Prediction of Parkinson’s Disease Depression Using LIME-Based Stacking Ensemble Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Data Prepocessing
2.3. Splitting Dataset
2.4. Feature Selection
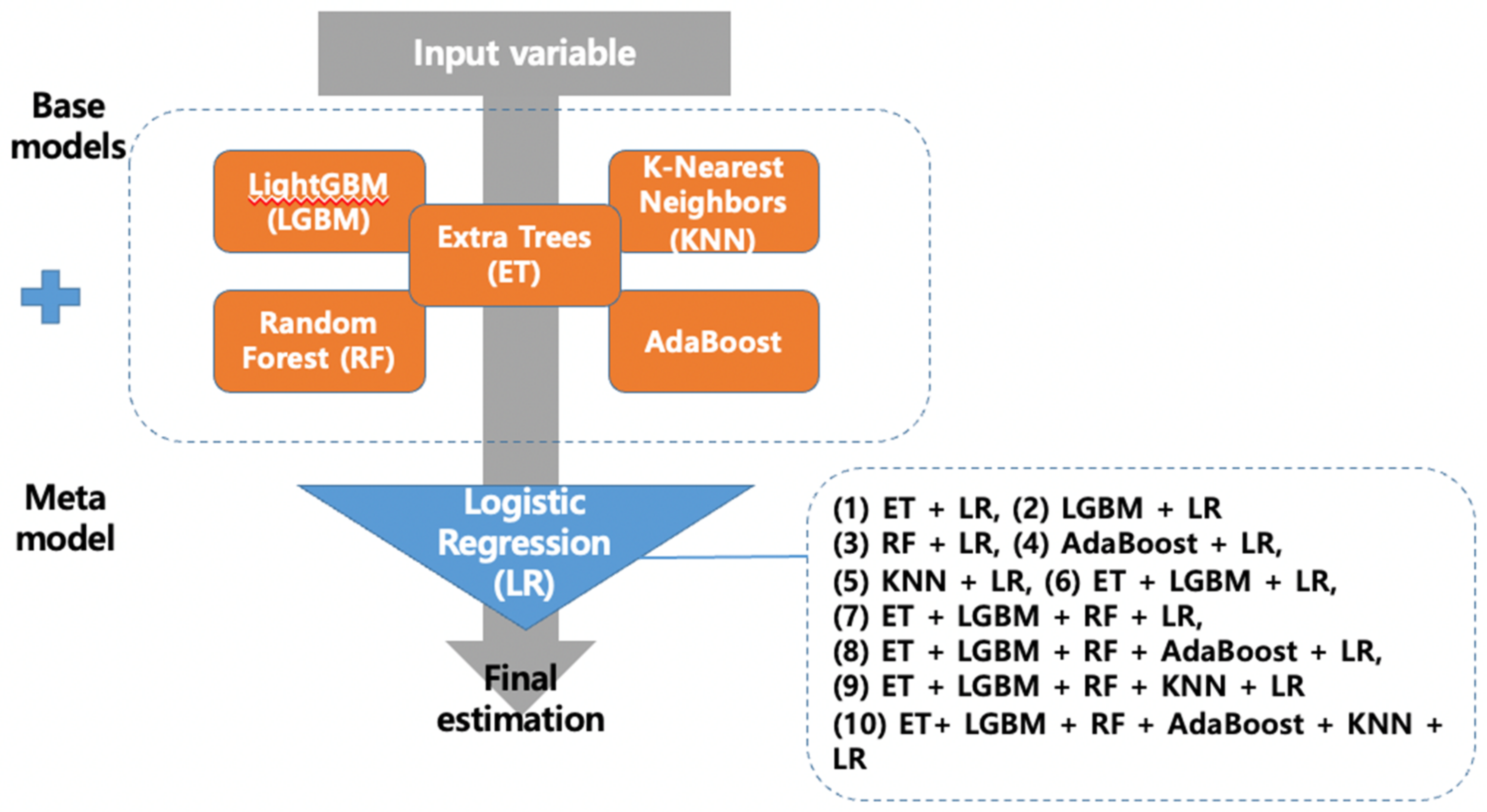
2.5. Development of Stacking Ensemble Model
2.5.1. Base Model: Light Gradient Boosting Machine (LGBM)
2.5.2. Base Model: K-Nearest Neighbors
| Algorithm 1: Pseudocode for KNN training |
| Require: Initialize k, func, target, data |
| Require: Initialize neighbors = [] |
| Train first weak decision tree model |
| for Each observation data do |
