Abstract
Millions of people died in the COVID-19 pandemic, which pressured hospitals and healthcare workers into keeping up with the speed and intensity of the outbreak, resulting in a scarcity of ICU beds for COVID-19 patients. Therefore, researchers have developed machine learning (ML) algorithms to assist in identifying patients at increased risk of requiring an ICU bed. However, many of these studies used state-of-the-art ML algorithms with arbitrary or default hyperparameters to control the learning process. Hyperparameter optimization is essential in enhancing the classification effectiveness and ensuring the optimal use of ML algorithms. Therefore, this study utilized an improved Hunger Games Search Optimization (HGSO) algorithm coupled with a robust extreme gradient boosting (XGB) classifier to predict a COVID-19 patient’s need for ICU transfer. To further mitigate the random initialization inherent in HGSO and facilitate an efficient convergence toward optimal solutions, the Metropolis–Hastings (MH) method is proposed for integration with HGSO. In addition, population diversity was reintroduced to effectively escape local optima. To evaluate the efficacy of the MH-based HGSO algorithm, the proposed method was compared with the original HGSO algorithm using the Congress on Evolutionary Computation benchmark function. The analysis revealed that the proposed algorithm converges better than the original method and exhibits statistical significance. Consequently, the proposed algorithm optimizes the XGB hyperparameters to further predict the need for ICU transfer for COVID-19 patients. Various evaluation metrics, including the receiver operating curve (ROC), precision–recall curve, bootstrap ROC, and recall vs. decision boundary, were used to estimate the effectiveness of the proposed HGSOXGB model. The model achieves the highest accuracy of 97.39% and an area under the ROC curve of 99.10% compared with other classifiers. Additionally, the important features that significantly affect the prediction of ICU transfer need using XGB were calculated.
Keywords:
COVID-19; ICU prediction; eXtreme gradient boosting; hunger games search optimization; Metropolis–Hastings MSC:
68T20
1. Introduction
The recent coronavirus outbreak caused a global pandemic. SARS-CoV-2, a novel coronavirus, was first detected in Wuhan, China, in 2019. Recognizing its potential to trigger a worldwide public health crisis, the World Health Organization (WHO) declared this coronavirus disease (COVID-19) a global epidemic on 11 March 2020 [1]. The rise of COVID-19 led to a global health and financial catastrophe. As of 16 August 2023, the number of COVID-19 patients identified worldwide has crossed 760 million, with nearly seven million fatalities [2]; however, the real numbers are assumed to be considerably higher because of testing limitations. The first coronavirus case was detected in Bangladesh on 7 March 2020. According to the country’s Ministry of Health, the total number of COVID-19-infected patients in Bangladesh has increased by more than two million, with the total death toll surpassing twenty-nine thousand as of 16 August 2023 [3]. Although the COVID-19 pandemic has officially ended, novel variants of the virus still threaten public health.
The infection rates are rapidly increasing in Bangladesh and the world. Temperature and humidity have been identified as important factors in modeling COVID-19 mortality rates [4]. Common symptoms experienced by most COVID-19-infected patients include cold-like symptoms, continuous coughing, sore throats, a loss or alteration of taste and/or smell, fatigue, headaches, aches and pains, diarrhea, and sometimes other respiratory infections, as well as fever [5]. Note that the virus seems to be transmitted through droplets, direct contact, aerosols, blood, fecal–oral contact, from animals to humans, and even from mothers to children. Although COVID-19 has been recorded in patients of almost all ages, a higher death rate has been observed in older individuals who already suffer from cardiovascular disease, hypertension, chronic lung and kidney disease, diabetes, and other disorders. The initial step in effectively managing the allocation of critical care resources is identifying patients who are unlikely to require critical care and encouraging them to self-quarantine at home. Patients with severe conditions, such as respiratory failure and pneumonia, must be admitted to the ICU as soon as possible. However, 10–20% of total cases require ICU admission, whereas 3–10% require intubation and 2–5% die [6]. The remaining 80–90% of COVID-19 cases are mild or asymptomatic [7].
During a pandemic, the effective management of limited critical care resources and ICU beds is crucial [8]. Concerns have been raised worldwide regarding the scarcity of intensive care unit (ICU) beds for COVID-19 patients, particularly in developing countries. For instance, in Bangladesh, only 1200 ICU beds are available (including in public and private hospitals), indicating a significant shortage of critical healthcare resources. Nevertheless, prioritizing COVID-19 patients for specialized care with emergency medical support, particularly in an ICU, can significantly reduce fatality rates. Hence, frontline healthcare workers are forced to care for the “sickest of the sick” patients because of the immense pressure on the ICU admission queue. Owing to limited medical and human resources, healthcare professionals may rely on cutting-edge technologies and machine learning (ML) models to monitor and assess high-risk patients.
Several studies have addressed the issue of predicting patient outcomes in the context of COVID-19. For instance, Cheng et al. [9] employed a random forest (RF) model trained on time-series data, including major signs and patient laboratory reports as the input variables, and achieved an accuracy of 76.2% in predicting actual ICU admissions. Wollenstein-Betech et al. [10] took a comprehensive approach, employing four different classification methods, an RF, sparse support vector machine (SVM), logistic regression, and extreme gradient boosting (XGB) in each model to predict hospitalization, death, and the need for ICU care. Agieb [11] focused on predicting ICU necessity for COVID-19 patients using ML models—naive Bayes, K-nearest neighbor (K-NN), and SVM—trained on the features extracted from patients’ X-ray images. In a different vein, Weikert et al. [12] applied a deep-learning-based, fully automated extraction method for cardiothoracic CT metrics to predict the management of COVID-19 patients, including their ICU needs. Heo et al. [13] proposed an integer-based scoring method to identify COVID-19 patients requiring intensive care support. Age, dyspnea, sex, initial body temperature, history of kidney disease, hemoptysis, and the ADL scale were criteria for the scoring system. A study by Palomo et al. [14] adopted a slightly different approach and emphasized the bed demand for COVID-19 patients utilizing queuing models to show occupancy scenarios based on various patient arrival patterns.
