Deep Neural Network-Based Simulation of Sel’kov Model in Glycolysis: A Comprehensive Analysis
Abstract
:1. Introduction
2. Mathematical Model and Deep Neural Network
Sel’kov Glycolysis Model
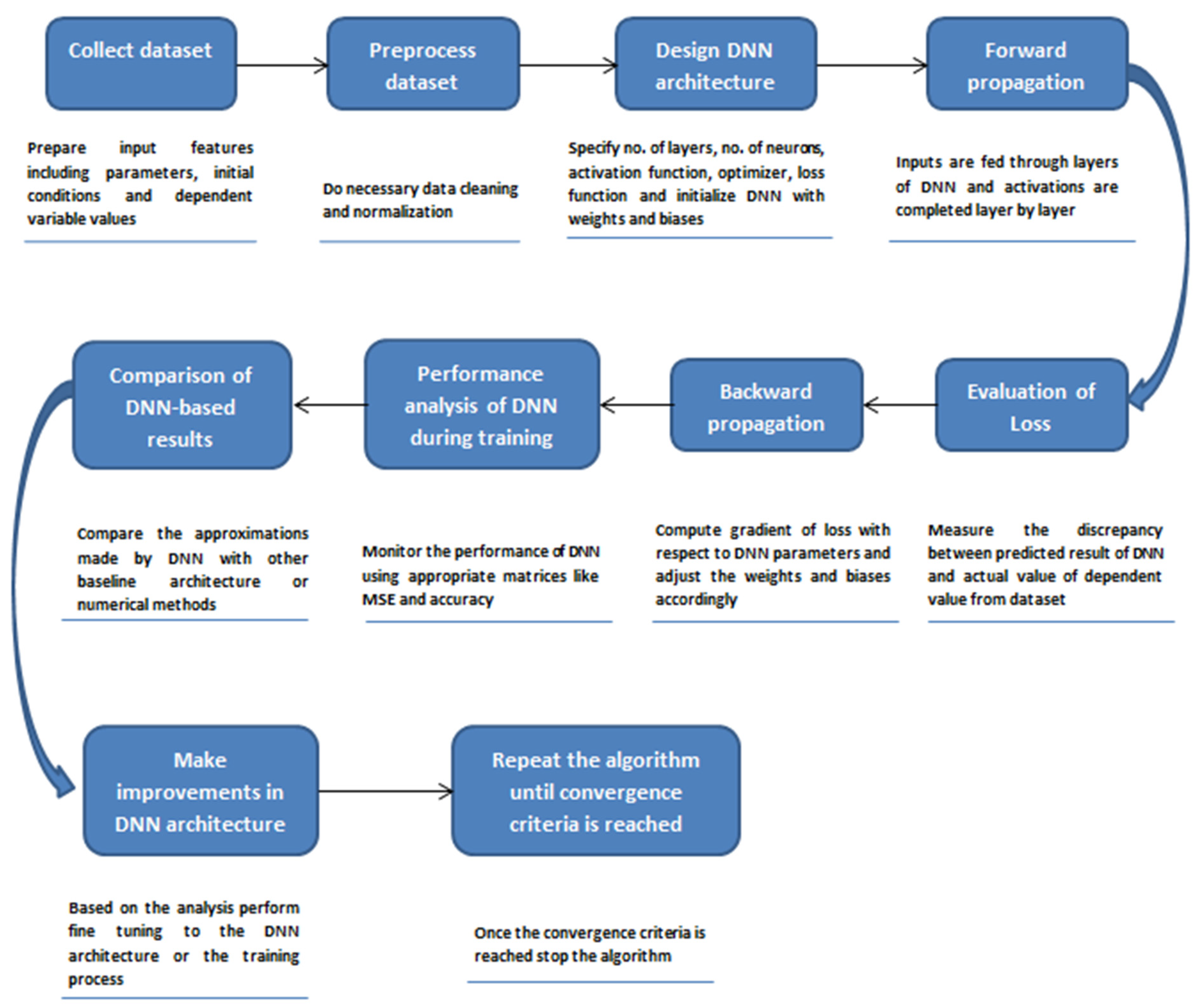
3. Methodology
3.1. Data Preparation
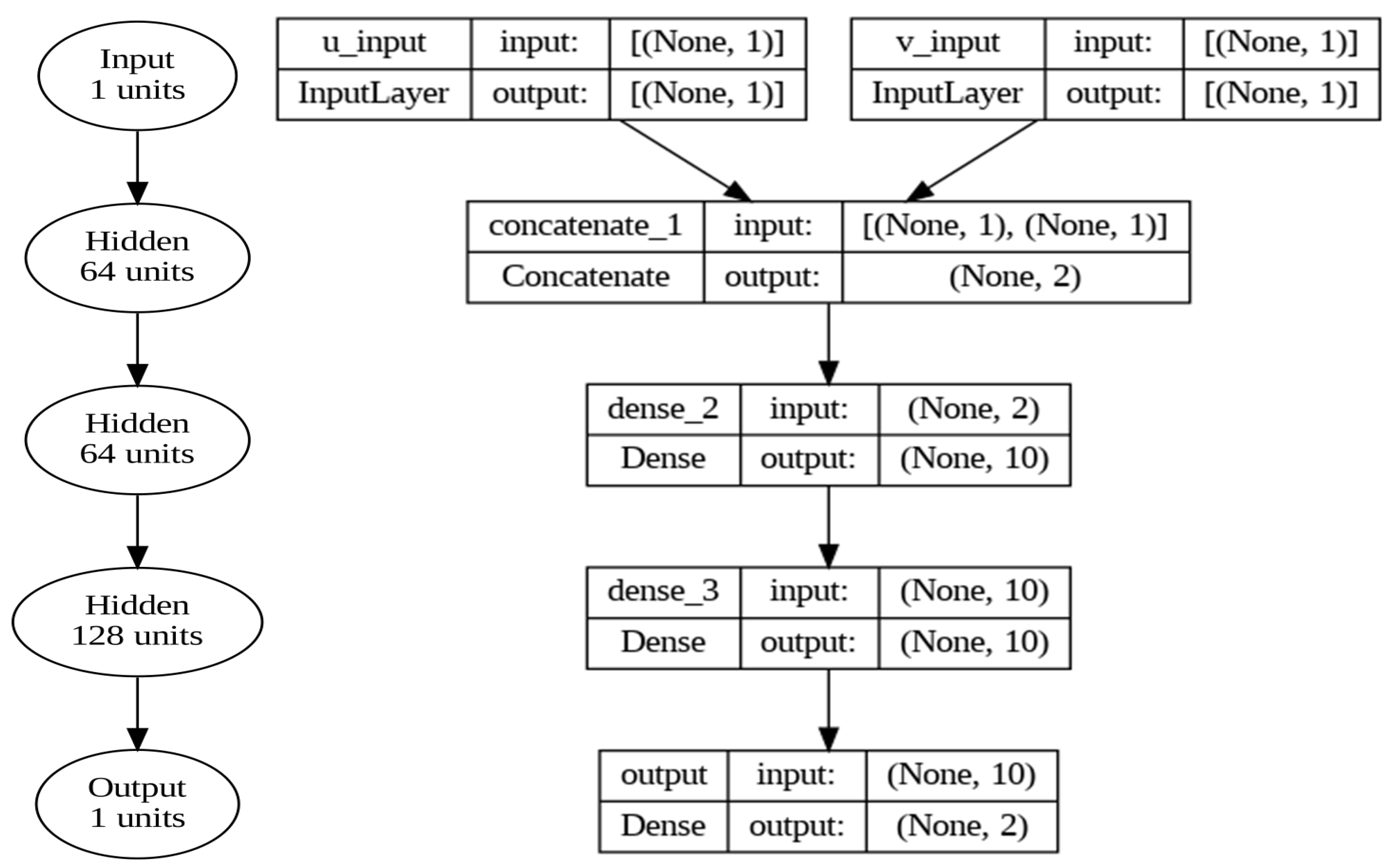
3.2. Neural Network Architecture Design
3.3. Training of the Model
- n = the number of sample points in the dataset,
- = true values of the sample
- = predicted values of the sample
3.4. Analysis of the Model’s Performance
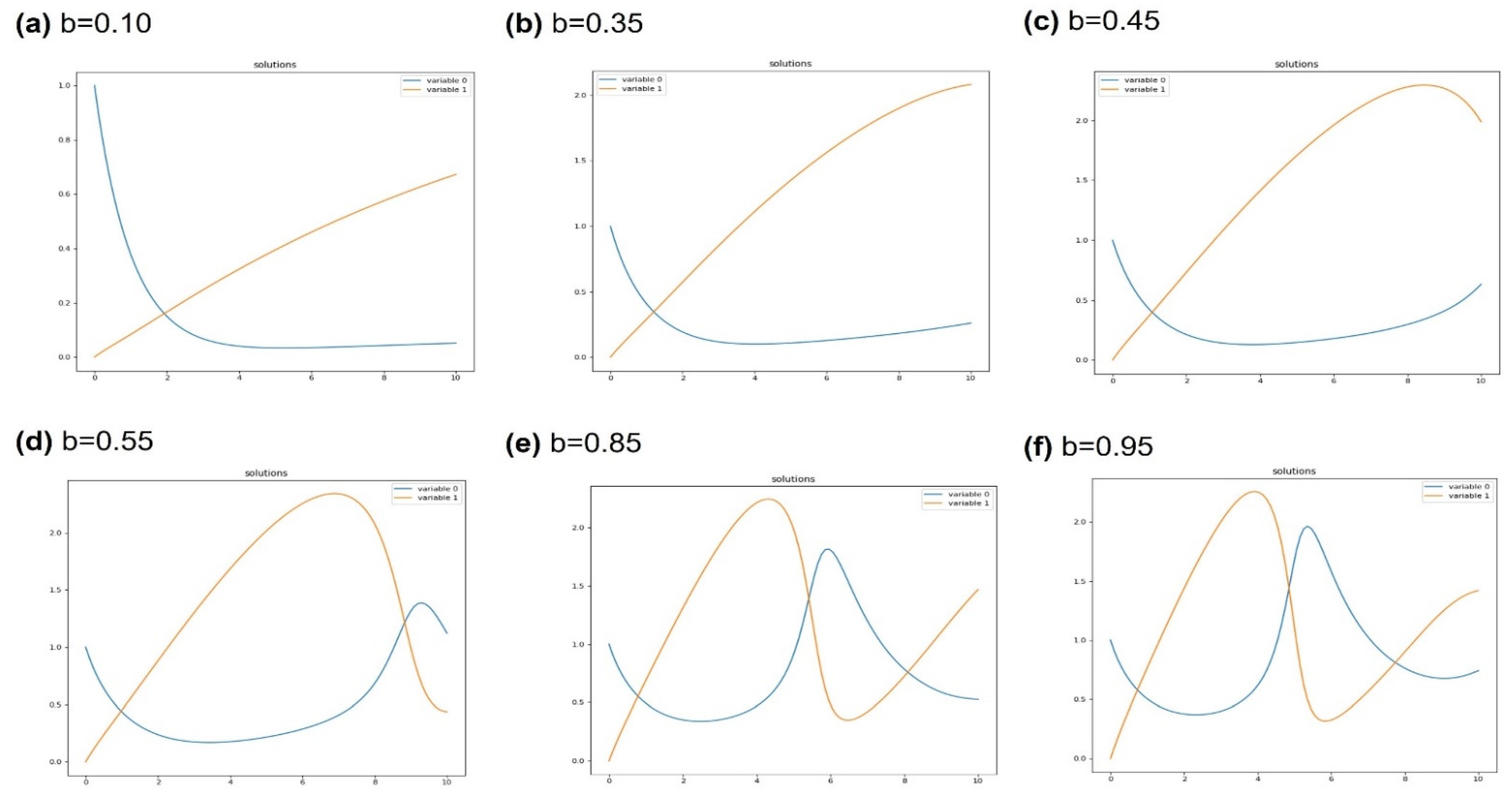
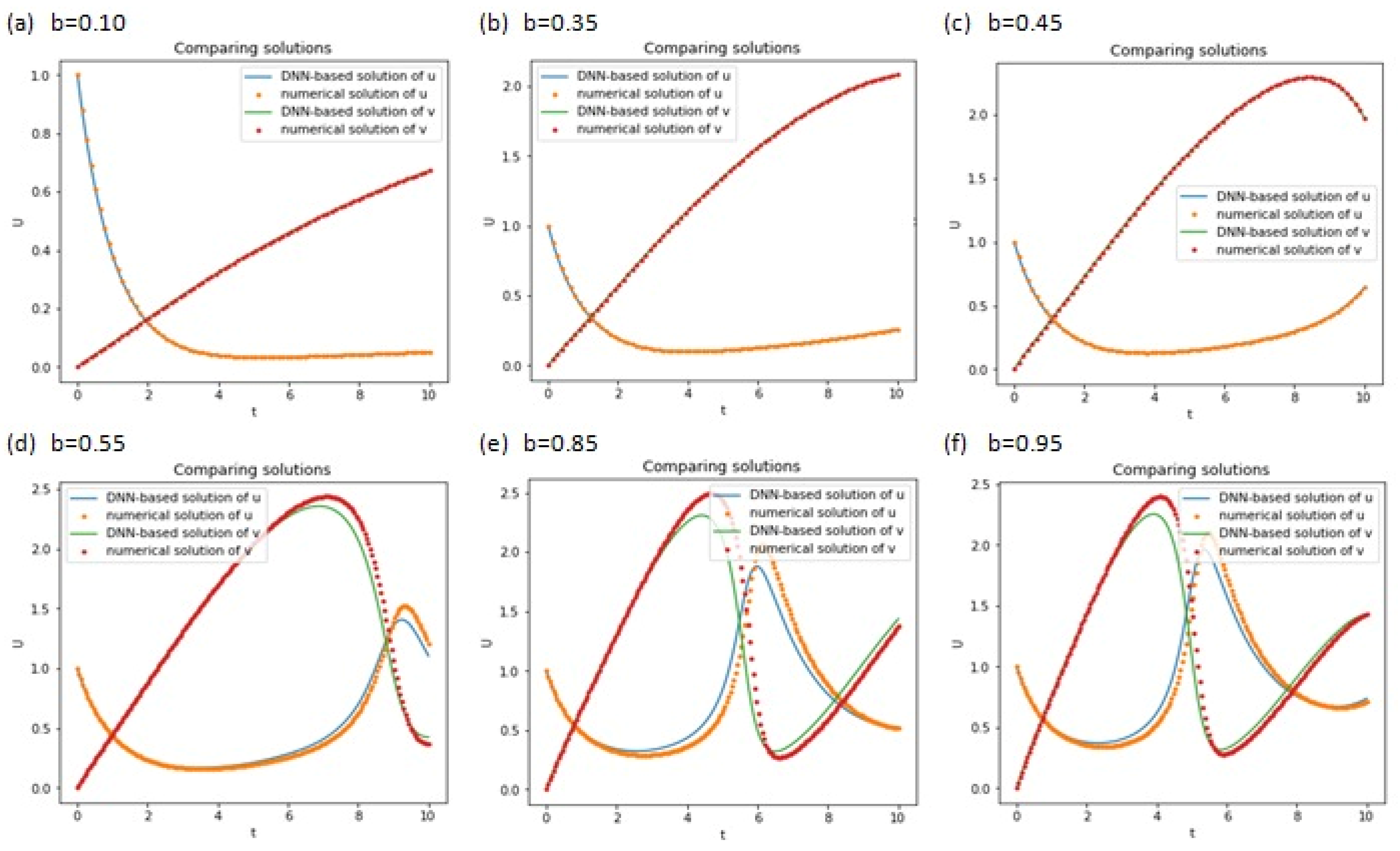
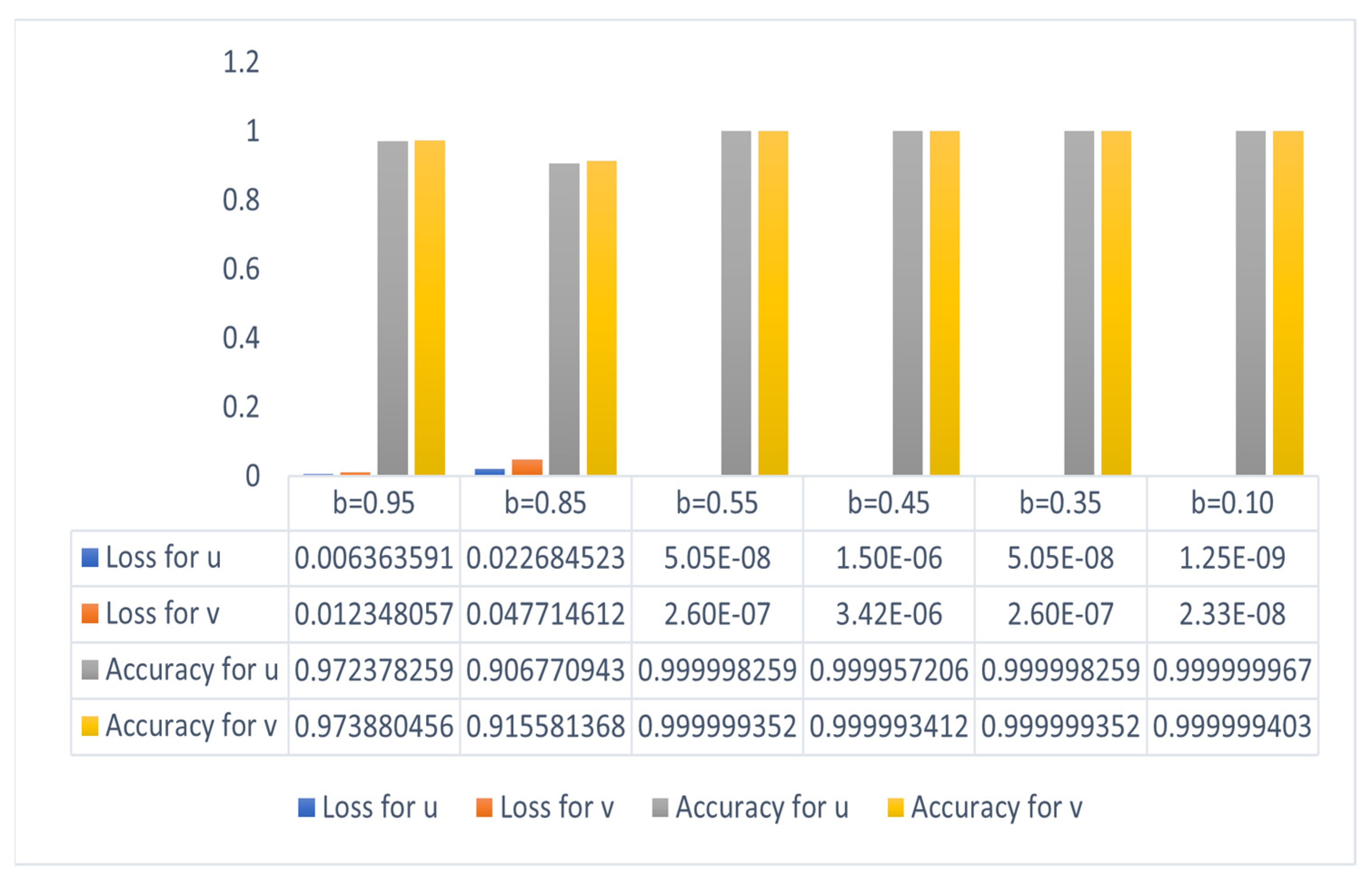
4. Results and Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- El-Safty, A.; Tolba, M.F.; Said, L.A.; Madian, A.H.; Radwan, A.G. A study of the nonlinear dynamics of human behavior and its digital hardware implementation. J. Adv. Res. 2020, 25, 111–123. [Google Scholar] [CrossRef]
- Peters, W.S.; Belenky, V.; Spyrou, K.J. Spyrou. Regulatory use of nonlinear dynamics: An overview. In Contemporary Ideas on Ship Stability; Elsevier: Amsterdam, The Netherlands, 2023; pp. 113–127. [Google Scholar]
