more thorough comparison, particularly across various datasets.
Estimating the parameters of bivariate XLindley models involves utilizing both maximum likelihood estimation (MLE) and Bayesian estimation methods. This consideration accounts for both complete and censored samples within a Type II framework.
Evaluating the estimators of novel bivariate models involves employing numerical techniques such as the Metropolis–Hastings algorithm.
Investigating two types of confidence intervals, namely asymptotic intervals and Bayesian credible intervals, for estimating the unknown parameters.
Assessing the fit of the bivariate model involves the use of various goodness-of-fit measures.
This study holds significant importance for two critical sectors in any country: the industrial and medical sectors.
In the industrial sector, tasks involving metal sheet manipulation, such as perforating procedures and measurements related to hole diameter and sheet thickness, are essential in metalworking and manufacturing processes. Burr formation, a common occurrence in these processes, is particularly significant for engineers and scientists due to its implications for product quality, manufacturing efficiency, innovation, and cost reduction.
In the healthcare sector, our research focuses on two key areas: diabetic nephropathy and infection recurrence in kidney patients. Understanding the progression of diabetic nephropathy is crucial, as it provides insights into key risk factors and aids in predicting renal complications, thus enabling the optimization of treatment protocols. Similarly, in the analysis of infection recurrence, survival analysis plays a pivotal role in evaluating infections at catheter insertion sites, guiding the optimization of infection management. This knowledge empowers healthcare professionals to refine treatment strategies, make well-informed decisions, and enhance patient outcomes in both diabetic nephropathy and infection recurrence scenarios.
The rest of this paper is organized as follows:
Section 2 provides a review of the copula function, along with discussions on the Frank, Gumbel, and Clayton copulas. The FBXL, GBXL, and CBXL distributions are defined in
Section 3. In
Section 4, maximum likelihood estimation and Bayesian estimation were performed.
Section 5 delves into the discussion of confidence intervals. In
Section 6, the suitability of the new models is demonstrated through a simulation study.
Section 7 discusses the applications of iron material jobs data, diabetic nephropathy data, and kidney patient data. Finally, in
Section 8, the conclusion and some remarks regarding the FBXL, GBXL, and CBXL distributions are presented.
2. Copula Function
In this section, the general definition of Archimedean copulas is presented, and the Frank, Gumbel, and Clayton copulas are discussed.
The copula is a convenient approach to describe a bivariate distribution with a dependence structure. The utilization of copulas is motivated by their numerous advantages in analyzing the interdependence between random variables. The copula functions present a flexible framework that allows for capturing diverse dependence patterns, encompassing both linear and non-linear relationships, tail dependencies, and asymmetries. By adopting a “vine copula” approach, copulas effectively separate the modeling of marginal distributions from the modeling of dependence. Additionally, the copulas facilitate the estimation of conditional dependencies, enabling the investigation of the interdependence among the remaining variables. This feature is valuable in risk management, portfolio optimization, and the pricing of complex financial products. Compared to traditional correlation-based approaches, copula models offer a more robust alternative by providing a comprehensive and accurate representation of dependence, particularly in the presence of non-linear relationships or non-normal distributions.
The Sklar theorem, introduced by Sklar [
37], plays a crucial role in copula theory. According to this theorem, for two random variables
X and
Y with marginal cumulative distribution functions (cdfs) denoted as
and
, and marginal probability density functions (pdfs) represented as
and
, respectively, let
represent the copula cdf, and
denote the copula pdf. Then, the joint cdf and joint pdf of (
X,
Y) based on the copula can be expressed as follows:
and
respectively, where
is a parameter that measures the dependence between the marginals. For further information regarding copulas, please refer to Nelsen [
2] and Joe [
38]. Vectors
and
represent the parameters for variables
X and
Y, respectively. Various copula functions have been developed using Equations (
1) and (
2), including the Frank, Gumbel, Clayton, and other copula models.
