1. Introduction
Non-negative matrix factorization (NMF) [
1,
2] is a linear dimensionality reduction technique that has become popular for its ability to automatically extract sparse and easily interpretable factors [
3], detect similarity between subsets of patients sharing local patterns of gene expression data [
4], cluster the rows or columns of a matrix [
5], perform filtering and deconvolution tasks [
6,
7], and interpret social phenomena through topic modeling [
8], to name just a few applications.
Let
be a non-negative matrix. For a given rank c such that
, a non-negative factorization of
M is given by:
with:
,
,
,
.
The non-negative matrices
and
are called the
components of the factorization, and the tensor product above is the regular product between
and the transpose of
. We use the tensor product notation because it symmetrizes the respective roles of
W and
H, and also because it is easily generalized to the context of tensor factorization. In order to simplify the notation, in what follows we drop the indices denoting the matrix size when there is no ambiguity. Each row of
M can be seen as a single observation of
f different features or attributes, stored in
f distinct columns of
M. Equation (
1) allows us to express the observations in
M in a more parsimonious way by reducing the original number
f of features to a smaller number
c. The tradeoff for this simplified representation is the presence of the factorization error term represented by matrix
E.
Let us refer to the
c synthetic features whose values populate the rows of matrix
W as “meta-features”. The transition from the condensed representation in the space of meta-features to the “physical” representation is provided by the matrix
H, which plays the role of a transition matrix. A single observation from
M is represented by its ith row
, and each column of
H, say
k, yields the decomposition of
as a non-negative linear combination of
c meta-features
, with
being the
column of
H, such that the following relation holds:
Non-negative matrix factorization has a geometric interpretation that makes it easier to understand: up to a certain approximation error coded in the matrix E, each observation vector belongs to the non-negative simplex generated by the c vectors. Non-negative matrix factorization can also be seen as a projection on this simplex, with the distance from the matrix to the simplex being the approximation error . If we add null columns to W and H and obtain and with the same number of rows and c’> c columns, the tensor product of these matrices remains unchanged: , hence . It follows that when the factorization rank increases, the approximation error is non-increasing. As an immediate consequence, an exact factorization at rank c remains exact at any larger rank . If one was only dealing with exact factorizations, the optimal factorization rank could be defined as the lowest rank for which the factorization is exact. Unfortunately, the additional noise makes the situation more complicated: there is no obvious information to extract from the non-increasing series of approximation errors . Therefore, an important question is whether the selected rank leads to a “meaningful” decomposition, since choosing a rank that is too high amounts to adjusting the noise, while choosing a rank that is too low leads to oversimplification of the signal.
In [
9], several methods were evaluated, e.g., Velicer’s Minimum Average Partial, Minka’s Laplace–PCA, Bayesian Information Criterion (BIC), and Brunet’s cophenetic correlation coefficient (
) [
10]. Another method is the minimum description length approach that selects the rank that ensures the best possible encoding of the data [
11]. For this approach, the encoding also covers the noise, so a low signal-to-noise ratio tends to inflate the effects of the noise. Cross-validation can also be used to select the rank that provides the most accurate imputation of missing data [
12]. Depending on the method used, significantly different ranks may be selected [
13]. Of course, in principle, using the results of all available methods together should help with the selection of the proper rank. Unfortunately, simple heuristic rules to do that have not been found yet, which led to the idea of using deep learning approaches.