- Mahdy, A.M.S. A numerical method for solving the nonlinear equations of Emden-Fowler models. J. Ocean. Eng. Sci. 2022, in press. [CrossRef]
- Yeongjun, L.; Lee, T.-W. Organic synapses for neuromorphic electronics: From brain-inspired computing to sensorimotor nervetronics. Acc. Chem. Res. 2019, 52, 964–974. [Google Scholar]
- Adina, T.M.-T.; Shortland, P. The Nervous System, E-Book: Systems of the Body Series; Elsevier Health Sciences: Amsterdam, The Netherlands, 2022. [Google Scholar]
- Money, S.; Aiyer, R. Musculoskeletal system. Adv. Anesth. Rev. 2023, 341, 152. [Google Scholar] [CrossRef]
- Morris, J.L.; Nilsson, S. The circulatory system. In Comparative Physiology and Evolution of the Autonomic Nervous System; Routledge: London, UK, 2021; pp. 193–246. [Google Scholar]
- Lakrisenko, P.; Stapor, P.; Grein, S.; Paszkowski; Pathirana, D.; Fröhlich, F.; Lines, G.T.; Weindl, D.; Hasenauer, J. Efficient computation of adjoint sensitivities at steady-state in ODE models of biochemical reaction networks. PLOS Comput. Biol. 2023, 19, e1010783. [Google Scholar] [CrossRef]
- Fu, Z.; Xi, S. The effects of heavy metals on human metabolism. Toxicol. Mech. Methods 2020, 30, 167–176. [Google Scholar] [CrossRef]