| distance = Euclidean distance (data[: −1], target) |
| calculate Euclidean distance |
| append neighbors |
| end for |
| pick the top K closest training data |
| take the most common label of these labels |
| return labels |
2.5.3. Base Model: Random Forest
2.5.4. Base Model: Extra Trees
2.5.5. Base Model: AdaBoost
- Train weak learner using distribution
- Get weak hypothesis
- Aim: select with low weighted error:
- Choose
- Update, for i = 1, …, m:
2.5.6. Meta Model: Logistic Regression
2.6. Model Evaluation
2.7. Hyperparameters Fine-Tuning by Sckit-Optimize Library
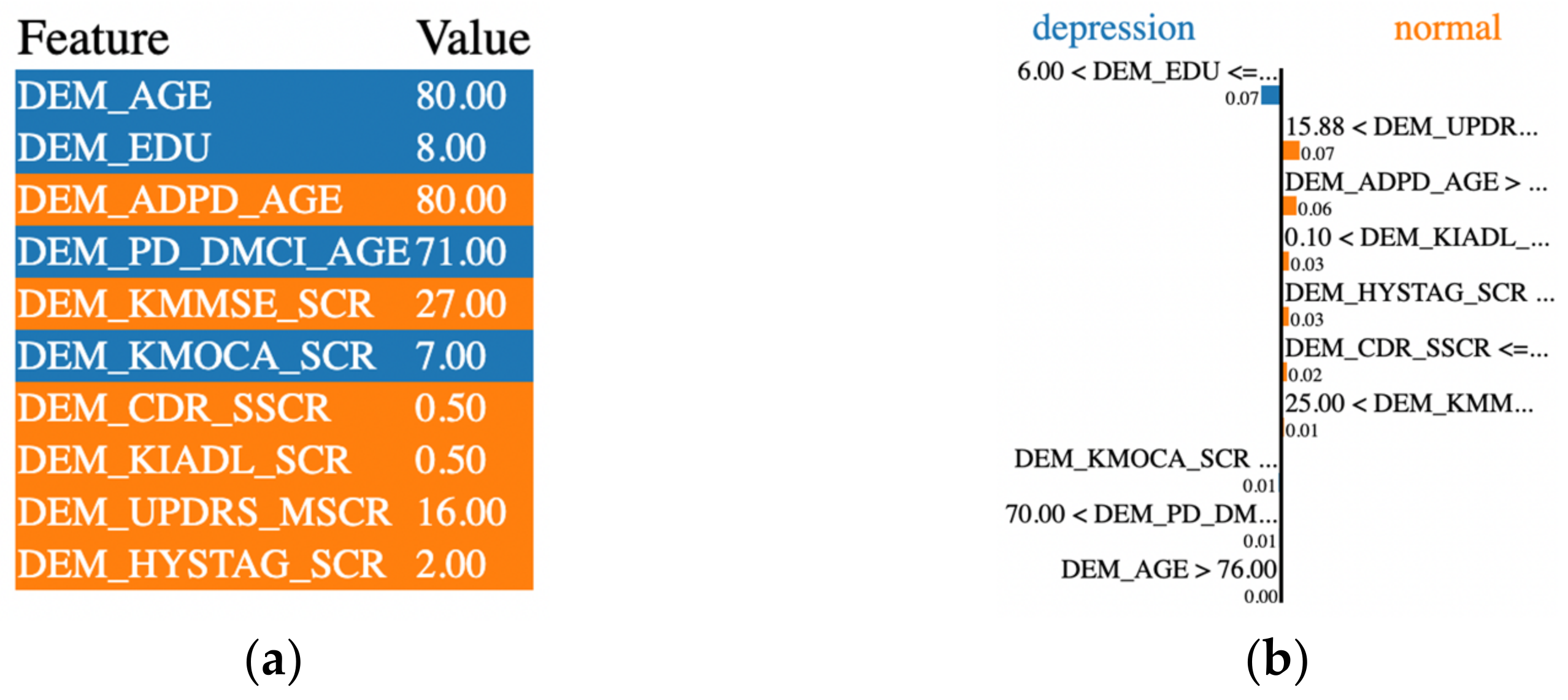
2.8. Local Interpretable Model-Agnostic Explanations (LIME)
3. Results
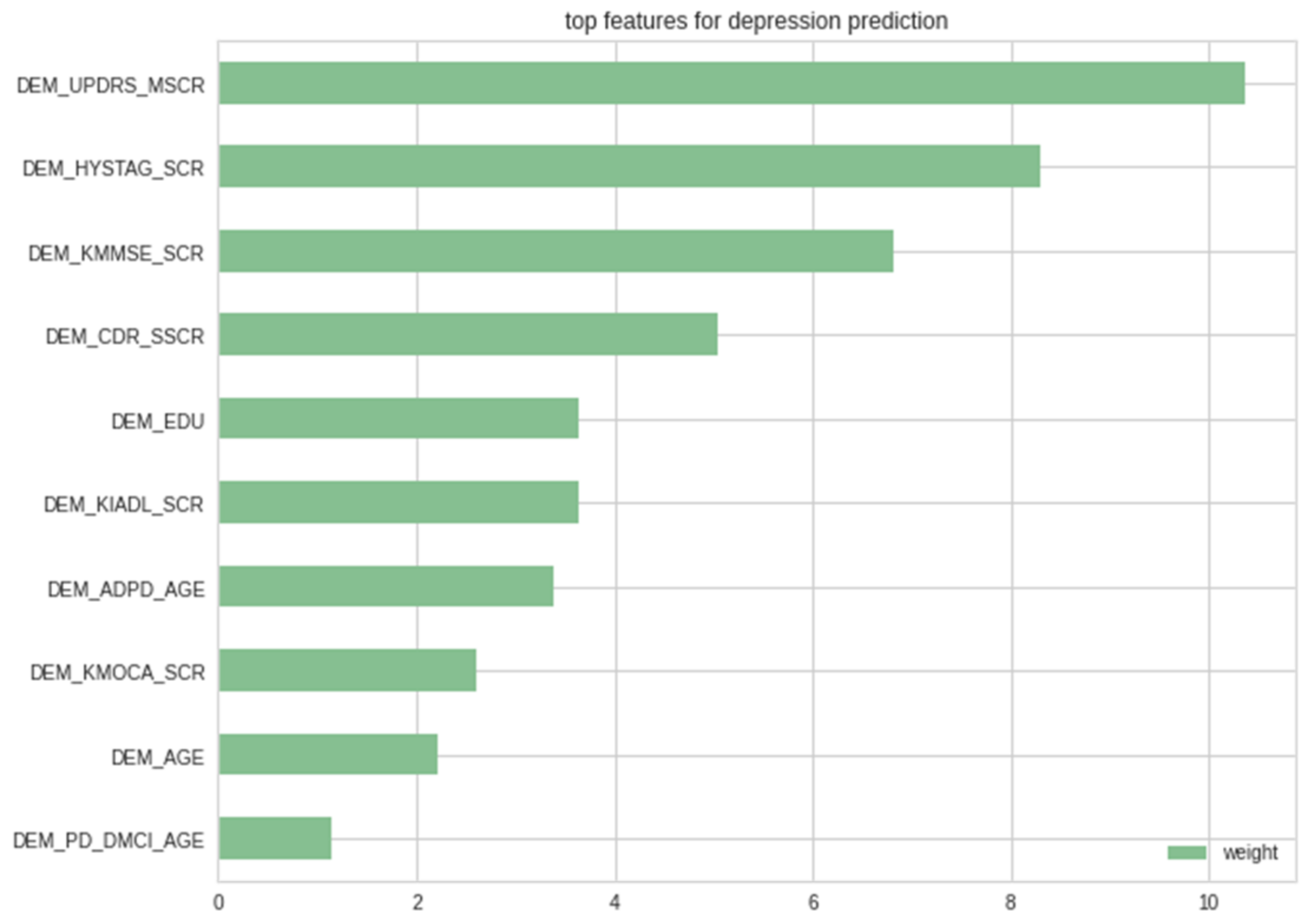
3.1. Selected Features Assessment
3.2. Hyperparameters Tuned Models Comparison
- LGBM: bagging_fraction = 0.8374829161718105, bagging_freq = 6, feature_fraction = 0.5841143936824905, learning_rate = 0.025450870095720408, min_child_samples = 2, min_split_gain = 0.1853520160149429, n_estimators = 234, num_leaves = 242, random_state = 3317, reg_alpha = 0.0032933153051456932, reg_lambda = 3.2851517043202704 × 10−6.
