Dual Space Latent Representation Learning for Image Representation
Abstract
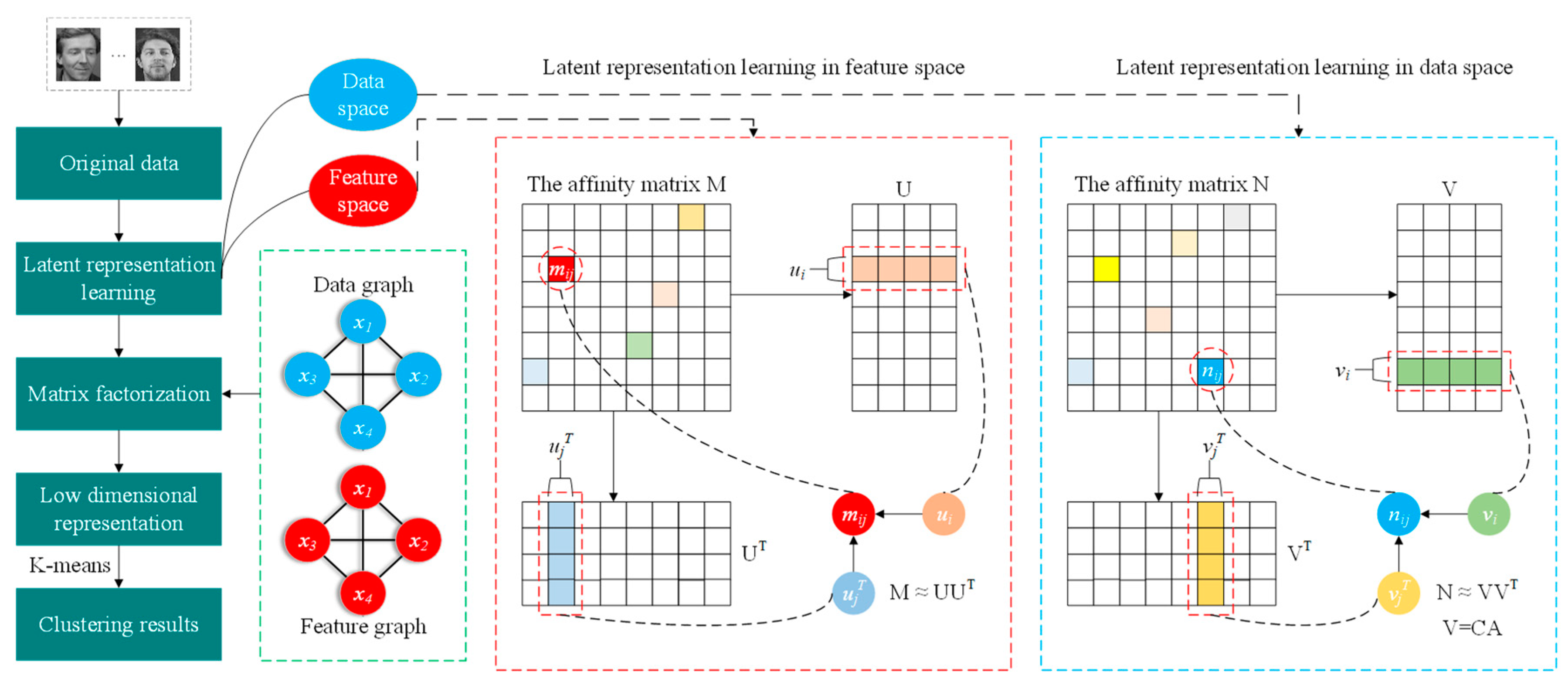
:1. Introduction
- (1)
- The dual latent representation mechanism is embedded into the semi-supervised NMF framework by the affinity matrix to explicitly exploit global interconnection information in dual space, which can reduce the adverse impacts caused by noise.
- (2)
- To steadily describe local data structures, the dual graph is introduced into latent representation learning to further fully investigate the coherent information structure in dual space.
- (3)
- To achieve a sparse representation of matrix factorization, the ℓ2,1-norm is incorporated into the basis matrix in the proposed framework, which can simplify the measurement process and improve the clustering performance.
2. Related Work
2.1. CNMF
2.2. DNMF
2.3. SODNMF
2.4. LRLMR
2.5. Latent Representation
3. Proposed Method
3.1. Dual Space Latent Representation Learning and Dual Graph Regularization
3.2. Objective Function
3.3. Optimization
3.4. Convergence Analysis
4. Experiments
4.1. Results on the Synthetic Dataset
4.2. Results on Public Datasets Benchmarks
4.2.1. Datasets
4.2.2. Compared Algorithms
- PCA [47]: Principal component analysis generates a low-rank subspace with the major information of the data by the projection of the direction of maximum variance.