- Wu, G. Nutrition and metabolism: Foundations for animal growth, development, reproduction, and health. In Recent Advances in Animal Nutrition and Metabolism; Springer: Berlin/Heidelberg, Germany, 2022; pp. 1–24. [Google Scholar]
- Basu, A.; Bhattacharjee, J.K. When Hopf meets saddle: Bifurcations in the diffusive Selkov model for glycolysis. Nonlinear Dyn. 2023, 111, 3781–3795. [Google Scholar] [CrossRef]
- Dhatt, S.; Chaudhury, P. Study of oscillatory dynamics in a Selkov glycolytic model using sensitivity analysis. Indian J. Phys. 2022, 96, 1649–1654. [Google Scholar] [CrossRef]
- Pankratov, A.; Bashkirtseva, I. Stochastic effects in pattern generation processes for the Selkov glycolytic model with diffusion. AIP Conf. Proceeding 2022, 2466, 090018. [Google Scholar]
- Montavon, G.; Samek, W.; Müller, K.-R. Methods for interpreting and understanding deep neural networks. Digit. Signal Process. 2018, 73, 1–15. [Google Scholar] [CrossRef]
- Ul Rahman, J.; Faiza, M.; Akhtar, A.; Sana, D. Mathematical modeling and simulation of biophysics systems using neural network. Int. J. Mod. Phys. B 2023, 2450066. [Google Scholar] [CrossRef]