The aforementioned studies demonstrated moderate accuracies in predicting ICU admission requirements using contemporary ML algorithms equipped with default hyperparameters. However, the classification and prediction performance can be significantly accelerated by optimizing the hyperparameters of ML algorithms using state-of-the-art optimization techniques [15,16,17,18]. Motivated by the need for hyperparameter optimization, this study adopts a novel HGSO algorithm [19], a recently introduced performance-based swarm optimization technique that demonstrates remarkable performance even when compared with an extensive collection of 23 reputable optimization functions and innovative algorithms, including the IEEE CEC 2014 benchmark test suite [20,21]. In addition, the Metropolis–Hastings (MH) algorithm was used instead of random initialization to adaptively initialize the hyperparameters [22,23]. Moreover, adaptive variational control (VC2) was required for robust optimization and to balance exploration and exploitation and thus converge to optimal solutions. Additionally, to escape the local optima effectively, the reintroduction of diversity is required to facilitate a broader explanation of the solution space. Considering these requirements, this study proposes an MH-based HGSO technique that introduces several novel contributions, which are summarized as follows.
- A novel HGSO algorithm, combined with a new fitness function, is adapted for the first time to predict the need for ICU transfer for COVID-19 patients using an XGB classifier (see Algorithm 1).
- Unlike the random initialization used in traditional HGSO, this study uses the MH algorithm to adaptively initialize the hyperparameter values (see Algorithm 2).
- This study employs an adaptive VC2 operator and reintroduces diversity to find the optimal solution and prevent premature convergence, respectively (see Section 2.4).
- This study identifies the most significant predictors for ICU transfer using XGB, providing valuable information for national policymakers (see Section 3.8).
- Comparing the proposed HGSOXGB model with cutting-edge classifiers reveals that the HGSOXGB obtains the highest accuracy (see Section 3.4 and Section 3.9).
| Algorithm 1 XGB classifier implementation algorithm |
Input: D-dimensional feature and target with N samples Output: The probability of outcome of unknown test dataset x, where for all , is the number of classes
|
| Algorithm 2 Proposed population initialization algorithm using the MH method |
Input: The number of XGB hyperparameter in the population (N), the number of XGB hyperparameter (H), and the lower () and upper () limits of each XGB hyperparameter. Output: Initial population,
|
2. Materials and Methods
Figure 1 shows the workflow of the proposed model. As illustrated, the columns containing 60% of missing values and unnecessary outliers were eliminated, and the remainder of the dataset underwent min–max normalization to convert all values to the same scale. Subsequently, the normalized dataset was divided into training and testing sets. To mitigate class imbalance, a large quantity of synthetic data was generated for the minority class inside the training set using the Synthetic Minority Oversampling Technique (SMOTE). Subsequently, the resulting balanced dataset was employed to train the ML models. The hyperparameters of the ML classifiers were finetuned using the HGSO algorithm. However, when comparing the performance of the original HGSO model with that of the CEC benchmark function (briefly discussed in Appendix A), we observed that the convergence of the HGSO algorithm requires further improvement. Another issue with the original HGSO is the random initialization of the hyperparameter values. Therefore, the MH algorithm was utilized to adaptively initialize the hyperparameter values, thereby accelerating the convergence speed of the HGSO algorithm. The model performance was evaluated using previously separated test data in the following steps. Various statistical analyses were performed in addition to feature importance calculations. Finally, the necessity of ICU care for COVID-19 patients was efficiently predicted and presented.
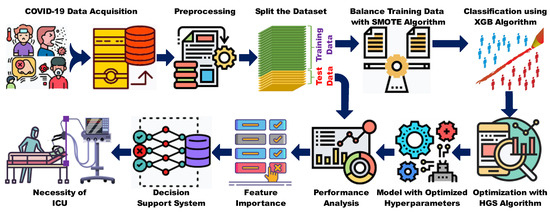
Figure 1.
Workflow diagram of the proposed system.
2.1. Data Source
A large COVID-19 dataset was collected from a publicly available repository of SARS-CoV-2 patient information in Mexico [24]. It contains 7233257 individual observations in 40 columns. The data contain demographic information, including age, nationality, location, origin, ethnicity, prior medical conditions, diabetes, asthma, hypertension, obesity, chronic obstructive pulmonary disease (COPD), chronic renal failure, immunosuppression, pregnancy, other prior diseases, and smoking information. In addition, they contained the dates of the first symptoms observed by the patient, the patient’s arrival in the ICU, and the patient’s death, and include columns that show whether the patient was hospitalized, had pneumonia, or required ICU care, as well as the results of the antigen test for SARS-CoV-2.
2.2. Data Preprocessing
Data preprocessing plays a pivotal role in machine learning classification. However, the raw dataset used in the proposed approach was impossible to work with because its column names were in Spanish and the data were considerably noisy, with nearly 60% missing values. Moreover, many columns in the dataset were irrelevant and did not influence the classification outcomes. In addition, the numerical values of the dataset were obtained at different scales. Given these challenges, achieving optimal results using raw datasets was impractical. Hence, data preprocessing was crucial for converting the data into a meaningful format and improving the ML performance. The steps taken to preprocess the dataset are outlined below.
- 1.
- Column translation: initially, the Spanish names of each column were translated into English.
- 2.
- Missing value removal: the columns containing almost 60% missing values were eliminated from the dataset because the interpretation and quality of the proposed approach can degrade if this large amount of missing values remains in the input dataset.
- 3.
- Removal of unnecessary columns: because the proposed approach aims to anticipate ICU necessity for COVID-19 patients, the unnecessary columns—origin, sector, date of admission to the ICU, etc.—were removed from the dataset, as they are unrelated to ICU prediction. The columns (features) containing outliers were also eliminated from the dataset.
- 4.
- Removing demographic variables: the columns of the demographic variables, such as ethnicity, region, nationality, location, and origin, were eliminated from the dataset such that the dataset can be utilized in all other countries.