- RF: bootstrap = False, criterion = ‘entropy’, max_depth = 11, max_features = 0.4, min_impurity_decrease = 1 × 10−9, min_samples_leaf = 6, min_samples_split = 10, n_estimators = 300.
- ET: criterion = ‘entropy’, max_depth = 11, max_features = 1.0, min_impurity_decrease = 1.1410366199157523 × 10−7, min_samples_split = 3, n_estimators = 300.
- AdaBoost: learning_rate = 0.06748784395891723, n_estimators = 300.
- KNN: n_neighbors = 4, weights = ‘distance’.
3.3. Comparing the Accuracy of Stacking Models Predicting the Depression of PWPD
3.4. Evaluation of LIME-Based Stacking Ensemble Model
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rossi, A.; Berger, K.; Chen, H.; Leslie, D.; Mailman, R.B.; Huang, X. Projection of the Prevalence of Parkinson’s Disease in the Coming Decades: Revisited. Mov. Disord. 2017, 33, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Baek, J.Y.; Lee, E.; Jung, H.-W.; Jang, I.-Y. Geriatrics Fact Sheet in Korea 2021. Ann. Geriatr. Med. Res. 2021, 25, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Reijnders, J.S.A.M.; Ehrt, U.; Weber, W.E.J.; Aarsland, D.; Leentjens, A.F.G. A Systematic Review of Prevalence Studies of Depression in Parkinson’s Disease. Mov. Disord. 2007, 23, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Wichowicz, H.M.; Sławek, J.; Derejko, M.; Cubała, W.J. Factors Associated with Depression in Parkinson’s Disease: A Cross-Sectional Study in a Polish Population. Eur. Psychiatry 2006, 21, 516–520. [Google Scholar] [CrossRef]
- Global Parkinson’s Disease Survey Steering Committee. Factors Impacting on Quality of Life in Parkinson’s Disease: Results from an International Survey. Mov. Disord. 2002, 17, 60–67. [Google Scholar] [CrossRef]
- Byeon, H. Predicting the Severity of Parkinson’s Disease Dementia by Assessing the Neuropsychiatric Symptoms with an SVM Regression Model. Int. J. Environ. Res. Public Health 2021, 18, 2551. [Google Scholar] [CrossRef]
- Avuçlu, E.; Elen, A. Evaluation of Train and Test Performance of Machine Learning Algorithms and Parkinson Diagnosis with Statistical Measurements. Med. Biol. Eng. Comput. 2020, 58, 2775–2788. [Google Scholar] [CrossRef]
- Byeon, H. Comparing Ensemble-Based Machine Learning Classifiers Developed for Distinguishing Hypokinetic Dysarthria from Presbyphonia. Appl. Sci. 2021, 11, 2235. [Google Scholar] [CrossRef]
- Byeon, H. Exploring Factors Associated with the Social Discrimination Experience of Children from Multicultural Families in South Korea by Using Stacking with Non-Linear Algorithm. Int. J. Adv. Comput. Sci. Appl. 2021, 12, 125–130. [Google Scholar] [CrossRef]
- Byeon, H. Can the Prediction Model Using Regression with Optimal Scale Improve the Power to Predict the Parkinson’s Dementia? World J. Psychiatry 2022, 12, 1031–1043. [Google Scholar] [CrossRef]
- Albreiki, B.; Zaki, N.; Alashwal, H. A Systematic Literature Review of Student’ Performance Prediction Using Machine Learning Techniques. Educ. Sci. 2021, 11, 552. [Google Scholar] [CrossRef]
- Mutlag, W.K.; Ali, S.K.; Aydam, Z.M.; Taher, B.H. Feature Extraction Methods: A Review. J. Phys. Conf. Ser. 2020, 1591, 012028. [Google Scholar] [CrossRef]
- Cawley, G.C.; Nicola, L.C. Talbot. On Over-fitting in Model Selection and Subsequent Selection Bias in Performance Evaluation. J. Mach. Learn. Res. 2010, 11, 2079–2107. [Google Scholar]