- K-Multiple Means (KMM) [48]: It is an improved K-means by introducing the local centroids into K-means.
- LRLMR [26]: Latent representation learning is embedded in feature selection to extract global interconnection information between data instances to guide feature selection.
- CNMF [21]: Semi-supervision is embedded into NMF as a robust constraint to improve the algorithm performance by a small amount of label data.
- DNMF [18]: The local manifold information of dual space is preserved by constructing a dual graph regularization.
- DSNMF [20]: The local manifold structures of dual space are preserved and retain the sparsity simultaneously.
- NMFAN [30]: The local manifold information is exploited by constructing an adaptive graph regularization term to obtain the optimal neighborhood graph.
- EWRNMF [33]: An adaptive weight NMF that guides the matrix factorization by assigning adaptive weights to each data instance.
- SODNMF [25]: A semi-supervised NMF with dual graph regularization terms, sparse constraints and biorthogonal constraints.
4.2.3. Experimental Settings
4.2.4. Performance
- (1)
- GNMF, DNMF, and DSNMF are three NMF variants with graph regularization terms. They outperformed LRLMR, KMM, and PCA on most datasets except for Lung_dis and ORL. The reason is that the graph models are constructed to retain the local manifold structure, which can achieve promising clustering results.
- (2)
- CNMF is superior to GNMF on the ORL, Yale32, and Yale64 datasets, which reflects the merits of semi-supervised NMF in improving clustering accuracy with less label information.
- (3)
- LRLMR achieves better performance than PCA, KMM, and CNMF on most test datasets since LRLMR exploits interconnection information by latent representation learning, which can further enhance the discriminative of the model.
- (4)
- NMFAN and EWRNMF are two adaptive methods that construct an adaptive graph and adaptive weights, respectively. Unsatisfactory results are achieved on most datasets because they fail to fully explore interconnection information and label information.
- (5)
- Compared with other methods, DLRGNMF achieves the best performance, especially on COIL20; the increases in ACC values are 19.09%, 21.25%, 16.65%, 19.98%, 6.91%, 2.61%, 1.83%, 19.37%, 17.90%, and 1.72% with respect to PCA, KMM, LRLMR, CNMF, GNMF, DNMF, DSNMF, NMFAN, EWRNMF, and SODNMF. DLRGNMF embeds dual space latent representation learning to guide matrix decomposition and reduce noise. Therefore, it learns more information about interconnection and achieves the best clustering results.
- (6)
- In terms of running time, DLRGNMF is a little slower than some compared methods on some datasets because latent representation learning inevitably entails more operations, whereas it outperforms SODNMF on Jaffe50, UMIST, and warpPIE10P datasets due to rapid convergence.
4.2.5. Intuitive Presentation
4.2.6. Ablation Study
4.2.7. Convergence Study
4.2.8. Parameter Sensitivity Experiment
4.2.9. Noise Test
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Roweis, S.; Saul, L. Nonlinear dimensionality reduction by locally linear embedding. Science 2000, 290, 2323–2326. [Google Scholar] [CrossRef]
- Lee, D.; Seung, H. Algorithms for non-negative matrix factorization. Adv. Neural Inf. Process. Syst. 2000, 13, 556–562. [Google Scholar]