An Archimedean copula with a strict generator is represented by the following form:
where the generator function
in an Archimedean copula must adhere to specific conditions outlined by Nelsen [
2]. Firstly,
should be a continuous, strictly decreasing, and convex function that maps the interval
onto
. Secondly, it is required that
equals infinity, and, finally,
should be equal to zero. Archimedean copulas have been widely applied due to several factors:
Their straightforward construction process.
The extensive assortment of copula families within this category.
The numerous favorable properties exhibited by copulas in this class.
For additional information, refer to Nelsen [
2]. Other well-known completely monotone generators of Archimedean copulas include the Frank, Gumbel, and Clayton copulas.
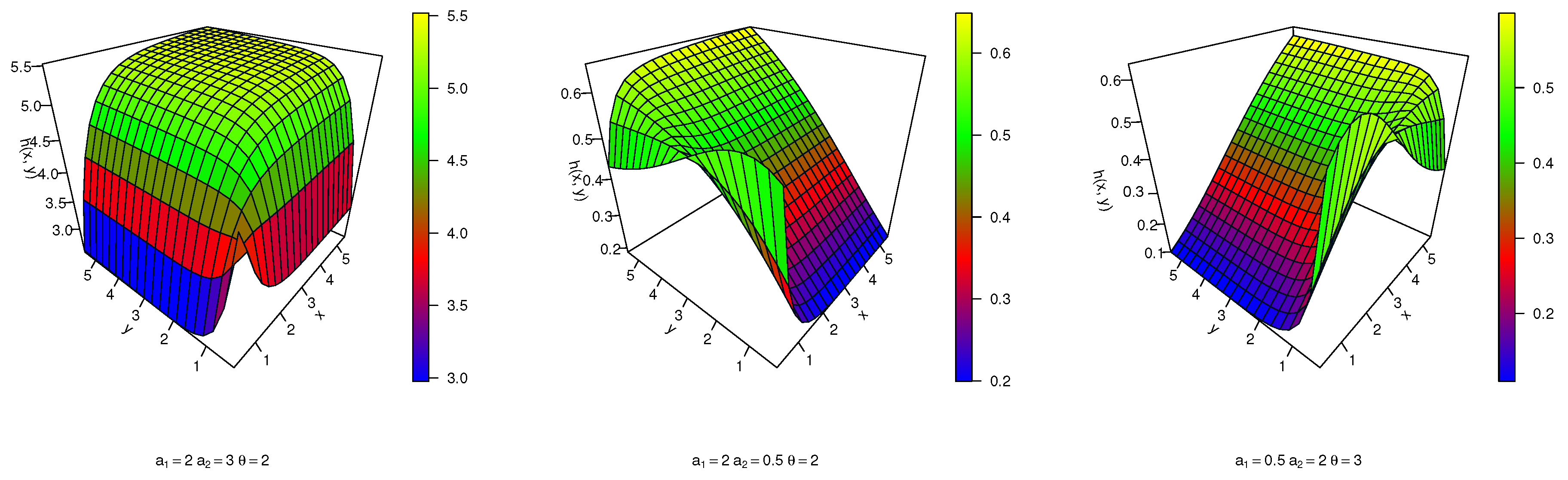
In this paper, three families of Archimedean copulas are considered: the Frank copula, the Gumbel copula, and the Clayton copula. These copulas are utilized for constructing bivariate distributions and are among the most commonly used Archimedean copulas.
2.1. Frank Copula
The Frank copula was introduced by Frank [
39], and several statistical properties of this copula family were discussed in Nelsen [
40] and Genest [
41]. It belongs to the class of Archimedean copulas, and its generator function is represented as
where
is the dependence parameter. Consequently, the cdf of the Frank copula with copula parameter
, where the symbol ∖ denotes set minus, is obtained as follows:
The copula density function for the Frank copula is given as follows:
where
. The Frank copula achieves the Frechet–Hoeffding upper bound as
approaches infinity and the Frechet–Hoeffding lower bound as
approaches negative infinity. Additionally, it has the capability to represent both negative and positive dependence, as highlighted by Nelsen [
2]. The Frank copula is renowned for its flexibility in modeling both positive and negative dependencies, making it a versatile choice for various applications, as discussed in Joe [
38]. Its parameter
directly governs the strength of tail dependence, offering intuitive interpretation, as described by Nelsen [
2]. However, its symmetric nature may not accurately capture the asymmetrical dependencies present in some datasets. Additionally, while it can model tail dependence to some extent, extreme tail dependence may not be fully captured by the Frank copula, as observed in Ashkar and Aucoin [
11].