- Kuhn, M.; Johnson, K. Feature Engineering and Selection; Chapman and Hall/CRC Data Science Ser.; Chapman & Hall/CRC: Boca Raton, FL, USA, 2019; ISBN 978-1-315-10823-0. [Google Scholar]
- Friedman, J.H. Greedy Function Approximation: A Gradient Boosting Machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.Y. LightGBM: A Highly Efficient Gradient Boosting Decision Tree. In Proceedings of the 31st Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017; Volume 30. [Google Scholar]
- Breiman, L. Random Forest. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Svetnik, V.; Liaw, A.; Tong, C.; Culberson, J.C.; Sheridan, R.P.; Feuston, B.P. Random Forest: A Classification and Regression Tool for Compound Classification and QSAR Modeling. J. Chem. Inf. Comput. Sci. 2003, 43, 1947–1958. [Google Scholar] [CrossRef]
- GALTON, F. Vox Populi. Nature 1907, 75, 450–451. [Google Scholar] [CrossRef]
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely Randomized Trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar] [CrossRef]
- Dezhen, Z.; Kai, Y. Genetic Algorithm Based Optimization for AdaBoost. In Proceedings of the 2008 International Conference on Computer Science and Software Engineering, Wuhan, China, 12–14 December 2008. [Google Scholar] [CrossRef]
- Kleinbaum, D.G.; Klein, M. Logistic Regression; Statistics for Biology and Health Ser.; Springer: Berlin/Heidelberg, Germany, 2010; ISBN 978-1-4419-1742-3. [Google Scholar]
- Roscher, R.; Bohn, B.; Duarte, M.F.; Garcke, J. Explainable Machine Learning for Scientific Insights and Discoveries. IEEE Access 2020, 8, 42200–42216. [Google Scholar] [CrossRef]
- Nguyen, H.V.; Byeon, H. Explainable Deep-Learning-Based Depression Modeling of Elderly Community after COVID-19 Pandemic. Mathematics 2022, 10, 4408. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. Why should I trust you? Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar]
- Yadav, D.C.; Pal, S. To Generate an Ensemble Model for Women Thyroid Prediction Using Data Mining Techniques. Asian Pac. J. Cancer Prev. 2019, 20, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Poon, P.K.-C.; Wang, S.Y.; Woodbridge, D.M. Depression Level Prediction in People with Parkinson’s Disease during the COVID-19 Pandemic. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Mexico, 1–5 November 2021. [Google Scholar] [CrossRef]
- Zhang, F.; Petersen, M.; Johnson, L.; Hall, J.; O’Bryant, S.E. Recursive Support Vector Machine Biomarker Selection for Alzheimer’s Disease. J. Alzheimer’s Dis. 2021, 79, 1691–1700. [Google Scholar] [CrossRef] [PubMed]
- Perrin, A.J.; Nosova, E.; Co, K.; Book, A.; Iu, O.; Silva, V.; Thompson, C.; McKeown, M.J.; Stoessl, A.J.; Farrer, M.J.; et al. Gender Differences in Parkinson’s Disease Depression. Parkinsonism Relat. Disord. 2017, 36, 93–97. [Google Scholar] [CrossRef]
- Cummings, J.L. Depression and Parkinson’s Disease: A Review. Am. J. Psychiatry 1992, 149, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Marsh, L. Depression and Parkinson’s Disease: Current Knowledge. Curr. Neurol. Neurosci. Rep. 2013, 13, 409. [Google Scholar] [CrossRef]
- Riedel, O.; Heuser, I.; Klotsche, J.; Dodel, R.; Wittchen, H.U.; GEPAD Study Group. Occurrence Risk and Structure of Depression in Parkinson Disease with and without Dementia: Results from the GEPAD Study. J. Geriatr. Psychiatry Neurol. 2009, 23, 27–34. [Google Scholar] [CrossRef]