Upon completing these steps, 18 fields were retained: intubated (identifying whether the patient required intubation), pneumonia, otras_com (identifying if the patient had other diseases), COPD, cardiovascular, inmusupr (indicating if the patient was immunosuppressed), another case (noting patient contact with other SARS CoV-2 cases), smoking, asthma, renal_cornica, age, pregnancy, obesity, diabetes, hypertension, sex, antigen_result (identifying the result of the antigen sample analysis), and ICU (determining if the patient required admission to an ICU). Following all previous steps, min–max normalization was performed to convert all data values to a scale ranging from 0 to 1, as the features may not be near the normal distribution. Furthermore, after successfully completing data preprocessing and before applying the ML algorithm, a sufficient amount of synthetic data was generated for the minority class of the training set (obtained through the train–test split) as part of data balancing.
2.3. Selection of Machine Learning Classifier
Owing to their remarkable success and effectiveness, ML and deep learning algorithms have gained substantial attention in various domains, including in the prediction of ICU needs for COVID-19 patients. Although deep learning has attained significant prominence in certain tasks, ML algorithms offer robust results owing to their superior generalization capabilities and the transparency of their outcomes. Researchers globally employ diverse ML algorithms to predict COVID-19 patients; however, XGB, a powerful, supervised ML algorithm, has exhibited robustness across a wide range of real-world applications [25]. For instance, Yan et al. [26] employed XGB to predict the mortality attributed to COVID-19. In addition, XGB was utilized in another study conducted by Awal et al. [27] to predict COVID-19 patients using inpatient facility data, leveraging its recursive tree-based decision system. Notably, XGB can assess the significance of each feature based on its cumulative usage in the decision-making process, thereby providing valuable insights into the feature importance. This information aids in identifying discriminative features, particularly those associated with clinically relevant parameters. Considering these factors, the proposed approach employed XGB, with its hyperparameters optimized using the HGSO technique. Equation (1) expresses the XGB objective function used to measure the model performance:
where denotes the parameters trained using the given data, is the regularization term, and L is the training loss. The objective function defines the regularization term for a decision tree, as outlined in Equation (2):
where denotes the number of leaves in a decision tree, denotes the complexity of each leaf, is a regularization parameter, and is the score of the leaves. Optimizing the objective function of XGB enables the change in model performance after a certain node split to be evaluated. If the performance of the decision tree in the XGB improves, the change is approved. Algorithm 1 shows the steps followed for XGB implementation.
To enhance the model performance, several hyperparameters must be optimized. The hyperparameters used in the proposed system are as follows:
- learning_rate: represents the shrinkage of each tree’s contribution.
- max_depth: specifies the maximum depth of the tree.
- Gamma: sets the minimum loss reduction required for further leaf node splitting.
- min_child_weight: defines the minimum instance load for a child node.
- colsample_by_tree: denotes the subsample of columns used to train each tree.
- subsample: determines the column ratio for tree building.
- alpha: represents the regularization weight term.
- eval_metric: specifies the validation metric for validation data.
Tuning these hyperparameters in the COVID-19 dataset can enhance the performance of the ICU prediction models. In this study, various popular ML algorithms—Gaussian naïve Bayes (GNB), linear discriminant analysis (LDA), RF, KNN, light gradient boosting machine (LGBM), gradient boosting classifier (GBC), and linear regression (LR)—were utilized for comparison. Subsequently, their performances were evaluated and compared. Additional mathematical details about these classifiers can be found in standard textbooks and the literature [27,28].
2.4. Need for Hyperparameter Optimization and Proposed HGSO
The necessity for optimizing the specific hyperparameters used by the XGB classifier in this study arises from the fact that classification metrics, such as accuracy, sensitivity, and specificity, rely extensively on selecting appropriate hyperparameters. This requirement promotes the framing of this study as an optimization problem, as demonstrated in Equation (3):
where denotes the hyperparameters listed in Table 1. The optimization problem is defined as follows: “select the optimal hyperparameters of XGB for which is the minimum”. In this study, the kappa score loss obtained from a -fold cross-validation of the training dataset was recommended as the fitness function , as expressed in Equation (4):

Table 1.
The architecture of the HGSOXGB model with optimized hyperparameters.
Hyperparameter optimization is required [29]. The HGSO technique, a metaheuristic algorithm, was employed to optimize the hyperparameters to precisely determine the necessity for ICU admission based on some preconditions of COVID-19 patients. This study adapted HGSO, a recent optimization technique that outperforms state-of-the-art optimization techniques; however, the original HGSO suffers from a random initialization issue, which often prolongs the convergence time and does not ensure that the highest global optimum is found. The existing problem with the traditional HGSO algorithm was addressed using the MH algorithm [23] to mitigate the population initialization problem of the traditional HGSO algorithm. The improved HGSO approach yields an enhanced convergence, requires fewer iterations, and achieves optimal values. Algorithm 2 illustrates the initialization process of the MH method. In this algorithm, the input parameters include the number of XGB hyperparameters in the population N, the number of XGB hyperparameters H, and the upper () and lower () bounds of each hyperparameter. The algorithm proceeded by allocating memory (line 1), initializing the first individuals randomly (line 2), generating an individual candidate, calculating the acceptance probability (lines 4 and 5), and accepting or rejecting the candidate based on a random number (lines 7–11). Finally, the MH-method-based initial parameters were adapted and returned (line 14).