2.2. Gumbel Copula
The Gumbel copula, initially introduced by Gumbel [
42], is commonly referred to as the Gumbel family by many authors. However, to avoid confusion with another Archimedean copula family associated with Gumbel’s name and its presence in Hougaard’s work [
43], Hutchinson and Lai [
44] named it the Gumbel–Hougaard family. This copula is classified as an Archimedean copula and is characterized by its generator function,
, with
and
. The cdf of the Gumbel copula with copula parameter
is defined as follows:
Setting to 1 indicates the presence of the independence copula, since . When , the Gumbel copula attains the Frechet–Hoeffding lower bound, implying a higher level of dependence in the upper tail of the copula distribution. The applicability of this copula function is limited to scenarios involving positive dependence. Furthermore, all members of this copula family are absolutely continuous.
The copula density function for the Gumbel copula is given as follows:
The Gumbel copula is a widely used copula model that is commonly employed to capture extreme value dependence, as discussed in Joe [
38].
2.3. Clayton Copula
The Clayton copula is a member of the Archimedean copula class and is characterized by its generator function,
, where
and
. This copula is commonly known as the Clayton copula and has been introduced by Clayton [
45], Cook and Johnson [
46], and Oakes [
47]. The cdf of the Clayton copula with copula parameter
is as follows:
As
approaches infinity, the Clayton copula achieves the Frechet–Hoeffding upper bound. This indicates that the Clayton copula exhibits lower tail dependence, implying a stronger dependence in the lower tail of the copula distribution.The copula density function for the Clayton copula is given as follows:
It is important to note that, as the parameter
approaches infinity, the Clayton copula exhibits perfect positive dependence. It boasts a straightforward parameterization, enhancing ease of interpretation, as noted in Nelsen [
2]. However, its ability to capture extreme tail dependence may be limited, and it may not be as flexible as other copula functions in modeling complex dependence structures, as discussed by Genest et al. [
11].
6. Simulation Study
In this section, a Monte Carlo simulation is conducted to compare bivariate XL distribution under different copulas. The simulation involves point estimation, interval estimation, and dependence measures. To generate random variables, we utilize the technique outlined by Nelsen [
2] for sampling from a designated joint bivariate distribution through the conditional distribution method. We will explore the generation of bivariate XL distribution under various copulas using both the conditional distribution method and iterative algorithms.
Firstly, we generate random variables Q and W independently from a uniform distribution with a lower bound of 0 and an upper bound of 1.
Secondly, we derive the conditional distribution of X given Y for each distribution using copulas:
In the Gumbel copula,
and
In the Clayton copula,
Here
u represents the cumulative distribution function (cdf) of the XL distribution with parameter
, while
v represents the cdf of another XL distribution with parameter
. When the real value of the parameters
is considered as
,
,
,
, we replicated Type II censored samples 5,000 times using various n (number of test items) and r (number of effective test items) options, for different choices of n such as
n = 35 and 100, as well as different censored samples such as
r = 25, 32, 35 for
n = 35 and
r = 70, 85, 100 for
n = 100. The number of iterations (sample) is 5000 iteration in the simulation study. The acquired mean squared error (MSE) and mean bias (MB) values are used to compare the likelihood and Bayes estimates of
, and
.
R software (version 4.3.0) was used to find the MLEs and 95% CIs for unknown parameters by the Newton Raphson iterative algorithm. This likely involved installing specific packages called “maxLik” and “AdequacyModel” in R, with the “maxlike” and “goodness.fit” functions, respectively. To estimate the unknown parameters using a Bayesian approach by the Metropolis–Hasting iterative algorithm, the “coda” and “HDInterval” packages in R (also version 4.3.0) were used. This method involved simulating 12,000 samples using MCMC to obtain Bayesian point estimates and their HPD intervals. It is important to note that the initial 2000 simulations were discarded (considered “burn-in”) to avoid including them in the analysis since they might not be representative of the final converged state.