- Rehman, S.; Akhtar, H.; Ul Rahman, J.; Naveed, A.; Taj, M. Modified Laplace based variational iteration method for the mechanical vibrations and its applications. Acta Mech. Autom. 2022, 16, 98–102. [Google Scholar] [CrossRef]
- Zarnan, J.A.; Hameed, W.M.; Kanbar, A.B. New Numerical Approach for Solution of Nonlinear Differential Equations. J. Hunan Univ. Nat. Sci. 2022, 49, 163–170. [Google Scholar] [CrossRef]
- Kremsner, S.; Steinicke, A.; Szölgyenyi, M. A deep neural network algorithm for semilinear elliptic PDEs with applications in insurance mathematics. Risks 2020, 8, 136. [Google Scholar] [CrossRef]
- Li, Y.; Wei, H.; Han, Z.; Huang, J.; Wang, W. Deep learning-based safety helmet detection in engineering management based on convolutional neural networks. Adv. Civ. Eng. 2020, 2020, 9703560. [Google Scholar] [CrossRef]
- Sahu, A.; Rana, K.P.S.; Kumar, V. An application of deep dual convolutional neural network for enhanced medical image denoising. Med. Biol. Eng. Comput. 2023, 61, 991–1004. [Google Scholar] [CrossRef] [PubMed]