The HGSO is an evolutionary and population-based competitive optimization algorithm that enhances exploratory and exploitative behaviors, and its tractability is challenging. Moreover, it addresses challenges posed by multimodal and local optima [19]. This draws inspiration from the cooperative behavior observed in animals; the search process is analogous to an animal’s response to hunger levels. In this context, evolutionary approaches are employed to modify and adapt processes to achieve high-quality outcomes and expedite convergence. The algorithm exhibits two characteristics during the search steps: exploration and exploitation. During the exploration stage, the randomness of the search space must be ensured to explore it comprehensively [19]. During the subsequent exploitation stage, the algorithm narrows its focus to a specific region in the feature space identified during the prior exploitation stage [19]. In the lives of animals, hunger is responsible for motivation and motivates behavior and decisions. Equation (5) expresses the proposed HGSO algorithm:
where t represents the current iterations, denotes the position of the XGB hyperparameter for the top individual in this iteration, and denote the weights associated with hunger, indicates the position of the hyperparameter of each individual, and l describes the experimental parameter for algorithm development. indicates how an individual performs a random search for food around its current position. The quantity denotes the range of movement of the current individual hyperparameters during the current iteration. The parameter E controls the degree of variation across all hyperparameter positions, and serves as a range control parameter. Algorithm 3 outlines the steps followed in the proposed HGSOXGB framework. The HGSOXGB algorithm uses random numbers which belong to .
| Algorithm 3 Proposed HGSOXGB algorithm |
Input: The H-dimensional hyperparameters of XGB applied on training data, maximum iteration (T), number of search agents or individuals (N), upper () and lower () bounds of H, sum of hungry of all individuals (), l. Output: HGSOXGB model and best-fit value
|
Unlike the original HGSO algorithm, the current approach integrates the VC2 operator to achieve a balanced exploration of the entire solution space and exploitation to refine the optimal solutions. This algorithm can better switch between diverse and concentrated searching, thereby enhancing the probability of obtaining optimal solutions. Additionally, the optimization process may occasionally lead search agents to converge to local optima. To effectively counter this, diversity in the population space was reintroduced by reinitializing 20% of the population every iteration using the proposed MH-based population initialization method (Line 14). The diversity outlined here helps the algorithm escape the local optima and converge towards a global solution. Furthermore, in some situations, the agent positions can turn into non-number (NaN) values owing to numerical instability and extreme values, rendering them unproductive for the ongoing search process. To ensure the contribution of each agent to the optimization, agents with NaN values in their positions are identified and subsequently reinitialized using Algorithm 2 (Line 15). For the convenience of the research community, a Python implementation of the algorithm is available at https://github.com/awalece04ku/HGSO_ICU, accessed on 5 September 2023.
2.5. Model Performance Analysis
To evaluate the effectiveness of the proposed ML scheme, various performance evaluation metrics—the accuracy (ACC), F1-score, Matthew’s correlation coefficient (MCC), kappa score, sensitivity (recall), and area under the curve (AUC)—were determines, along with an assessment of the confusion matrix. In addition, novel statistical parameters—Cramer’s V, phi-squared, classification success index (CSI), Pearson’s C, and Scott’s Pi—obtained from Pycm, a Python library for multiclass confusion matrices [30], were used for evaluation. Equations (6) and (10) summarize these evaluation measures.
Cramer’s V: it expresses the relationship between two nominal variables and ranges from 0 to 1:
where is the phi coefficient and r and c are the numbers of rows and columns, respectively.
Phi-squared: it shows the relationship between two binary variables:
where is the chi-squared statistic and n is the total number of samples in the dataset.
CSI: it is an overall measure defined by averaging the individual classification success index (ICSI), ranging from −1 to 1, over all classes:
where ICSI is 1 minus the sum of the positive predictive value and true positive rate, and is the number of classes.
Pearson’s C: it is known as the Pearson coefficient, ranging from −1 to 1, and represents whether two variables are dependent on each other.
Scott’s Pi: it explains how two nominal data points are internally reliable:
where is the observed agreement and is the expected agreement.
Moreover, an ROC curve, a precision–recall curve, a bootstrap ROC curve, and the recall rate vs. decision boundary were created to assess the classifier’s performance.
2.6. Feature Importance Using XGB
An analysis of the feature set revealed that certain features were less important or even irrelevant in predicting the need for ICU admission. The dominant features were sorted using XGB, and a graph was generated to display the feature importance values in descending order. The graph lists the features on the Yaxis, and the corresponding importance values are shown on the Xaxis. The XGB algorithm utilizes the “gain” metric to assess the relative significance of each feature. Furthermore, “gain” plays a crucial role in determining the optimal node split during tree construction in the training phase. The average gain was subsequently used to derive the final importance score for each feature. Equation (11) gives the mathematical formulation for calculating the gain:
where , , , are the first-, first-, second-, and second-order gradient of the left, right, left, and right nodes, respectively. and represent penalty and regularization parameters, respectively.
3. Results and Discussion
Having developed the proposed MH-based HGSO algorithm, a preliminary benchmark analysis was conducted to assess its performance before analyzing its viability for ICU prediction.
3.1. Initial Benchmark Analysis
The effectiveness of the proposed HGSO algorithm was tested on four benchmark functions: Sphere, Ackley, Levy, and Schaffer. Subsequently, it was compared with the original HGSO algorithm using the same number of generations and iterations, resulting in the proposed HGSO algorithm outperforming the original algorithm. Appendix B provides the complete analysis of the benchmark functions from the Congress on Evolutionary Computation (CEC) benchmark and their graphical illustrations.
3.2. ICU Prediction Results
The preceding section demonstrated the superior performance of the HGSO algorithm on the CEC benchmark dataset. Therefore, the optimal values of the XGB hyperparameter, which significantly influences ICU prediction, were determined using HGSO, as presented in Table 1. Note that the same training and testing datasets were employed across all classifiers to ensure the consistency of the analysis.
3.3. Confusion Matrix
The principal diagonal cells of each confusion matrix, visualized in Figure 2, represent the percentage of correct classifications achieved by the proposed approach compared with state-of-the-art classification algorithms, i.e., GNB, LDA, KNN, RF, LGBM, GBC, and LR. In particular, as depicted in Figure 2a, out of 6758 ICU-no instances, 6527 were accurately classified, with only 231 misclassifications; in contrast, among 6768 ICU-yes instances, 6646 were correctly classified, with 122 incorrect classifications, achieving an impressive accuracy of 97.39%. Figure 2b–h shows the accuracy scores of the other leading classification algorithms. For instance, LGBM secured the second position in correctly classifying the need for ICU beds for COVID-19 patients, and GNB exhibited the lowest accuracy among the algorithms. Consequently, the proposed HGSOXGB was concluded to outperform traditional classifiers.