- Lee, D.; Seung, H. Learning the parts of objects by non-negative matrix factorization. Nature 1999, 401, 788–791. [Google Scholar]
- Xu, Z.; King, I.; Lyu, M.; Jin, R. Discriminative semi-supervised feature selection via manifold regularization. IEEE Trans. Neural Netw. 2010, 21, 1033–1047. [Google Scholar]
- Lipovetsky, S. PCA and SVD with nonnegative loadings. Pattern Recognit. 2009, 42, 68–76. [Google Scholar] [CrossRef]
- Sandler, R.; Lindenbaum, M. Nonnegative matrix factorization with earth mover’s distance metric for Image analysis. IEEE Trans. Pattern Anal. Mach. Intell. 2011, 33, 1590–1602. [Google Scholar]
- Tu, D.; Chen, L.; Lv, M.; Shi, H.; Chen, G. Hierarchical online NMF for detecting and tracking topic hierarchies in a text stream. Pattern Recognit. 2018, 76, 203–214. [Google Scholar] [CrossRef]
- Chen, W.; Zhao, Y.; Pan, B.; Chen, B. Supervised kernel nonnegative matrix factorization for face recognition. Neurocomputing 2016, 205, 165–181. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, G.; Wang, K.; Liu, H.; Yin, Y. Robust multi-feature collective non-negative matrix factorization for ECG biometrics. Pattern Recognit. 2022, 123, 108376. [Google Scholar] [CrossRef]
- Zheng, Z.; Yang, J.; Zhu, Y. Initialization enhancer for non-negative matrix factorization. Eng. Appl. Artif. Intell. 2007, 20, 101–110. [Google Scholar] [CrossRef]
- Zdunek, R.; Sadowski, T. Segmented convex-hull algorithms for near-separable NMF and NTF. Neurocomputing 2019, 331, 150–164. [Google Scholar] [CrossRef]
- Chen, M.; Gong, M.; Li, X. Feature Weighted Non-Negative Matrix Factorization. IEEE Trans. Cybern. 2021, 53, 1093–1105. [Google Scholar]
- Jacob, Y.; Denoyer, L.; Gallinari, P. Learning latent representations of nodes for classifying in heterogeneous social networks. In Proceedings of the 7th ACM International Conference on Web Search and Data Mining, New York, NY, USA, 24 February 2014; pp. 373–382. [Google Scholar]
- Samaria, F.; Harter, A. Parameterisation of a stochastic model for human face identification. In Proceedings of the 2nd IEEE Workshop on Applications of Computer Vision, Sarasota, FL, USA, 1 January 1994; pp. 138–142. [Google Scholar]
- Huang, J.; Nie, F.; Huang, H.; Ding, C. Robust manifold nonnegative matrix factorization. ACM Trans. Knowl. Disc. Data 2014, 8, 1–21. [Google Scholar] [CrossRef]
- Cai, D.; He, X.; Han, J.; Huang, T.S. Graph regularized nonnegative matrix factorization for data representation. IEEE Trans. Pattern Anal. Mach. Intell. 2011, 33, 1548–1560. [Google Scholar]
- Liu, H.; Yang, Z.; Yang, J.; Wu, Z.; Li, X. Local coordinate concept factorization for image representation. IEEE Trans. Neural Netw. Learn. Syst. 2014, 25, 1071–1082. [Google Scholar]
- Shang, F.; Jiao, L.; Wang, F. Graph dual regularization nonnegative matrix factorization for co-clustering. Pattern Recognit. 2012, 45, 2237–2250. [Google Scholar] [CrossRef]
- Hoyer, P. Non-negative matrix factorization with sparseness constraints. J. Mach. Learn. Res. 2004, 5, 1457–1469. [Google Scholar]
- Meng, Y.; Shang, R.; Jiao, L.; Zhang, W.; Yuan, Y.; Yang, S. Feature selection based dual-graph sparse non-negative matrix factorization for local discriminative clustering. Neurocomputing 2018, 290, 87–99. [Google Scholar] [CrossRef]