- Aarsland, D.; Påhlhagen, S.; Ballard, C.G.; Ehrt, U.; Svenningsson, P. Depression in Parkinson Disease—Epidemiology, Mechanisms and Management. Nat. Rev. Neurol. 2011, 8, 35–47. [Google Scholar] [CrossRef]
- Dobkin, R.D.; Rubino, J.T.; Friedman, J.; Allen, L.A.; Gara, M.A.; Menza, M. Barriers to Mental Health Care Utilization in Parkinson’s Disease. J. Geriatr. Psychiatry Neurol. 2013, 26, 105–116. [Google Scholar] [CrossRef]
- Havlikova, E.; van Dijk, J.P.; Nagyova, I.; Rosenberger, J.; Middel, B.; Dubayova, T.; Gdovinova, Z.; Groothoff, J.W. The Impact of Sleep and Mood Disorders on Quality of Life in Parkinson’s Disease Patients. J. Neurol. 2011, 258, 2222–2229. [Google Scholar] [CrossRef]
- Lawrence, B.J.; Gasson, N.; Kane, R.; Bucks, R.S.; Loftus, A.M. Activities of Daily Living, Depression, and Quality of Life in Parkinson’s Disease. PLoS ONE 2014, 9, e102294. [Google Scholar] [CrossRef]
- Byeon, H. Development of a Depression in Parkinson’s Disease Prediction Model Using Machine Learning. World J. Psychiatry 2020, 10, 234–244. [Google Scholar] [CrossRef]
| Variables | Description | Field Type |
|---|---|---|
| DEM_SEX | Gender | Categorical: Male (1), Female (2) |
| DEM_AGE | Age | Continuous: ( ) years old |
| DEM_EDU | Training period | Continuous: ( ) years |
| DEM_HAND | Dominant hand | Categorical: Right (1), Left (2), Both (3) |
| DEM_SMOKE | Smoking experience | Categorical: No (1), Smoking in the past (2), Smoking in the current (3) |
| DEM_COFFEE | Whether or not drink coffee | Categorical: No (1), Drinking in the past (2), Drinking in the current (3) |
| DEM_AGRICULCHEM | Pesticide exposure | Categorical: No (1), Exposure in the past (2), Exposure in the current (3) |
| DEM_COINTOXI | Carbon monoxide poisoning | Categorical: No (1), Yes (2) |
| DEM_MN | Manganese poisoning | Categorical: No (1), Yes (2) |
| DEM_HEADINJ | Head injury | Categorical: No (1), Yes (2) |
| DEM_CVA | Stroke | Categorical: No (1), Yes (2) |
| DEM_DM | Diabetes | Categorical: No (1), Yes (2) |
| DEM_HT | Hypertension | Categorical: No (1), Yes (2) |
| DEM_LP | Hyperlipidemia | Categorical: No (1), Yes (2) |
| DEM_AF | Atrial fibrillation | Categorical: No (1), Yes (2) |
| DEM_PATIENT_TY | Patient type | Categorical: Parkinson disease with dementia, PD-D (1), Parkinson disease with mild cognitive impairment, PD-MCI (2), Parkinson disease with normal cognition, PD-NC |
| DEM_DISEASEACC | Comorbidities | Categorical: No (1), Yes (2) |
| DEM_ADPD_AGE | First diagnosis age | Continuous: ( ) years old |
| DEM_PD_DMCI_AGE | PD-D or PD-MCI first diagnosis age | Continuous: ( ) years old |
| DEM_PDFAM | Family history of PD | Categorical: No (1), Yes (2) |
| DEM_ADDEMFAM | Family history of Dementia | Categorical: No (1), Yes (2) |
| DEM_TREMOR | Tremor | Categorical: No (1), Yes (2) |
| DEM_RIGIDITY | Rigidity | Categorical: No (1), Yes (2) |
| DEM_AKBK | Bradykinesia/Akinesia | Categorical: No (1), Yes (2) |
| DEM_PI | Postural instability (PI) | Categorical: No (1), Yes (2) |
| DEM_LMC | Late motor complications | Categorical: No (1), Yes (2) |
| DEM_RBD | Rapid eye movement (REM) sleep behavior disorders | Categorical: No (1), Yes (2) |