- Pan, G.; Zhang, P.; Chen, A.; Deng, Y.; Zhang, Z.; Lu, H.; Zhu, A.; Zhou, C.; Wu, Y.; Li, S. Aerobic glycolysis in colon cancer is repressed by naringin via the HIF1A pathway. J. Zhejiang Univ. Sci. B 2023, 24, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ji, Y.; Zhao, W.; Niu, H.; Yang, X.; Jiang, X.; Zhang, Y.; Lei, J.; Yang, H.; Chen, R.; et al. Fructose-6-phosphate-2-kinase/fructose-2, 6-bisphosphatase regulates energy metabolism and synthesis of storage products in developing rice endosperm. Plant Sci. 2023, 326, 111503. [Google Scholar] [CrossRef]
- Hsu, S.-B.; Chen, K.-C. Ordinary Differential Equations with Applications; World Scientific: Singapore, 2022; Volume 23. [Google Scholar]
- Tian, Y.; Su, D.; Lauria, S.; Liu, X. Recent advances on loss functions in deep learning for computer vision. Neurocomputing 2022, 497, 129–158. [Google Scholar] [CrossRef]
- Ul Rahman, J.; Ali, A.; Ur Rehman, M.; Kazmi, R. A unit softmax with Laplacian smoothing stochastic gradient descent for deep convolutional neural networks. In Proceedings of the Intelligent Technologies and Applications: Second International Conference, INTAP 2019, Bahawalpur, Pakistan, 6–8 November 2019; Springer: Singapore, 2020. Revised Selected Papers 2. [Google Scholar]