- Liu, H.; Wu, Z.; Cai, D.; Huang, T. Constrained nonnegative matrix factorization for image representation. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 1299–1311. [Google Scholar] [CrossRef]
- Peng, S.; Ser, W.; Chen, B.; Lin, Z. Robust semi-supervised nonnegative matrix factorization for image clustering. Pattern Recognit. 2021, 111, 107683. [Google Scholar] [CrossRef]
- Sun, J.; Wang, Z.; Sun, F.; Li, H. Sparse dual graph-regularized NMF for image co-clustering. Neurocomputing 2018, 316, 156–165. [Google Scholar] [CrossRef]
- Babaee, M.; Tsoukalas, S.; Babaee, M.; Rigoll, G.; Datcu, M. Discriminative non-negative matrix factorization for dimensionality reduction. Neurocomputing 2016, 173, 212–223. [Google Scholar] [CrossRef]
- Meng, Y.; Shang, R.; Jiao, L.; Zhang, W. Dual-graph regularized non-negative matrix factorization with sparse and orthogonal constraints. Eng. Appl. Artif. Intell. 2018, 69, 24–35. [Google Scholar] [CrossRef]
- Tang, C.; Bian, M.; Liu, X.; Li, M.; Zhou, H.; Wang, P.; Yin, H. Unsupervised feature selection via latent representation learning and manifold regularization. Neural Netw. 2019, 117, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Ding, D.; Yang, X.; Xia, F.; Ma, T.; Liu, H.; Tang, C. Unsupervised feature selection via adaptive hypergraph regularized latent representation learning. Neurocomputing 2020, 378, 79–97. [Google Scholar] [CrossRef]
- Shang, R.; Wang, L.; Shang, F.; Jiao, L.; Li, Y. Dual space latent representation learning for unsupervised feature selection. Pattern Recognit. 2021, 114, 107873. [Google Scholar] [CrossRef]
- Ye, J.; Jin, Z. Feature selection for adaptive dual-graph regularized concept factorization for data representation. Neural Process. Lett. 2017, 45, 667–668. [Google Scholar] [CrossRef]
- Huang, S.; Xu, Z.; Wang, F. Nonnegative matrix factorization with adaptive neighbors. In Proceedings of the 2017 International Joint Conference on Neural Networks (IJCNN), Anchorage, AK, USA, 14 May 2017; pp. 486–493. [Google Scholar]
- Luo, P.; Peng, J.; Guan, Z.; Fan, J. Dual-regularized multi-view non-negative matrix factorization. Neurocomputing 2018, 294, 1–11. [Google Scholar] [CrossRef]
- Xing, Z.; Wen, M.; Peng, J.; Feng, J. Discriminative semi-supervised non-negative matrix factorization for data clustering. Eng. Appl. Artif. Intell. 2021, 103, 104289. [Google Scholar] [CrossRef]
- Shen, T.; Li, J.; Tong, C.; He, Q.; Li, C.; Yao, Y.; Teng, Y. Adaptive weighted nonnegative matrix factorization for robust feature representation. arXiv 2022, arXiv:2206.03020. [Google Scholar] [CrossRef]
- Li, H.; Gao, Y.; Liu, J.; Zhang, J.; Li, C. Semi-supervised graph regularized nonnegative matrix factorization with local coordinate for image representation. Signal Process. Image Commun. 2022, 102, 116589. [Google Scholar] [CrossRef]
- Chavoshinejad, J.; Seyedi, S.; Tab, F.; Salahian, N. Self-supervised semi-supervised nonnegative matrix factorization for data clustering. Pattern Recognit. 2023, 137, 109282. [Google Scholar] [CrossRef]
- Li, S.; Li, W.; Lu, H.; Li, Y. Semi-supervised non-negative matrix tri-factorization with adaptive neighbors and block-diagonal learning. Eng. Appl. Artif. Intell. 2023, 121, 106043. [Google Scholar] [CrossRef]
- Lu, M.; Zhao, X.; Zhang, L.; Li, F. Semi-supervised concept factorization for document clustering. Inf. Sci. 2016, 331, 86–98. [Google Scholar] [CrossRef]