| DEM_KMMSE_SCR | Korean mini mental state examination | Continuous: _ points/30 points |
| DEM_KMOCA_SCR | Korean montreal cognitive assessment | Continuous: _points/30 points |
| DEM_CDR_GSCR | Global Clinical dementia rating (CDR) score | Continuous: ( ) points/5 points |
| DEM_CDR_SSCR | Clinical dementia rating score (sum of boxes) | Continuous: ( ) points/5 points |
| DEM_DEMENTIA | Dementia based on DSM-IV (Diagnostic and statistical manual of mental disorders IV) | Categorical: No (1), Dementia (2) |
| DEM_KIADL_SCR | Korean instrumental activities of daily living score | Continuous: ( ) points/5 points |
| DEM_UPDRS_MSCR | Motor Untitled Parkinson disease rating scale (UPDRS) score | Continuous: ( ) points/108 points |
| DEM_HYSTAG_SCR | Hoehn and Yahr staging score | Continuous: ( ) points/5 points |
| DEM_DEPRESSION | Depression determined by BDI (Beck’s Depression Inventory) or GDS (Geriatric Depression Score) | Categorical: Yes (0), No (1) |
| Model | Accuracy | Recall | Precision | F1-Score | |
|---|---|---|---|---|---|
| All features | LGBM | 0.7155 | 0.8045 | 0.7394 | 0.7658 |
| KNN | 0.6971 | 0.7904 | 0.7342 | 0.7561 | |
| ET | 0.6964 | 0.8058 | 0.7263 | 0.7576 | |
| RF | 0.6912 | 0.8128 | 0.7130 | 0.7542 | |
| AdaBoost | 0.6819 | 0.7474 | 0.7266 | 0.7315 | |
| REFCV’s features | LGBM | 0.7400 | 0.8199 | 0.7606 | 0.7858 |
| RF | 0.7395 | 0.8442 | 0.7534 | 0.7897 | |
| ET | 0.7348 | 0.8359 | 0.7495 | 0.7842 | |
| AdaBoost | 0.7110 | 0.7808 | 0.7535 | 0.7561 | |
| KNN | 0.6969 | 0.7987 | 0.7283 | 0.7557 |
| Model | Accuracy | Recall | Precision | F1-Score |
|---|---|---|---|---|
| ET | 0.7690 | 0.8853 | 0.7672 | 0.8165 |
| LGBM | 0.7586 | 0.8442 | 0.7716 | 0.8023 |
| RF | 0.7440 | 0.8686 | 0.7438 | 0.7983 |
| AdaBoost | 0.7400 | 0.8449 | 0.7624 | 0.7927 |
| KNN | 0.7355 | 0.7987 | 0.7702 | 0.7821 |
| Stacking Model | Accuracy | Recall | Precision | F1-Score |
|---|---|---|---|---|
| (1) ET + LR | 0.7448 | 0.8936 | 0.7373 | 0.8028 |
| (2) LGBM + LR | 0.7540 | 0.8609 | 0.7598 | 0.8034 |
| (3) RF + LR | 0.7107 | 0.8763 | 0.7065 | 0.7792 |
| (4) AdaBoost + LR | 0.5962 | 1 | 0.5962 | 0.7468 |
| (5) KNN + LR | 0.7017 | 0.8628 | 0.7112 | 0.7764 |
| (6) ET + LGBM + LR | 0.7686 | 0.8692 | 0.7743 | 0.8141 |
| (7) ET + LGBM + RF + LR | 0.7736 | 0.8692 | 0.7795 | 0.8172 |
| (8) ET + LGBM + RF + AdaBoost + LR | 0.7736 | 0.8692 | 0.7795 | 0.8172 |
| (9) ET + LGBM + RF + KNN + LR | 0.7590 | 0.8609 | 0.7763 | 0.8075 |
| (10) ET + LGBM + RF + AdaBoost + KNN + LR | 0.7590 | 0.8609 | 0.7763 | 0.8075 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, H.V.; Byeon, H. Prediction of Parkinson’s Disease Depression Using LIME-Based Stacking Ensemble Model. Mathematics 2023, 11, 708. https://doi.org/10.3390/math11030708
Nguyen HV, Byeon H. Prediction of Parkinson’s Disease Depression Using LIME-Based Stacking Ensemble Model. Mathematics. 2023; 11(3):708. https://doi.org/10.3390/math11030708
Chicago/Turabian StyleNguyen, Hung Viet, and Haewon Byeon. 2023. "Prediction of Parkinson’s Disease Depression Using LIME-Based Stacking Ensemble Model" Mathematics 11, no. 3: 708. https://doi.org/10.3390/math11030708
APA StyleNguyen, H. V., & Byeon, H. (2023). Prediction of Parkinson’s Disease Depression Using LIME-Based Stacking Ensemble Model. Mathematics, 11(3), 708. https://doi.org/10.3390/math11030708