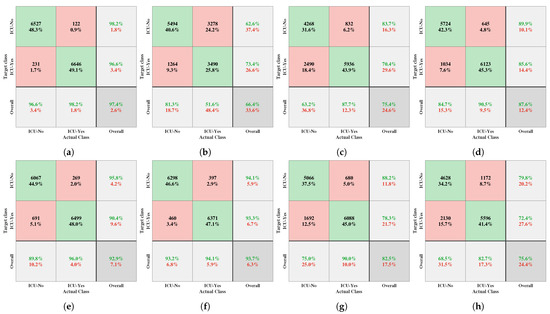
Figure 2.
Confusion matrices generated for the (a) HGSOXGB, (b) GNB, (c) LDA, (d) KNN, (e) RF, (f) LGBM, (g) GBC, and (h) LR methods. Note that in each confusion matrix, rows correspond to the target classes, columns denote the actual classes, diagonal cells highlight correct classifications, and off-diagonal cells indicate errors. Each cell displays both a count and percentage value. The rightmost column of the confusion matrix presents precision and false discovery rate metrics, providing insight into the accuracy of the predictions for each class. Furthermore, the bottom row of each matrix reflects the recall and false negative rates, indicating the accuracy for actual class instances. Ultimately, the bottom-right cell of each matrix encapsulates the overall accuracy of the model.
3.4. Comparative Performance Analysis of Different Classifiers
This study rigorously evaluated the performance of the proposed model against a set of cutting-edge classifiers, including GNB, LDA, KNN, RF, LGBM, GBC, and LR. A comprehensive evaluation was conducted using multiple robust metrics (the ACC, F_score, kappa score, MCC score, sensitivity, and AUC) to evaluate the effectiveness of each classifier (as detailed in Table 2). The tabulated data clearly show that the proposed HGSOXGB exhibits the best results in terms of all the classification metrics. For example, the accuracy score and AUC value of HGSOXGB were significantly higher than 97%, surpassing those of all other classifiers, whereas GNB remained at the bottom position with respect to the performance evaluation metrics. Table 3 compares the performance of HGSOXGB in terms of additional overall statistical parameters, illustrating that the proposed framework outperforms the original XGB algorithm.

Table 2.
Performance comparison with different classifiers.

Table 3.
Performance comparison with other models.
3.5. ROC, Precision–Recall, and Bootstrap ROC Curves
The ROC curve illustrated in Figure 3 clearly shows the superiority of the proposed HGSOXGB model compared with the other classifiers. Figure 3b presents the precision–recall curve, summarizing the relationship between the true positive rate and positive predictive value along the y- and x-axes, respectively, to predict the need for ICU admission for patients with COVID-19 using the proposed HGSOXGB model at varying probability thresholds. The bootstrapped ROC curve displayed in Figure 3c shows whether the bias in the training dataset is prevalent during the training phase. Employing N_boot = 100 for the bootstrapping, the estimation of a mean AUC exceeding 99% provided insights into the model’s robustness against potential training dataset skewness. Furthermore, calculating a 90% confidence interval for the bootstrap ROC curve provides an additional layer of statistical validity.
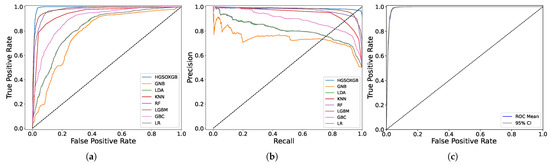
Figure 3.
The (a) ROC, (b) precision–recall, and (c) bootstrap ROC curves for HGSOXGB.
3.6. Recall vs. Decision Boundary
The recall rate represents the number of ICU cases that must be predicted from a complete dataset. For this investigation, the classification task has been accomplished, given that “ICU-no” and “ICU-yes” were equally important with a decision boundary threshold of 0.5, as visualized in Figure 4a,b. The graphical illustration exhibits that the HGSOXGB approach had the highest recall rate, at approximately 96.58% (also displayed in Table 2), implying that over 96% of the time, the proposed approach can accurately classify the necessity for ICU admission for COVID-19 patients.
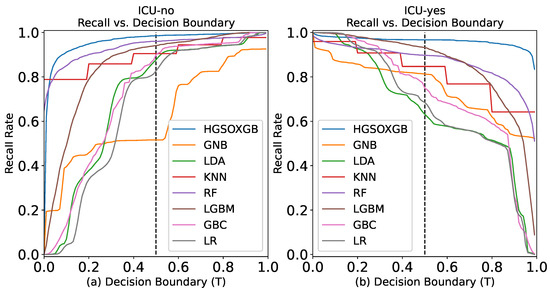
Figure 4.
Recall rate vs. decision boundary for (a) ICU-no class and (b) ICU-yes class.
3.7. Ten-Fold Cross-Validation Performance of the Proposed Model
This study includes the violin plot presented in Figure 5, which illustrates the cross-validation accuracy of the various models. The plot clearly shows that the proposed HGSOXGB outperforms the other models in terms of cross-validation accuracy. The white dots represent the median value, the thick gray bars in the center denote the interquartile range, and the upper and lower thin gray lines indicate the values 1.5 times the interquartile range. A wider section of the violin plot indicates a higher likelihood, whereas narrower sections indicate a lower probability. Furthermore, the analysis of variance (ANOVA) test indicated that the HGSOXGB results are statistically significant ().
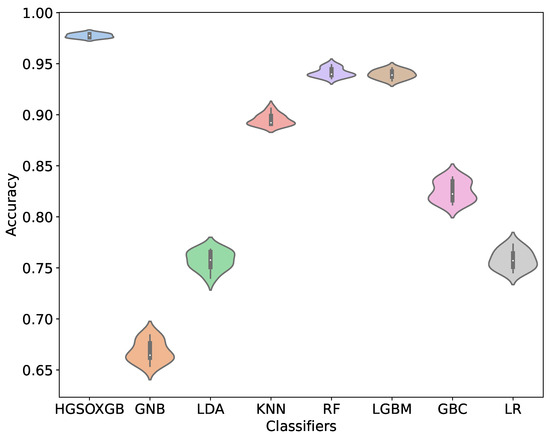
Figure 5.