Table 1,
Table 2,
Table 3 and
Table 4 show the obtained results based on the complete and censored samples (if
r =
n this is a complete sample and if
r <
n then these results are censored samples).
In terms of the least MSE, MB, the length of asymptotic confidence intervals (LACI), and coverage probability (CP) values, all provided estimates of the unidentified bivariate XL parameters , , and are outstanding.
As n (or r) increases, all estimated estimations perform as expected. When decreases, all estimates act in the same manner.
The MCMC estimates performed based on SE loss better against the MLE (are more favorable compared to the classical estimates) in terms of the smallest MSE and MB values when compared to the point estimation methods of , and , because , and parameters have gamma prior.
When , which typically yields the best results for all unknown parameters as expected, the approaches for estimating model parameters or reliability qualities perform the best among the derived estimates. This is an expected outcome because the acquired estimates were obtained using all of the data from the entire sample.
When comparing the bivariate XL distribution based on three copula functions, we find that in most cases the result of the bivariate XL based on the Clayton copula is better.
7. Application to Real Data
Iron material jobs data: The data were sourced from Dasgupta [
53]. For tasks involving iron sheets, we utilize a perforation process. Specifically, we drill four holes—two on each arm—into an L-shaped rectangular sheet with dimensions of 100 mm by 150 mm. This is accomplished swiftly using a 100-ton press operating at a speed of 250 strokes per hour, a process known as piercing. During each operation, two holes are created simultaneously. These L-shaped iron sheets with punctures are essential for use in the chassis of mini- or light-duty trucks. Following the piercing process, a burr forms around each hole on the metal sheet, creating a circular ridge. The high pressure used in piercing distorts the contact surface, causing the metal granules forming the burr to be unevenly raised around the hole’s rim. The burr size varies based on the metal grain characteristics and the applied piercing load. According to Skrotzki et al. [
54], factors such as composition, melting point, cooling rate, thermal and constitutional undercooling, and convection influence the grain structure and texture of metals. Subsequently, the burr is removed by chamfering with a drill. The burr measurements for the datasets were taken using a dial gauge with a minimum count of 20 microns (µm), or 0.02 millimeters. In the first dataset, which includes 50 observations of burr measurements (in millimeters), the hole diameter is 12 mm and the sheet thickness is 3.15 mm. In the second dataset, also comprising 50 observations, the hole diameter is 9 mm and the sheet thickness is 2 mm. For each set, one hole is selected and oriented according to a predetermined direction, and the diameter readings are taken with reference to that hole. These two datasets are associated with two different computers being compared. For more details on the iron material jobs data, see Dasgupta [
53]. X: “0.04, 0.02, 0.06, 0.12, 0.14, 0.08, 0.22, 0.12, 0.08, 0.26, 0.24, 0.04, 0.14, 0.16, 0.08, 0.26, 0.32, 0.28, 0.14, 0.16, 0.24, 0.22, 0.12, 0.18, 0.24, 0.32, 0.16, 0.14, 0.08, 0.16, 0.24, 0.16, 0.32, 0.18, 0.24, 0.22, 0.16, 0.12, 0.24, 0.06, 0.02, 0.18, 0.22, 0.14, 0.06, 0.04, 0.14, 0.26, 0.18, 0.16”. Y: “0.06, 0.12, 0.14, 0.04, 0.14, 0.16, 0.08, 0.26, 0.32, 0.22, 0.16, 0.12, 0.24, 0.06, 0.02, 0.18, 0.22, 0.14, 0.22, 0.16, 0.12, 0.24, 0.06, 0.02, 0.18, 0.22, 0.14, 0.02, 0.18, 0.22, 0.14, 0.06, 0.04, 0.14, 0.22, 0.14, 0.06, 0.04, 0.16, 0.24, 0.16, 0.32, 0.18, 0.24, 0.22, 0.04, 0.14, 0.26, 0.18, 0.16”.