- Chen, F.; Sondak, D.; Protopapas, P.; Mattheakis, M.; Liu, S.; Agarwal, D.; Di Giovanni, M. Neurodiffeq: A python package for solving differential equations with neural networks. J. Open Source Softw. 2020, 5, 1931. [Google Scholar] [CrossRef]
- Rahman, J.; Ul, F.M.; Dianchen Lu, D. Amplifying Sine Unit: An Oscillatory Activation Function for Deep Neural Networks to Recover Nonlinear Oscillations Efficiently. arXiv, 2023; arXiv:2304.09759. [Google Scholar]
- Roy, S.K.; Manna, S.; Dubey, S.R.; Chaudhuri, B.B. LiSHT: Non-parametric linearly scaled hyperbolic tangent activation function for neural networks. In International Conference on Computer Vision and Image Processing, Nagpur, India, 4–6 November 2022; Springer Nature: Cham, Switzerland, 2022. [Google Scholar]
- Qi, J.; Du, J.; Siniscalchi, S.M.; Ma, X.; Lee, C.-H. On mean absolute error for deep neural network based vector-to-vector regression. IEEE Signal Process. Lett. 2020, 27, 1485–1489. [Google Scholar] [CrossRef]
- Tianle, C.; Gao, R.; Hou, J.; Chen, S.; Wang, D.; He, D. A gram-gauss-newton method learning overparameterized deep neural networks for regression problems. arXiv 2019, arXiv:1905.11675. [Google Scholar]