In [
14], the starting idea was that several metrics may be potentially useful for rank selection. The authors included the approximation error as well as penalized versions like the AIC or BIC criteria. NMF clustering stability metrics, such as the silhouette coefficient, were also included. Because none of these metrics can indicate on their own which rank should be selected by simply observing the values taken for different ranks (a graph referred to as a scree-plot), the authors have entrusted a neural network, taking some candidate metrics as inputs, with the task of determining a (non-linear) function of the metrics that can be used to select the correct rank. Because this network has to work well for many types of matrices, it needs to be trained on a large and diverse set of matrices. These matrices are constructed such that their rank is known, and this rank is used as a label in the training set. The network presented by the authors worked well for matrices of the same type as those from the training set, but it did not perform well when applied to different types of matrices. The lack of generalizability to non-synthetic matrices is due to the inherent limitation of using a simulation framework, which, even if well designed, it is not able to reproduce all types of matrices that may be encountered in real life. Nevertheless, it is of interest to determine which specific metrics or combinations of metrics are most useful for selecting the correct factorization rank for synthetic datasets, even if each individual metric is not sufficient on its own. Unfortunately, the results from [
14] do not provide any insights with regard to this.
The estimates of the NMF components are inherently stochastic [
15]: the NMF estimates are obtained through iterative updating rules that offer no guarantee of converging to a global minimum, making them sensitive to initial “guessed” values. Yang et al. [
16] propose to alleviate this problem by mutually complementing NMF and Deep Learning. However, their approach is essentially based on a multilayer NMF, first proposed in [
17], and far beyond the scope of this work, which is focused on a single-layer NMF. By contrast, the cophenetic correlation coefficient,
, fully exploits the stochastic nature of the NMF estimates without requiring any training: for a given factorization rank,
provides an estimate of the consistency of the co-clustering of observations or features that can be derived from the NMF components over multiple runs of the algorithm, each using a random initialization [
10]. The factorization rank is chosen to maximize this consistency. The wide use of
in practice, thanks to its availability in common NMF packages [
18,