Diabetic nephropathy data: Grover et al. [
55] have discussed this data. Two datasets were used in our investigation. The first dataset comes from Dr. Lal’s Path Lab, a reputable NABL-certified path lab, and contains retrospective data on 132 type 2 diabetic patients who had their diabetes diagnosed in accordance with ADA guidelines. A house-to-house survey was used to reach out to these patients, and current pathology reports from the time of the diabetes diagnosis to the end of the study, in November 2007, were gathered. Each patient had information about the length of their diabetes as well as their Fasting Blood Glucose (FBG), Diastolic Blood Pressure (DBP), Systolic Blood Pressure (SBP), Low Density Lipoprotein (LDL), and SrCr readings recorded. The study automatically disregards the impact on the eyes, heart, and other organs because it exclusively examines renal complications caused by type 2 diabetes. We have also eliminated instances in which the development of diabetes was preceded by renal complications. Patients in our study who have had diabetes for the same amount of time have varying levels of renal health. SrCr is used to assess a patient’s renal health since it serves as a key indicator for DN risk prediction due to its rapid rise in value. Thus, using SrCr values, the data were divided into two groups: DN (SrCr 1.4mg/dl) and non-diabetic nephropathy (NDN) (SrCr 1.4 mg/dL). It was discovered at the conclusion of the study that only 60 (45.45%) of the 132 patients were DN cases and 72 (54.55%) were NDN cases. These data are: X: “7.4, 9, 10, 11, 12, 13,13.75, 14.92, 15.8286, 16.9333, 18, 19, 20, 21, 22, 23, 24, 26, 26.6” Y: “1.925, 1.5, 2, 1.6, 1.7, 1.7533, 1.54, 1.694, 1.8843, 1.8433, 1.832, 1.59, 1.7833, 1.2, 1.792, 1.5, 1.5033, 2, 2.14”.
Kidney patients: For 30 kidney patients using a portable dialysis unit, McGilchrist and Aisbett [
56] provided a dataset that included the times (in days) between the first and second recurrences of infection at the site of catheter insertion. The recurrence time is the period of time between infections. The first recurrence of infection is measured at the time a catheter is implanted, and the second recurrence of infection is measured as the time between the second catheter insertion and the second infection. This information illustrates the frequency of infection recurrence in kidney patients, where X denotes the frequency of the first occurrence and Y that of the second occurrence. The following data set is as follows: X: “8, 23, 22, 447, 30, 24, 7, 511, 53, 15, 7, 141, 96, 149, 536, 152, 402, 13, 39, 12, 113, 132, 34, 2, 130, 17, 185, 292, 22, 15”. Y: “16, 13, 28, 318, 12, 245, 9, 30, 196, 154, 333, 8, 38, 70, 25, 362, 24, 66, 46, 40, 201, 156, 30, 25, 26, 4, 117, 114, 159, 108”.