- Yong, H.; Huang, J.; Hua, X.; Zhang, L. Gradient centralization: A new optimization technique for deep neural networks. In Proceedings of the Computer Vision–ECCV 2020: 16th European Conference, Glasgow, UK, 23–28 August 2020; Springer International Publishing: Cham, Switzerland, 2020. Proceedings, Part I 16. [Google Scholar]
- Wright, L.G.; Onodera, T.; Stein, M.M.; Wang, T.; Schachter, D.T.; Hu, Z.; McMahon, P.L. Deep physical neural networks trained with backpropagation. Nature 2022, 601, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Ariff, N.A.M.; Ismail, A.R. Study of adam and adamax optimizers on alexnet architecture for voice biometric authentication system. In Proceedings of the 2023 17th International Conference on Ubiquitous Information Management and Communication (IMCOM), Seoul, Republic of Korea, 3–5 January 2023; IEEE: Piscataway, NJ, USA, 2023. [Google Scholar]
- Legaard, C.M.; Schranz, T.; Schweiger, G.; Drgoňa, J.; Falay, B.; Gomes, C.; Iosifidis, A.; Abkar, M.; Larsen, P.G. Constructing Neural Network Based Models for Simulating Dynamical Systems. ACM Comput. Surv. 2023, 55, 236. [Google Scholar] [CrossRef]