- Feng, X.; Jiao, Y.; Lv, C.; Zhou, D. Label consistent semi-supervised non-negative matrix factorization for maintenance activities identification. Eng. Appl. Artif. Intell. 2016, 52, 161–167. [Google Scholar] [CrossRef]
- Kuang, D.; Ding, C.; Park, H. Symmetric nonnegative matrix factorization for graph clustering. In Proceedings of the 2012 SIAM International Conference on Data Mining, Anaheim, CA, USA, 26–28 April 2012; pp. 106–117. [Google Scholar]
- Tang, L.; Liu, H. Relational learning via latent social dimensions. In Proceedings of the 15th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Paris, France, 28 June–1 July 2009; pp. 817–826. [Google Scholar]
- Cui, J.; Zhu, Q.; Wang, D.; Li, Z. Learning robust latent representation for discriminative regression. Pattern Recognit. Lett. 2018, 117, 193–200. [Google Scholar] [CrossRef]
- Li, H.; Zhang, J.; Liu, J. Class-driven concept factorization for image representation. Neurocomputing 2016, 190, 197–208. [Google Scholar] [CrossRef]
- Shang, R.; Zhang, Z.; Jiao, L.; Liu, C.; Li, Y. Self-representation based dual-graph regularized feature selection clustering. Neurocomputing 2016, 171, 1242–1253. [Google Scholar] [CrossRef]
- Shang, R.; Wang, W.; Stolkin, R.; Jiao, L. Subspace learning-based graph regularized feature selection. Knowl. Based Syst. 2016, 112, 152–165. [Google Scholar] [CrossRef]
- Nie, F.; Wu, D.; Wang, R.; Li, X. Self-weighted clustering with adaptive neighbors. IEEE Trans. Neural Netw. Learn. Syst. 2020, 31, 3428–3441. [Google Scholar] [CrossRef]
- Shang, R.; Zhang, Z.; Jiao, L.; Wang, W.; Yang, S. Global discriminative-based nonnegative spectral clustering. Pattern Recognit. 2016, 55, 172–182. [Google Scholar] [CrossRef]
- Gewers, F.; Ferreira, G.; Arruda, H.; Silva, F.; Comin, C.; Amancio, D.; Costa, L. Principal component analysis: A natural approach to data exploration. ACM Comput. Surv. 2021, 54, 1–34. [Google Scholar] [CrossRef]
- Nie, F.; Wang, C.; Li, X. K-multiple-means: A multiple-means clustering method with specified K clusters. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, KDD 2019, Anchorage, AK, USA, 4–8 August 2019; pp. 959–967. [Google Scholar]
- Sun, J.; Gai, X.; Sun, F.; Hong, R. Dual graph-regularized constrained nonnegative matrix factorization for image clustering. KSII Trans. Internet Inf. Syst. 2017, 11, 2607–2627. [Google Scholar] [CrossRef]


















| Methods | Semi-Supervised | Sparse Representation | Local Data Structure | Interconnection Information |
|---|---|---|---|---|
| NMF with sparseness constraints (2004) [19] | √ | × | × | × |
| Constrained NMF (CNMF) (2012) [21] | √ | × | × | × |
| Graph regularized NMF (GNMF) (2011) [16] | × | × | √ | × |
| Graph dual regularization NMF (DNMF) (2012) [18] | × | × | √ | × |
| Robust manifold NMF (RMNMF) (2014) [15] | × | × | √ | × |
| Local coordinate concept factorization (LCF) (2014) [17] | × | √ | √ | × |
| Discriminative NMF (2016) [24] | √ | × | × | × |
| Adaptive dual-graph regularized CF with FS (ADGCFFS) (2017) [29] | × | × | √ | × |
| NMF with adaptive neighbors (NMFAN) (2017) [30] | × | × | √ | × |
| Dual-graph sparse NMF (DSNMF) (2018) [20] | × | √ | √ | × |
| Dual-regularized multi-view NMF (DMvNM) (2018) [31] | × | × | √ | × |