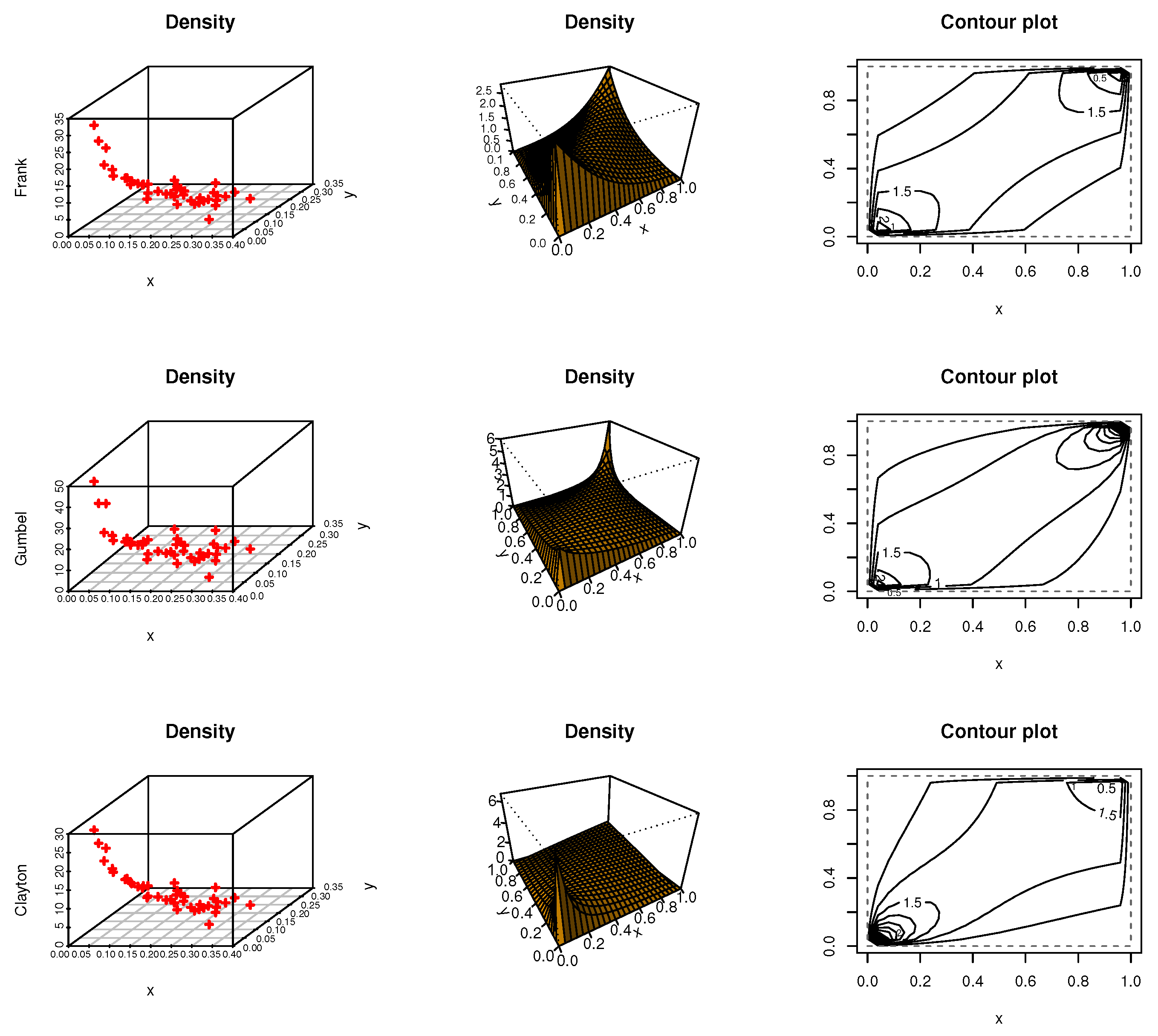
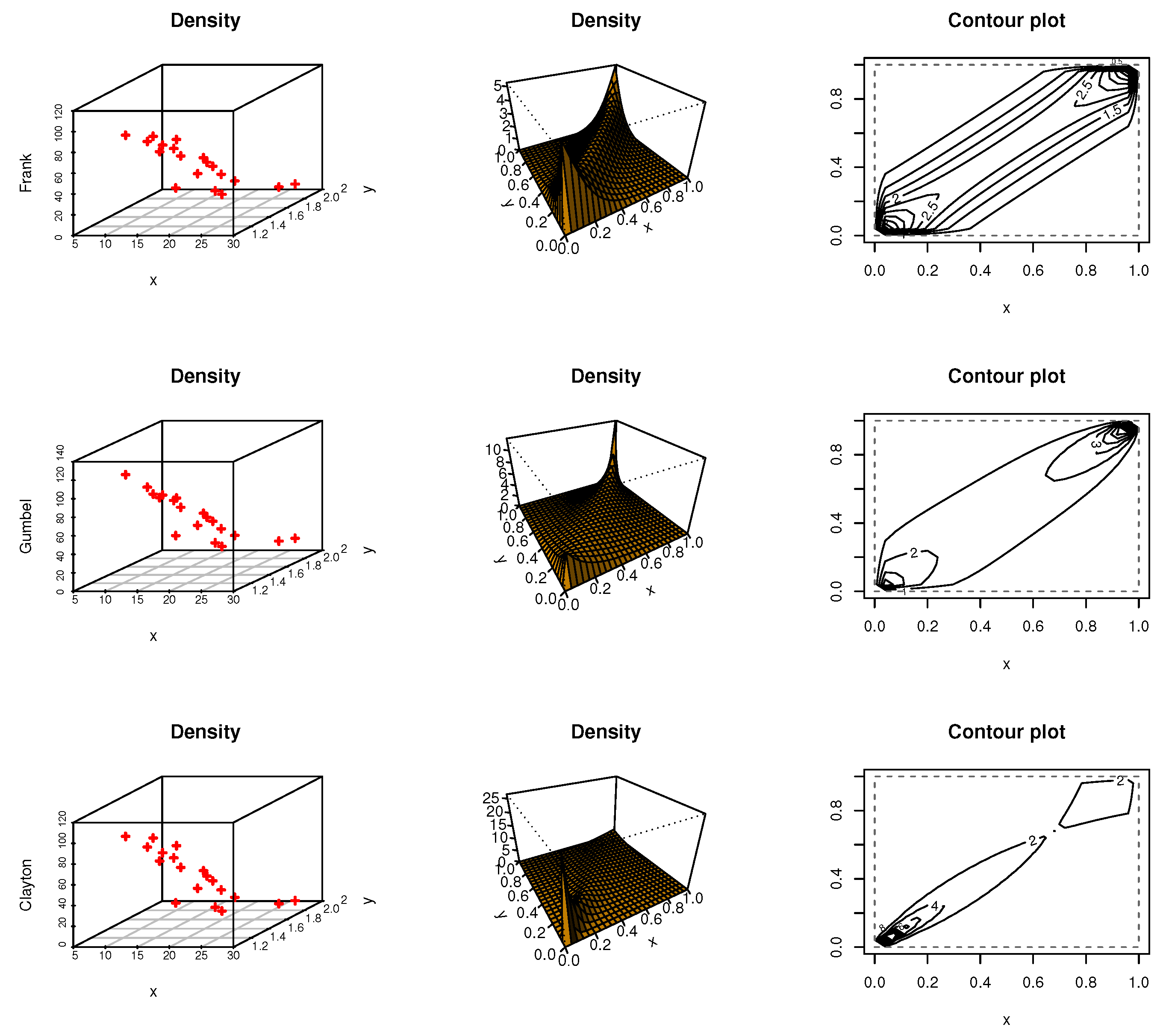
Figure 7 shows the correlation matrix for each dataset, kidney, iron material jobs, and diabetic nephropathy, respectively. All the data under study have a correlation value between 0.04 and 0.16. In addition,
Table 5 shows goodness-of-fit measures such as Anderson–Darling (AD), Cramer–von Mises criterion (CVM), and the estimated copula parameter by the non-parametric method to test whether or not these copula are a good fit for this data. By the results of goodness of fit test for each copula, we note that
p-values are more than 0.05, then these copula functions are a good fit for these datasets.
Figure 8,
Figure 9 and
Figure 10 discussed scatter, contour, and density plots for Frank, Gumbel, and Clayton for iron material jobs, diabetic nephropathy, and kidney patients data, respectively.
These included the Akaike information criterion (AIC), the Kolmogorov–Smirnov test, the corrected Akaike information criterion (CAIC), the Bayesian information criterion (BIC), and the Hannan–Quinn information criterion (HQIC). The three real data examples illustrate the suitability of the proposed distributions.
Table 6 discussed MLE, StEr, and goodness of fit for different datasets.
Here, we fit the FBXL, CBXL, and GBXL models on each dataset based on Type II censored samples. In
Table 7,
Table 8 and
Table 9, we provide the estimates and Standard Error (StEr) values for all parameters of each model. The Bayesian estimations have smaller StEr than MLE, Therefore, the Bayesian estimation is better than MLE. Also, when
r decreases then the StEr of the parameter model increases.