19], and its relatively good performance on synthetic matrices with orthogonal underlying factors [
9], has led us to use it as a reference metric while introducing a new criterion, which we call
. The latter also exploits the stochastic nature of the NMF estimates, this time by examining their stability relative to reference estimates obtained by non-negative double singular value decomposition (NNDSVD) [
20] up to some permutation. To evaluate the performance of our new criterion and compare it to the
criterion, we apply a CUSUM-based automatic detection algorithm to publicly available datasets and to synthetic matrices. In addition, we show that considering the ratio of
or
to the approximation error significantly decreases the rank selection error. Remarkably, our new criterion works very well with a broader class of matrices than those tested in [
9], namely matrices for which the underlying factors are not assumed to be orthogonal.
4. Conclusions
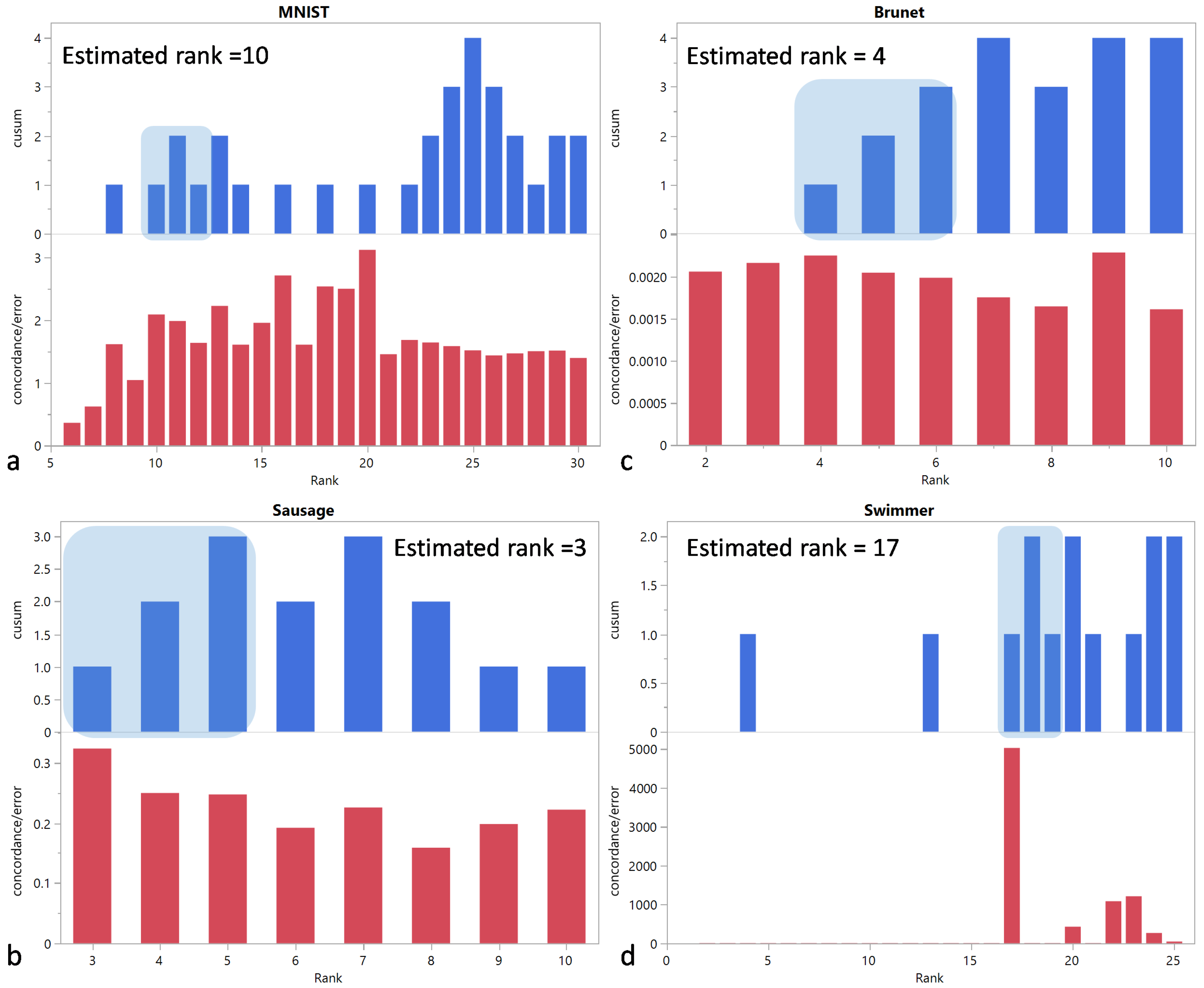
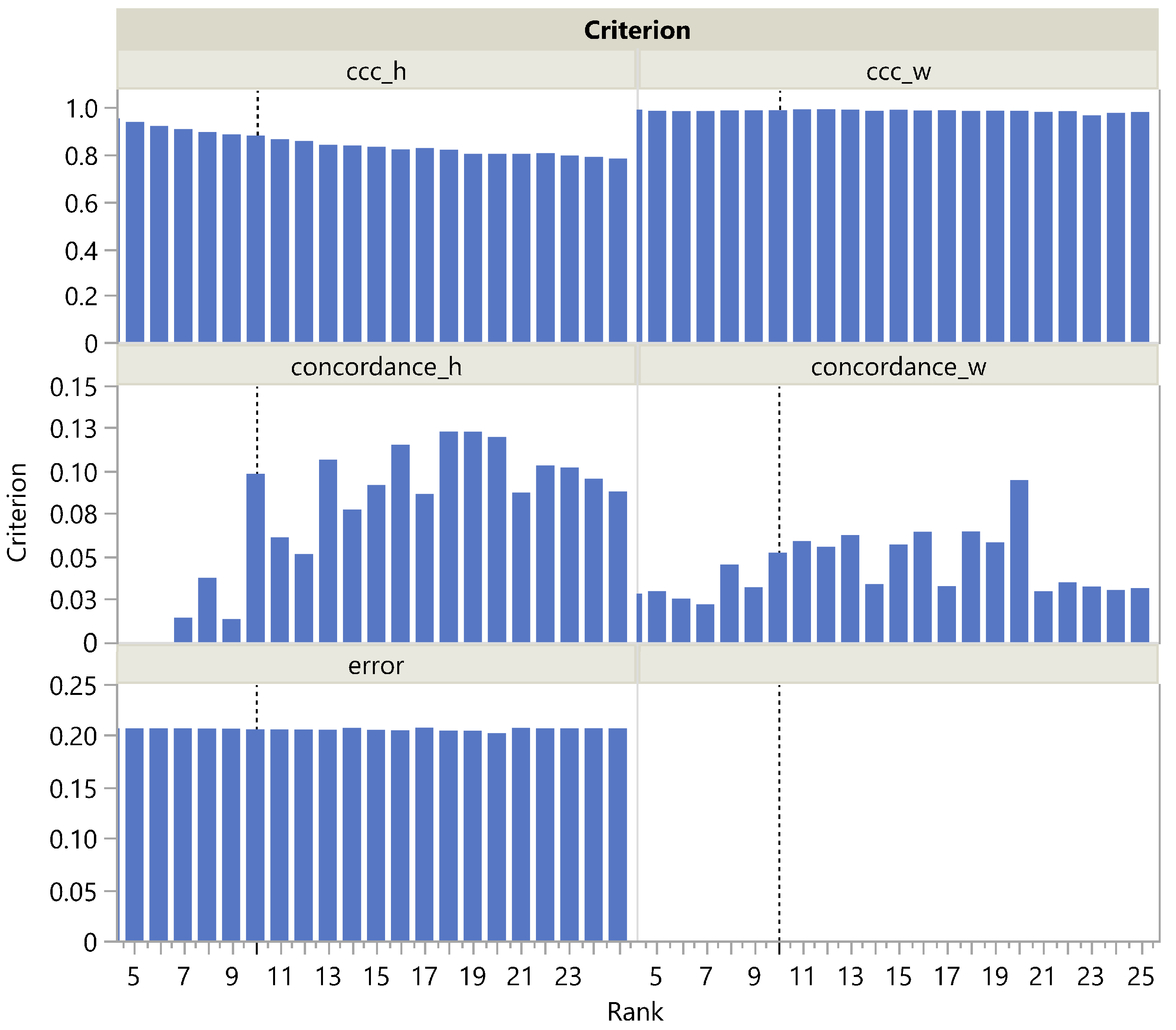
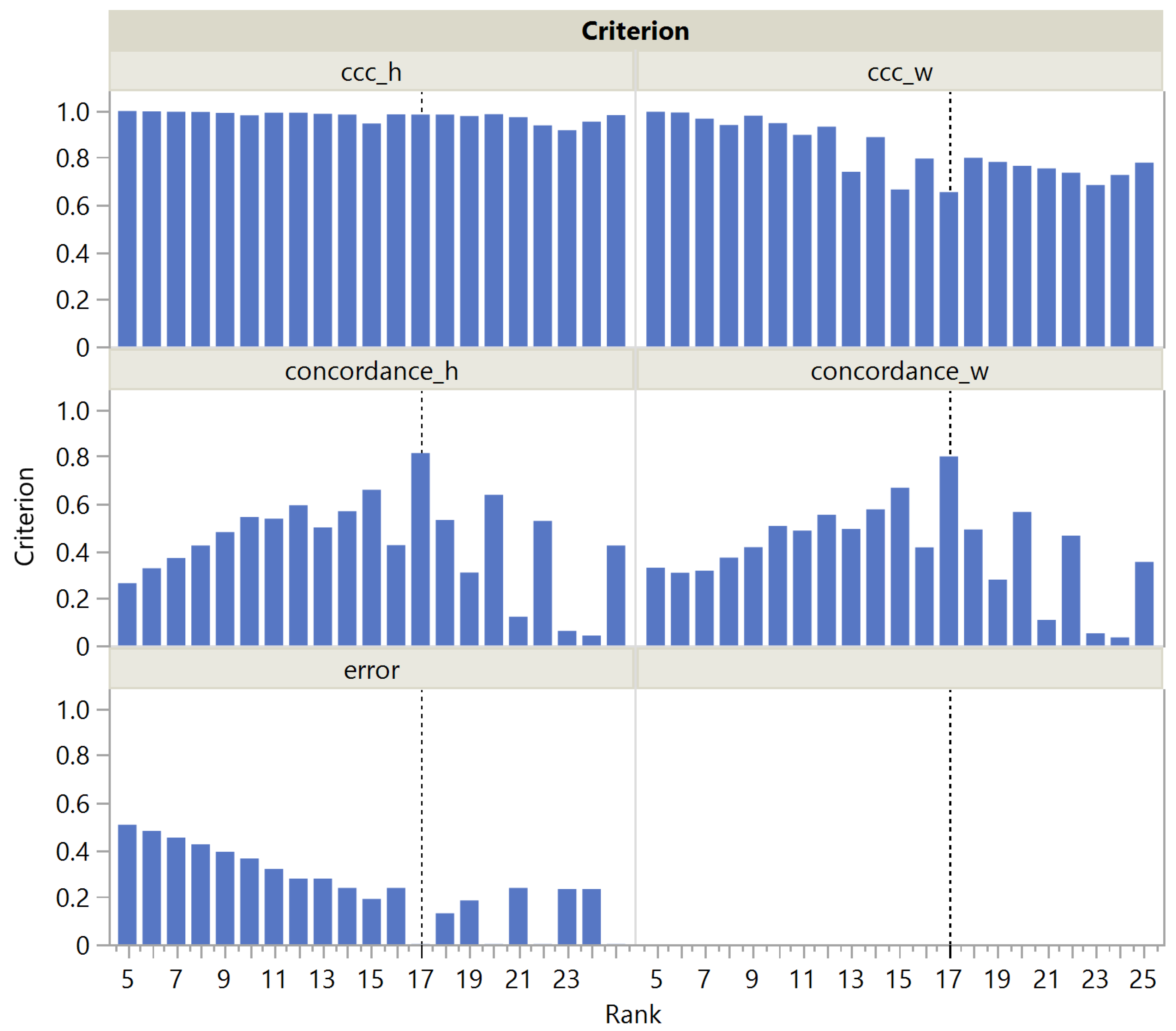
In this paper, we have introduced a simulation framework that allowed us to generate a broad class of matrices in terms of shape, correlation structure, and sparsity. Based on these artificial matrices and public datasets, we evaluated the performance of rank selection criteria