| Dual-graph regularized NMF with sparse and orthogonal constraints (SODNMF) (2018) [25] | √ | √ | √ | × |
| Correntropy-based semi-supervised NMF (CSNMF) (2021) [22] | × | × | √ | × |
| Discriminative semi-supervised NMF (DSSNMF) (2021) [32] | √ | × | × | × |
| Feature weighted NMF (FNMF) (2021) [12] | × | × | √ | × |
| Entropy weighted robust NMF (EWRNMF) (2022) [33] | × | × | × | × |
| Semi-supervised graph regularized NMF with local coordinate (SGLNMF) (2022) [34] | √ | √ | √ | × |
| Self-supervised semi-supervised NMF (S4NMF) (2023) [35] | √ | × | × | × |
| Semi-supervised NMF with adaptive neighbors and block-diagonal (ABNMTF) (2023) [36] | √ | × | √ | × |
| DLRGNMF (this study) | √ | √ | √ | √ |
| Algorithm 1 The optimization process of DLRGNMF |
|---|
| 1: Input: Data matrix , class number of samples c, neighborhood size k, balance parameters α, β, θ and γ, the maximum number of iterations , and the ratio of training samples . |
| 2: Initialization: Normalize the data matrix X, generate U, R, and A, and pick up per% as the label information from the original data to construct matrix C, iteration time t = 0; |
| 3: Construct the dual space latent representation learning; |
| 4: Construct the dual-graph regularized model; |
| 5: while not converged do 6: Update U using Equation (22); 7: Update R using Equation (23); 8: Update A using Equation (24); 9: Update C using Equation (25); 10: Update Q using Equation (17); 11: Update t by: t = t + 1, t ≤ NIter; 12: end while |
| 13: Output: The label constraint matrix C, the basis matrix U, the diagonal scaling matrix R, and the label auxiliary matrix A. |
| Datasets | Samples | Features | Classes | Type |
|---|---|---|---|---|
| COIL20 | 1440 | 1024 | 20 | Object image |
| JAFFE50 | 213 | 1024 | 10 | Face image |
| Lung_dis | 73 | 325 | 7 | Biological |
| ORL | 400 | 1024 | 40 | Face image |
| Soybean | 47 | 35 | 4 | Digital image |
| UMIST | 575 | 1024 | 20 | Face image |
| PIE10P | 210 | 2420 | 10 | Face image |
| Yale32 | 165 | 1024 | 15 | Face image |
| Yale64 | 165 | 4096 | 15 | Face image |
| Methods | COIL20 | JAFFE50 | Lung_dis | ORL | Soybean | UMIST | PIE10P | Yale32 | Yale64 |
|---|---|---|---|---|---|---|---|---|---|
| PCA | 64.54 ± 3.00 | 89.60 ± 1.25 | 82.53 ± 5.85 | 55.15 ± 2.38 | 73.09 ± 1.04 | 40.03 ± 1.44 | 29.19 ± 1.97 | 42.18 ± 2.98 | 52.94 ± 3.11 |
| KMM | 62.82 ± 3.09 | 85.92 ± 5.29 | 78.90 ± 5.34 | 52.11 ± 2.47 | 76.28 ± 14.83 | 41.32 ± 1.98 | 31.90 ± 1.95 | 39.03 ± 3.04 | 47.70 ± 2.61 |
| LRLMR | 66.49 ± 2.89 | 84.08 ± 4.52 | 82.19 ± 4.58 | 54.43 ± 3.04 | 89.15 ± 4.68 | 55.48 ± 2.92 | 48.74 ± 3.12 | 41.09 ± 2.85 | 50.94 ± 3.33 |
| CNMF | 63.83 ± 2.53 | 81.13 ± 6.75 | 51.99 ± 5.63 | 62.57 ± 2.92 | 80.96 ± 7.22 | 40.18 ± 1.78 | 41.52 ± 2.58 | 43.21 ± 3.00 | 54.36 ± 4.11 |
| GNMF | 74.26 ± 4.31 | 86.20 ± 4.74 | 68.22 ± 2.90 | 55.13 ± 2.33 | 88.72 ± 2.59 | 53.71 ± 4.09 | 78.14 ± 5.43 | 40.21 ± 1.75 | 51.52 ± 1.96 |
| DNMF | 77.69 ± 3.31 | 91.03 ± 5.57 | 68.49 ± 6.05 | 59.44 ± 1.73 | 89.26 ± 0.48 | 57.26 ± 4.29 | 77.88 ± 3.40 | 40.70 ± 2.59 | 53.52 ± 3.15 |
| DSNMF | 78.31 ± 2.50 | 90.85 ± 5.06 | 71.03 ± 0.92 | 58.76 ± 2.21 | 92.13 ± 1.96 | 56.36 ± 3.11 | 78.36 ± 3.70 | 42.58 ± 2.51 | 52.24 ± 2.04 |
| NMFAN | 64.32 ± 3.82 | 83.85 ± 4.30 | 49.86 ± 4.58 | 54.69 ± 2.46 | 82.34 ± 8.26 | 41.36 ± 2.13 | 43.17 ± 3.38 | 41.39 ± 3.29 | 51.79 ± 3.54 |