Violin plot of 10-fold cross-validation score of different models.
3.8. Feature Importance Using XGB
The dominant features that significantly affect the necessity for ICU care for COVID-19 patients were categorized using XGB. The feature importance plot of this study in Figure 6 reveals that “intubated” is the most important feature in predicting ICU requirement, followed by “pneumonia”. Note that features such as “otras_com”, “smoking”, “COPD”, and “asthma” substantially influence the prediction for ICU requirement for COVID-19 patients [13,31].
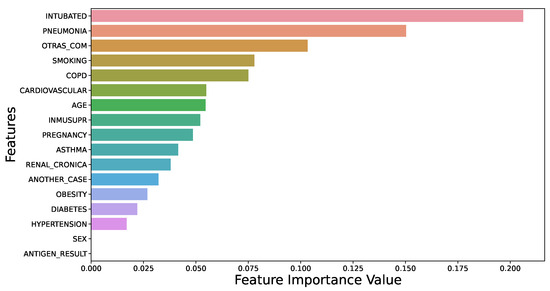
Figure 6.
Feature importance using XGB.
In addition to influencing ICU prediction, the XGB-based feature importance technique can assist national policymakers. For instance, when individuals exhibit symptoms such as colds, continuous coughing, sore throats, the loss or alteration of one’s sense of taste or smell, fatigue, headaches, aches and pains, diarrhea, and other respiratory infections, including fever, a coronavirus infection is suspected. In such cases, policymakers should encourage workers to engage in remote work to mitigate the potential spread of the virus within their workplaces and broader communities. Notably, novel variants remain a threat, although the pandemic is officially over. Therefore, the feature importance technique may contribute to broader public health endeavors to mitigate the impact of the virus.
3.9. Performance Comparison with Other Studies
The effectiveness of the proposed HGSOXGB model was compared with that of other contemporary studies using various performance indices. Some methods utilized the same dataset as in this study, whereas others were employed using their original dataset, and we attempted to replicate their approach and compare the proposed model accordingly. Table 4 presents the comparison results, which indicate that the proposed model outperforms existing methods.

Table 4.
Performance comparison with other studies.
3.10. Limitations and Future Works
The proposed framework was evaluated using a benchmark and one of the largest single-center datasets; thus, a multicenter dataset was required to achieve a broader generalization, although acquiring such a dataset is time-consuming. Another noteworthy aspect of using multicenter datasets is the opportunity to test across various datasets, facilitating valuable generalization. Additionally, although the proposed framework relies solely on clinical data, additional data sources such as genetic data or data containing social determinants of health (SDH)—non-medical factors that influence health outcomes—will be incorporated in the future to improve prediction accuracy. Another limitation of the proposed technique is its reliance on ML models. In the future, deep learning models will be implemented to evaluate the proposed technique and converge to the optimal solution for predicting ICU admissions. Note that the source code of the proposed framework has been released, and researchers are encouraged to contribute in this important area.
4. Conclusions
This study proposes an ML framework for predicting the need for ICU admission for COVID-19 patients based on clinical data using an XGB classifier with HGSO-optimized hyperparameters. Instead of randomly initializing the population, the MH method was adopted to enhance the performance of the proposed HGSO algorithm. The SMOTE algorithm was applied to the COVID-19 clinical data to balance the dataset. The proposed HGSOXGB achieved the highest accuracy and AUC values of 97.39% and 99.10%, respectively. Furthermore, XGB was used to identify pneumonia as the most crucial feature for predicting ICU requirements. Therefore, the dominant features categorized using the proposed XGB-based feature importance technique can significantly aid policymakers in imposing necessary rules during critical situations. This feature importance technique can then help safeguard communities from infections or pandemics caused by deadly viruses. Therefore, the findings of this study have the potential to predict individual ICU needs for COVID-19 patients and may be extended to other classification problems, such as asthma and diabetes.
Author Contributions
Conceptualization, F.T.P. and M.A.A.; Methodology, F.T.P., M.A.A. and M.S.H.; Software, M.A.A. and K.M.M.; Validation, M.A.A., M.S.H., K.M.M. and J.A.F.; Formal Analysis, F.T.P., M.A.A. and K.M.M.; Investigation, M.S.H., R.R., L.A., A.K. and M.A.S.; Resources, M.A.A. and J.A.F.; Data Curation, M.A.A. and K.M.M.; Writing, F.T.P., M.A.A. and M.S.H.; Writing—Review and Editing, M.A.A., M.S.H., R.R. and M.A.S.; Visualization, M.S.H. and M.A.S.; Supervision, M.A.A. and M.A.S.; Project Administration, M.A.A.; Funding Acquisition, L.A., A.K. and M.A.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Princess Nourah bint Abdulrahman University Researchers Supporting Project (number PNURSP2023R349), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The processed data, trained model, and codes related to this study are available at https://github.com/awalece04ku/HGSO_ICU (accessed on 5 September 2023).
Conflicts of Interest
The authors have no conflict of interest to declare.
Appendix A. Benchmark Functions
Benchmark functions demonstrate how well an optimization algorithm reaches the optimal solution within a defined search space. The effectiveness of the proposed HGSO algorithm depends on the CEC benchmark function. The mathematical abstraction of widely used functions is as follows:
- Sphere function: The Sphere function—a simple convex function with a single minimum at the origin—is widely used to test the optimization algorithm by serving as a starting point in evaluating the performance. Equation (A1) outlines the mathematics behind the Sphere function in n-dimensional space:where describes the ith dimension of the input vector. The mean value of the sphere function is zero, and the variance is one.
- Ackley function: The Ackley function, a two-dimensional function with multiple local optima and a global minimum, is widely used for testing optimization algorithms. Equation (A2) presents the mathematical formulation of the Ackley function:
- Levy function: The Levy function is a continuous probability distribution for a nonnegative random variable. It is used in the optimization technique to search for all local minima. Equation (A3) outlines the mathematical background of the levy function:where for all ; .