- Hong, Z.; Lu, Y.; Liu, B.; Ran, C.; Lei, X.; Wang, M.; Wu, S.; Yang, Y.; Wu, H. Glycolysis, a new mechanism of oleuropein against liver tumor. Phytomedicine 2023, 114, 154770. [Google Scholar] [CrossRef] [PubMed]
- Jamshaid Ul, R.; Makhdoom, F.; Lu, D. ASU-CNN: An Efficient Deep Architecture for Image Classification and Feature Visualizations. arXiv 2023, arXiv:2305.19146. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ul Rahman, J.; Danish, S.; Lu, D. Deep Neural Network-Based Simulation of Sel’kov Model in Glycolysis: A Comprehensive Analysis. Mathematics 2023, 11, 3216. https://doi.org/10.3390/math11143216
Ul Rahman J, Danish S, Lu D. Deep Neural Network-Based Simulation of Sel’kov Model in Glycolysis: A Comprehensive Analysis. Mathematics. 2023; 11(14):3216. https://doi.org/10.3390/math11143216
Chicago/Turabian StyleUl Rahman, Jamshaid, Sana Danish, and Dianchen Lu. 2023. "Deep Neural Network-Based Simulation of Sel’kov Model in Glycolysis: A Comprehensive Analysis" Mathematics 11, no. 14: 3216. https://doi.org/10.3390/math11143216
APA StyleUl Rahman, J., Danish, S., & Lu, D. (2023). Deep Neural Network-Based Simulation of Sel’kov Model in Glycolysis: A Comprehensive Analysis. Mathematics, 11(14), 3216. https://doi.org/10.3390/math11143216