| EWRNMF | 65.49 ± 2.72 | 83.22 ± 4.95 | 49.25 ± 7.63 | 53.09 ± 2.65 | 80.32 ± 7.46 | 41.37 ± 2.61 | 40.69 ± 2.60 | 42.27 ± 2.27 | 52.42 ± 3.22 |
| SODNMF | 78.40 ± 3.61 | 92.51 ± 4.58 | 73.56 ± 5.03 | 64.04 ± 2.18 | 94.15 ± 5.11 | 56.73 ± 5.54 | 79.40 ± 3.78 | 43.24 ± 1.72 | 55.36 ± 2.74 |
| DLRGNMF | 79.77 ± 2.89 | 93.33 ± 4.33 | 82.67 ± 4.06 | 65.04 ± 2.66 | 94.79 ± 5.37 | 58.23 ± 4.85 | 80.05 ± 4.17 | 45.12 ± 2.69 | 56.33 ± 2.23 |
| Methods | COIL20 | JAFFE50 | Lung_dis | ORL | Soybean | UMIST | PIE10P | Yale32 | Yale64 |
|---|---|---|---|---|---|---|---|---|---|
| PCA | 74.96 ± 1.45 | 86.85 ± 1.26 | 75.16 ± 3.72 | 73.42 ± 1.40 | 71.22 ± 0.17 | 59.78 ± 1.08 | 31.17 ± 2.47 | 47.26 ± 2.61 | 56.71 ± 2.69 |
| KMM | 74.04 ± 1.60 | 85.40 ± 3.60 | 72.04 ± 4.59 | 71.62 ± 1.38 | 68.74 ± 17.40 | 60.63 ± 1.38 | 36.14 ± 3.89 | 44.51 ± 2.89 | 51.88 ± 2.48 |
| LRLMR | 75.29 ± 1.15 | 83.19 ± 2.58 | 74.86 ± 4.47 | 73.25 ± 1.69 | 85.38 ± 6.30 | 67.77 ± 1.78 | 60.45 ± 3.10 | 45.03 ± 2.28 | 54.71 ± 2.01 |
| CNMF | 73.13 ± 1.53 | 80.57 ± 3.68 | 47.15 ± 4.59 | 72.64 ± 1.83 | 77.01 ± 7.28 | 58.40 ± 1.52 | 49.74 ± 3.23 | 49.40 ± 2.38 | 58.60 ± 3.41 |
| GNMF | 85.04 ± 2.41 | 83.31 ± 3.57 | 66.23 ± 1.74 | 73.38 ± 1.37 | 80.74 ± 2.49 | 68.81 ± 2.27 | 82.90 ± 3.19 | 45.52 ± 1.57 | 54.27 ± 1.53 |
| DNMF | 88.77 ± 1.33 | 91.14 ± 2.90 | 63.52 ± 7.34 | 74.19 ± 1.23 | 81.30 ± 0.86 | 74.86 ± 2.09 | 83.14 ± 1.58 | 44.88 ± 2.47 | 55.28 ± 1.79 |
| DSNMF | 88.80 ± 1.30 | 90.77 ± 1.98 | 67.65 ± 1.25 | 74.26 ± 1.20 | 84.95 ± 3.07 | 74.36 ± 1.95 | 82.59 ± 2.70 | 46.43 ± 1.84 | 54.39 ± 1.87 |
| NMFAN | 73.31 ± 2.04 | 82.08 ± 2.83 | 39.29 ± 4.19 | 73.61 ± 1.39 | 77.48 ± 7.57 | 59.65 ± 1.66 | 50.32 ± 2.62 | 45.46 ± 2.60 | 55.37 ± 3.10 |
| EWRNMF | 74.53 ± 1.95 | 80.85 ± 3.69 | 43.86 ± 7.09 | 71.89 ± 1.43 | 75.38 ± 5.43 | 60.52 ± 1.62 | 46.67 ± 2.69 | 47.15 ± 1.57 | 54.77 ± 2.87 |
| SODNMF | 88.79 ± 1.34 | 91.78 ± 2.24 | 69.72 ± 5.11 | 77.65 ± 1.45 | 89.35 ± 8.45 | 75.46 ± 2.52 | 83.42 ± 2.08 | 48.95 ± 1.57 | 57.52 ± 2.14 |
| DLRGNMF | 89.34 ± 0.98 | 92.13 ± 2.09 | 75.22 ± 2.28 | 78.37 ± 1.70 | 90.94 ± 8.46 | 75.67 ± 3.10 | 83.89 ± 2.26 | 50.28 ± 2.25 | 57.97 ± 1.97 |
| Methods | COIL20 | Jaffe50 | Lung_dis | ORL | Soybean | UMIST | PIE10P | Yale32 | Yale64 |
|---|---|---|---|---|---|---|---|---|---|
| PCA | 0.2064 | 0.0164 | 0.0218 | 0.0671 | 0.0044 | 0.0744 | 0.0320 | 0.0165 | 0.0233 |
| KMM | 4.6838 | 1.4697 | 0.9710 | 1.8979 | 0.0108 | 2.7622 | 8.8967 | 2.1180 | 26.5506 |
| LRLMR | 7.3728 | 0.7337 | 0.0119 | 5.8150 | 0.0009 | 1.2146 | 15.0992 | 2.7130 | 129.4183 |
| CNMF | 42.4159 | 2.5530 | 0.1713 | 4.9734 | 0.0118 | 8.2539 | 10.0721 | 2.1764 | 25.4597 |
| GNMF | 0.8345 | 0.0855 | 0.0444 | 0.4206 | 0.0156 | 0.3292 | 0.1360 | 0.0963 | 0.3016 |
| DNMF | 5.6850 | 0.6785 | 0.0845 | 1.2101 | 0.0071 | 1.6707 | 2.3783 | 0.6355 | 6.3090 |
| DSNMF | 5.7917 | 0.7314 | 0.0853 | 1.1919 | 0.0035 | 1.7108 | 2.5678 | 0.0328 | 0.2136 |
| NMFAN | 15.4842 | 0.5660 | 0.4548 | 6.8950 | 0.0303 | 2.8173 | 1.9255 | 0.5662 | 0.9083 |
| EWRNMF | 1.7153 | 0.2276 | 0.0262 | 0.5418 | 0.0045 | 0.6429 | 0.4273 | 0.1970 | 0.6678 |
| SODNMF | 45.7755 | 0.9343 | 0.0702 | 0.2572 | 0.0084 | 8.5589 | 10.2358 | 0.0950 | 0.9597 |
| DLRGNMF | 52.3244 | 0.4156 | 0.2653 | 0.3481 | 0.0293 | 5.9124 | 8.2403 | 0.1673 | 2.4980 |