- Schaffer function: The Schaffer function is a pair of multimodal functions commonly used as benchmark problems for testing optimization algorithms. Equation (A4) provides mathematical insights into the Schaffer function:where the upper and lower bounds of the hyperparameters are defined as .
Appendix B. Benchmark Analysis
In total, 300 generations and 30 populations were employed, and the proposed algorithm was benchmarked against the original HGSO algorithm. The results clearly show that the proposed algorithm exhibits a superior convergence for each function compared to the original algorithm (as shown in Figure A1). Notably, the same number of generations and population sizes was used to ensure fair comparisons in each case.
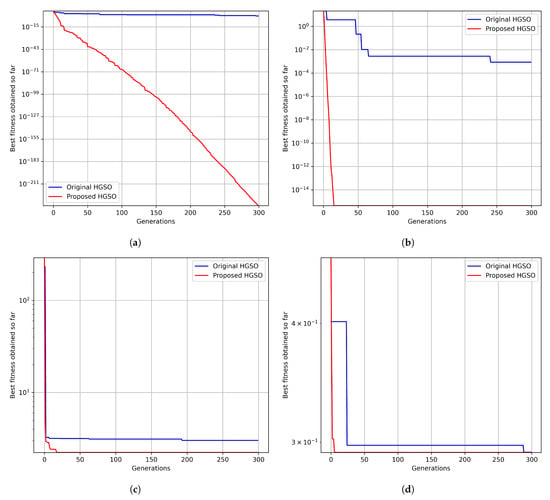
Figure A1.
Comparing the effectiveness of the proposed and original HGSO in the context of a convergence analysis for the CEC benchmark functions: (a) Sphere, (b) Ackley, (c) Levy, and (d) Schaffer.
In addition, only the performance of the proposed method was compared with that of the original HGSO. The study compared the original HGSO with many other optimization algorithms.
To further demonstrate the effectiveness of the optimization algorithm, the code was run 100 times for both the proposed and original methods. Subsequently, a Mann–Whitney U statistical significance test was conducted. In each case, the median value of the box plot was statistically different, as indicated by a p-value of less than 0.005 (Figure A2). The benchmark functions, along with their convergence plot and statistical significance codes, are available at https://github.com/awalece04ku/HGSO_ICU (accessed on 5 September 2023) for further justification.
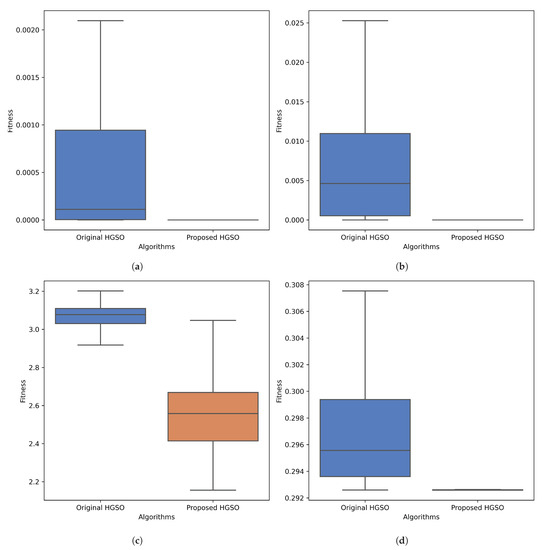
Figure A2.
Comparing the effectiveness of the proposed and original HGSO using box plots (with different p-values) for different CEC benchmark functions: the (a) Sphere function for , (b) Ackley function for , (c) Levy function for , and (d) Schaffer function for .
References
- Cucinotta, D.; Vanelli, M. WHO declares COVID-19 a pandemic. Acta Bio Med. Atenei Parm. 2020, 91, 157. [Google Scholar] [CrossRef]
- WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 16 August 2023).
- COVID-19 Dynamic Dashboard for Bangladesh. Available online: http://103.247.238.92/webportal/pages/covid19.php (accessed on 16 August 2023).
- Mousavi, M.; Salgotra, R.; Holloway, D.; Gandomi, A.H. COVID-19 time series forecast using transmission rate and meteorological parameters as features. IEEE Comput. Intell. Mag. 2020, 15, 34–50. [Google Scholar] [CrossRef]
- Dowdall, M.; Stewart, K. Differential diagnosis: Cold, flu or COVID-19? Pharm. J. 2020, 2–3. [Google Scholar] [CrossRef]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.; et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Pascarella, G.; Strumia, A.; Piliego, C.; Bruno, F.; Del Buono, R.; Costa, F.; Scarlata, S.; Agrò, F.E. COVID-19 diagnosis and management: A comprehensive review. J. Intern. Med. 2020, 288, 192–206. [Google Scholar] [CrossRef]
- Rahimi, I.; Chen, F.; Gandomi, A.H. A review on COVID-19 forecasting models. Neural Comput. Appl. 2021, 1–11. [Google Scholar] [CrossRef]
- Cheng, F.Y.; Joshi, H.; Tandon, P.; Freeman, R.; Reich, D.L.; Mazumdar, M.; Kohli-Seth, R.; Levin, M.A.; Timsina, P.; Kia, A. Using machine learning to predict ICU transfer in hospitalized COVID-19 patients. J. Clin. Med. 2020, 9, 1668. [Google Scholar] [CrossRef]
- Wollenstein-Betech, S.; Cassandras, C.G.; Paschalidis, I.C. Personalized predictive models for symptomatic COVID-19 patients using basic preconditions: Hospitalizations, mortality, and the need for an ICU or ventilator. Int. J. Med. Inform. 2020, 142, 104258. [Google Scholar] [CrossRef]
- Agieb, R. Machine learning models for the prediction the necessity of resorting to icu of COVID-19 patients. Int. J. Adv. Trends Comput. Sci. Eng. 2020, 9, 6980–6984. [Google Scholar] [CrossRef]
- Weikert, T.; Rapaka, S.; Grbic, S.; Re, T.; Chaganti, S.; Winkel, D.J.; Anastasopoulos, C.; Niemann, T.; Wiggli, B.J.; Bremerich, J.; et al. Prediction of patient management in COVID-19 using deep learning-based fully automated extraction of cardiothoracic CT metrics and laboratory findings. Korean J. Radiol. 2021, 22, 994. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.; Han, D.; Kim, H.J.; Kim, D.; Lee, Y.K.; Lim, D.; Hong, S.O.; Park, M.J.; Ha, B.; Seog, W. Prediction of patients requiring intensive care for COVID-19: Development and validation of an integer-based score using data from Centers for Disease Control and Prevention of South Korea. J. Intensive Care 2021, 9, 1–9. [Google Scholar] [CrossRef]