that exploit the stochastic nature of NMF estimates. An automatic evaluation on a very large set of artificial matrices was made possible by a simple algorithm inspired by CUSUM charts to determine which rank to select based on the screeplot of the criterion versus the tentative ranks.
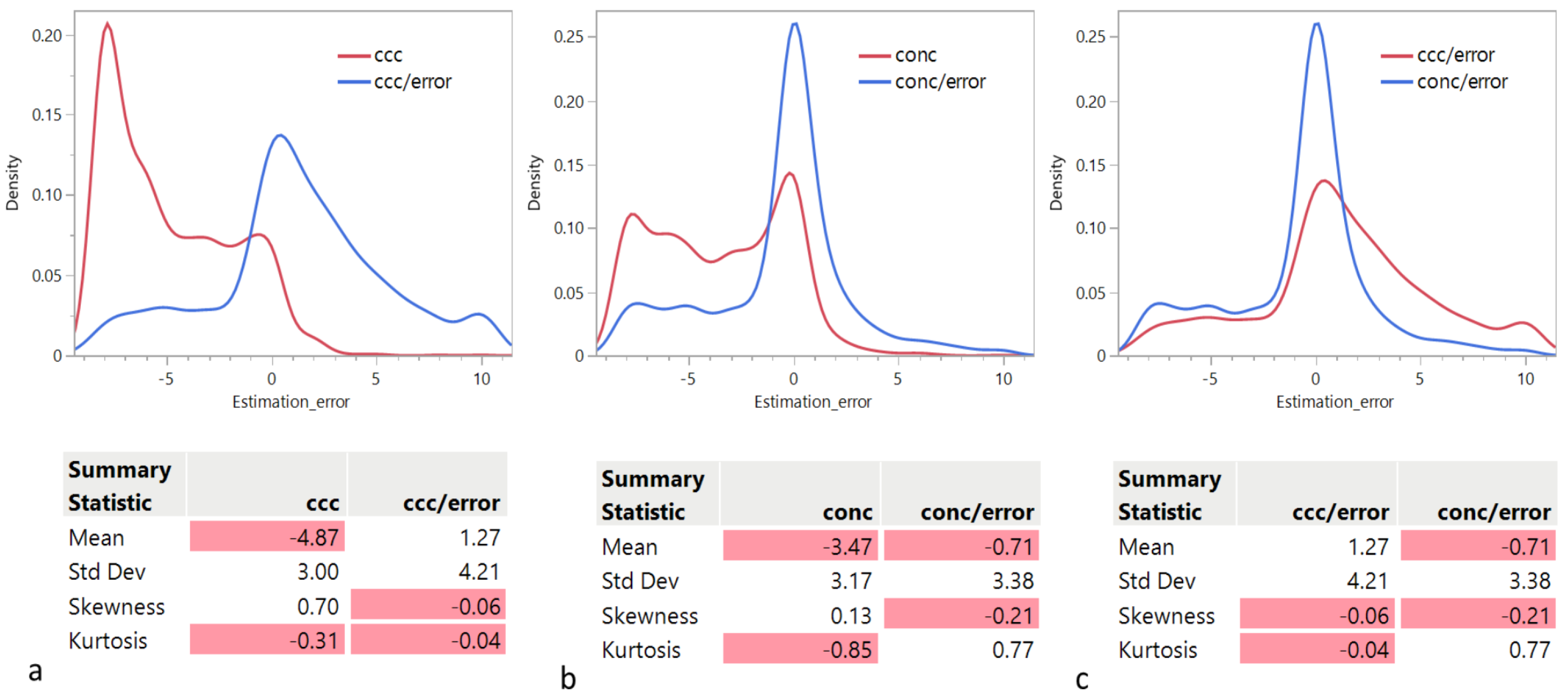
Given the wide use and acceptance of the cophenetic correlation coefficient criterion among data scientists, the fact that this criterion appears significantly biased toward the selection of lower ranks was a surprising finding. Considering its ratio to the approximation error , the distribution of the rank estimation error appears less skewed, but still biased, albeit in the opposite direction. Strikingly, only on the public Swimmer dataset succeeds in selecting the correct rank, unlike .
We have also proposed a new criterion, , that outperforms on both public and simulated datasets, both in its original form and in its ratio to the approximation form. In addition, we have shown that rank determination using is up to three times faster than with .
This work focuses on metrics that rely on the stochastic nature of the NMF estimates, as is the case for
and
. The relatively good performance of
on synthetic matrices with orthogonal underlying factors [
9], led us to use it as a reference metric when introducing the
criterion. Non-stochastic metrics, such as those from [
14] and many others, could have also been considered for comparative purposes. We have not explored that in the context of our current work because our research aims were: (i) to evaluate the performance of
and
on a larger class of synthetic matrices with non-orthogonal underlying factors (with the matrices being generated by using a novel simulation framework, which is one of the contributions of our study), and (ii) to evaluate the impact of combining a particular metric with the approximation error by considering their ratio. Nevertheless, the evaluation of the performance of other metrics on this larger class of matrices is an important area for future work.
Incidentally, this study shows that the NNDSVD initialization not only converges faster, as contended by Boustidis et al. [
20], but also provides reliable NMF estimates, in the sense that they can be used as a reference when calculating the deviation of estimates obtained with a random initialization.
The fact that the error term in the MNIST dataset does not decrease with rank highlights the limitation of matrix generation using known factors and adding noise, where the error term always tends to decrease with rank. This limitation may explain why deep-learning models trained on generated matrices, as in [
14], do not generalize well.