| Methods | Accuracy (%) | Normalized Mutual Information (%) | ||||
|---|---|---|---|---|---|---|
| 8 × 8 Noise | 12 × 12 Noise | 16 × 16 Noise | 8 × 8 Noise | 12 × 12 Noise | 16 × 16 Noise | |
| PCA | 41.24 ± 2.03 | 40.91 ± 2.71 | 39.76 ± 2.77 | 46.98 ± 2.29 | 47.02 ± 2.15 | 46.14 ± 1.98 |
| KMM | 35.24 ± 2.94 | 32.18 ± 3.38 | 31.52 ± 2.72 | 40.63 ± 3.87 | 37.70 ± 3.80 | 36.24 ± 3.10 |
| LRLMR | 40.21 ± 3.50 | 38.15 ± 3.19 | 35.76 ± 1.77 | 44.28 ± 3.03 | 42.06 ± 3.02 | 38.31 ± 2.37 |
| CNMF | 42.36 ± 3.97 | 42.24 ± 3.50 | 39.97 ± 3.71 | 48.46 ± 2.80 | 47.62 ± 2.55 | 46.31 ± 3.01 |
| GNMF | 40.09 ± 3.62 | 39.55 ± 2.29 | 39.09 ± 2.65 | 45.19 ± 2.92 | 44.45 ± 2.36 | 44.89 ± 2.76 |
| DNMF | 38.36 ± 2.65 | 37.42 ± 1.69 | 36.45 ± 1.99 | 44.26 ± 2.51 | 44.10 ± 1.38 | 42.36 ± 1.89 |
| DSNMF | 40.27 ± 2.16 | 39.42 ± 2.23 | 37.18 ± 1.71 | 45.97 ± 1.94 | 44.17 ± 2.10 | 42.61 ± 2.00 |
| NMFAN | 40.03 ± 2.78 | 39.21 ± 2.90 | 38.18 ± 3.66 | 45.22 ± 2.15 | 44.84 ± 2.82 | 44.15 ± 3.01 |
| EWRNMF | 39.79 ± 2.28 | 38.76 ± 3.14 | 38.39 ± 2.40 | 44.96 ± 1.83 | 44.06 ± 2.61 | 43.28 ± 2.03 |
| SODNMF | 42.94 ± 2.39 | 40.58 ± 2.51 | 38.48 ± 2.12 | 49.04 ± 1.86 | 47.11 ± 2.54 | 45.91 ± 1.65 |
| DLRGNMF | 44.58 ± 2.64 | 42.39 ± 1.88 | 40.24 ± 2.30 | 50.43 ± 2.51 | 47.88 ± 1.55 | 46.37 ± 1.68 |
| Methods | Accuracy (%) | Normalized Mutual Information (%) | ||||
|---|---|---|---|---|---|---|
| 8 × 8 Noise | 12 × 12 Noise | 16 × 16 Noise | 8 × 8 Noise | 12 × 12 Noise | 16 × 16 Noise | |
| PCA | 88.24 ± 5.38 | 86.67 ± 2.21 | 83.29 ± 4.36 | 88.40 ± 3.36 | 85.25 ± 2.87 | 81.83 ± 2.06 |
| KMM | 86.01 ± 5.02 | 83.83 ± 3.31 | 77.82 ± 5.52 | 87.16 ± 2.60 | 86.25 ± 2.72 | 81.64 ± 3.87 |
| LRLMR | 83.09 ± 3.49 | 82.90 ± 3.95 | 77.93 ± 3.34 | 82.23 ± 2.85 | 81.29 ± 2.79 | 77.17 ± 2.86 |
| CNMF | 83.10 ± 6.60 | 81.08 ± 5.64 | 78.97 ± 5.18 | 81.50 ± 4.98 | 80.07 ± 3.08 | 77.80 ± 3.64 |
| GNMF | 83.22 ± 3.93 | 81.71 ± 3.09 | 78.83 ± 6.41 | 84.40 ± 2.42 | 83.94 ± 2.21 | 78.13 ± 3.82 |
| DNMF | 90.96 ± 5.12 | 90.54 ± 3.76 | 84.25 ± 5.10 | 91.31 ± 2.31 | 90.32 ± 2.09 | 85.37 ± 2.80 |
| DSNMF | 90.12 ± 4.70 | 88.52 ± 4.70 | 84.77 ± 4.35 | 90.68 ± 2.26 | 89.07 ± 2.97 | 85.39 ± 2.79 |
| NMFAN | 83.31 ± 6.31 | 83.00 ± 4.92 | 79.69 ± 3.33 | 81.99 ± 3.46 | 80.60 ± 3.82 | 77.84 ± 3.33 |
| EWRNMF | 82.32 ± 5.02 | 82.51 ± 5.19 | 80.80 ± 5.49 | 80.88 ± 3.67 | 80.08 ± 3.65 | 79.27 ± 3.61 |
| SODNMF | 91.74 ± 3.28 | 90.14 ± 3.76 | 88.69 ± 4.90 | 91.17 ± 1.98 | 89.57 ± 2.11 | 87.84 ± 3.32 |
| DLRGNMF | 93.54 ± 3.14 | 93.57 ± 3.21 | 90.42 ± 4.07 | 92.25 ± 1.84 | 92.40 ± 1.80 | 89.29 ± 2.12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Ma, Z.; Li, H.; Wang, J. Dual Space Latent Representation Learning for Image Representation. Mathematics 2023, 11, 2526. https://doi.org/10.3390/math11112526
Huang Y, Ma Z, Li H, Wang J. Dual Space Latent Representation Learning for Image Representation. Mathematics. 2023; 11(11):2526. https://doi.org/10.3390/math11112526
Chicago/Turabian StyleHuang, Yulei, Ziping Ma, Huirong Li, and Jingyu Wang. 2023. "Dual Space Latent Representation Learning for Image Representation" Mathematics 11, no. 11: 2526. https://doi.org/10.3390/math11112526
APA StyleHuang, Y., Ma, Z., Li, H., & Wang, J. (2023). Dual Space Latent Representation Learning for Image Representation. Mathematics, 11(11), 2526. https://doi.org/10.3390/math11112526