- Palomo, S.; Pender, J.; Massey, W.A.; Hampshire, R.C. Flattening the Curve: Insights From Queueing Theory. PLoS ONE 2020, 18, 0286501. [Google Scholar] [CrossRef]
- Hashim, F.A.; Houssein, E.H.; Mabrouk, M.S.; Al-Atabany, W.; Mirjalili, S. Henry gas solubility optimization: A novel physics-based algorithm. Future Gener. Comput. Syst. 2019, 101, 646–667. [Google Scholar] [CrossRef]
- Heidari, A.A.; Mirjalili, S.; Faris, H.; Aljarah, I.; Mafarja, M.; Chen, H. Harris hawks optimization: Algorithm and applications. Future Gener. Comput. Syst. 2019, 97, 849–872. [Google Scholar] [CrossRef]
- Kudela, J. The Evolutionary Computation Methods No One Should Use. arXiv 2023, arXiv:2301.01984. [Google Scholar] [CrossRef]
- Xu, L.; Yan, W.; Ji, J. The research of a novel WOG-YOLO algorithm for autonomous driving object detection. Sci. Rep. 2023, 13, 3699. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, H.; Heidari, A.A.; Gandomi, A.H. Hunger games search: Visions, conception, implementation, deep analysis, perspectives, and towards performance shifts. Expert Syst. Appl. 2021, 177, 114864. [Google Scholar] [CrossRef]
- Erlich, I.; Rueda, J.L.; Wildenhues, S.; Shewarega, F. Evaluating the mean-variance mapping optimization on the IEEE-CEC 2014 test suite. In Proceedings of the 2014 IEEE Congress on Evolutionary Computation (CEC), Beijing, China, 6–11 July 2014; pp. 1625–1632. [Google Scholar] [CrossRef]
- Hu, Z.; Bao, Y.; Xiong, T. Partial opposition-based adaptive differential evolution algorithms: Evaluation on the CEC 2014 benchmark set for real-parameter optimization. In Proceedings of the 2014 IEEE Congress on Evolutionary Computation (CEC), Beijing, China, 6–11 July 2014; pp. 2259–2265. [Google Scholar] [CrossRef]
- Chib, S.; Greenberg, E. Understanding the Metropolis-hastings Algorithm. Am. Stat. 1995, 49, 327–335. [Google Scholar] [CrossRef]
- Cuevas, E.; Escobar, H.; Sarkar, R.; Eid, H.F. A new population initialization approach based on Metropolis–Hastings (MH) method. Appl. Intell. 2022, 53, 16575–16593. [Google Scholar] [CrossRef]
- Open Data General Directorate of Epidemiology. Available online: https://www.gob.mx/salud/documentos/datos-abiertos-152127 (accessed on 1 October 2021).
- Chen, T.; Guestrin, C. XGBoost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar] [CrossRef]
- Yan, L.; Zhang, H.T.; Goncalves, J.; Xiao, Y.; Wang, M.; Guo, Y.; Sun, C.; Tang, X.; Jing, L.; Zhang, M.; et al. An interpretable mortality prediction model for COVID-19 patients. Nat. Mach. Intell. 2020, 2, 283–288. [Google Scholar] [CrossRef]
- Awal, M.A.; Masud, M.; Hossain, M.S.; Bulbul, A.A.M.; Mahmud, S.H.; Bairagi, A.K. A novel bayesian optimization-based machine learning framework for COVID-19 detection from inpatient facility data. IEEE Access 2021, 9, 10263–10281. [Google Scholar] [CrossRef]
- Awal, M.A.; Hossain, M.S.; Debjit, K.; Ahmed, N.; Nath, R.D.; Habib, G.M.; Khan, M.S.; Islam, M.A.; Mahmud, M.P. An Early Detection of Asthma Using BOMLA Detector. IEEE Access 2021, 9, 58403–58420. [Google Scholar] [CrossRef]
- Islam, M.S.; Awal, M.A.; Laboni, J.N.; Pinki, F.T.; Karmokar, S.; Mumenin, K.M.; Al-Ahmadi, S.; Rahman, M.A.; Hossain, M.S.; Mirjalili, S. HGSORF: Henry Gas Solubility Optimization-based Random Forest for C-Section prediction and XAI-based cause analysis. Comput. Biol. Med. 2022, 147, 105671. [Google Scholar] [CrossRef] [PubMed]
- Haghighi, S.; Jasemi, M.; Hessabi, S.; Zolanvari, A. PyCM: Multiclass confusion matrix library in Python. J. Open Source Softw. 2018, 3, 729. [Google Scholar] [CrossRef]
- Kavadi, D.P.; Patan, R.; Ramachandran, M.; Gandomi, A.H. Partial derivative nonlinear global pandemic machine learning prediction of covid 19. Chaos Solitons Fractals 2020, 139, 110056. [Google Scholar] [CrossRef]
- Zhao, Z.; Chen, A.; Hou, W.; Graham, J.M.; Li, H.; Richman, P.S.; Thode, H.C.; Singer, A.J.; Duong, T.Q. Prediction model and risk scores of ICU admission and mortality in COVID-19. PLoS ONE 2020, 15, e0236618. [Google Scholar] [CrossRef]
- Famiglini, L.; Bini, G.; Carobene, A.; Campagner, A.; Cabitza, F. Prediction of ICU admission for COVID-19 patients: A Machine Learning approach based on Complete Blood Count data. In Proceedings of the 2021 IEEE 34th International Symposium on Computer-Based Medical Systems (CBMS), Aveiro, Portugal, 7–9 June 2021; pp. 160–165. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).