Robust Evaluation and Comparison of EEG Source Localization Algorithms for Accurate Reconstruction of Deep Cortical Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Algorithms for Evaluation
2.2. Simulation Model for Algorithm Evaluation
2.3. Generation of Simulated Source and EEG Signals
2.4. Evaluation Metrics for Algorithm Performance
- Normalized mean square error (MSE) [41]:
- Distribution discrepancy (DD):
- Relative mean square error (RMSE) [32]:
3. Results
3.1. Mathematical Verification on Algorithm Methodologies
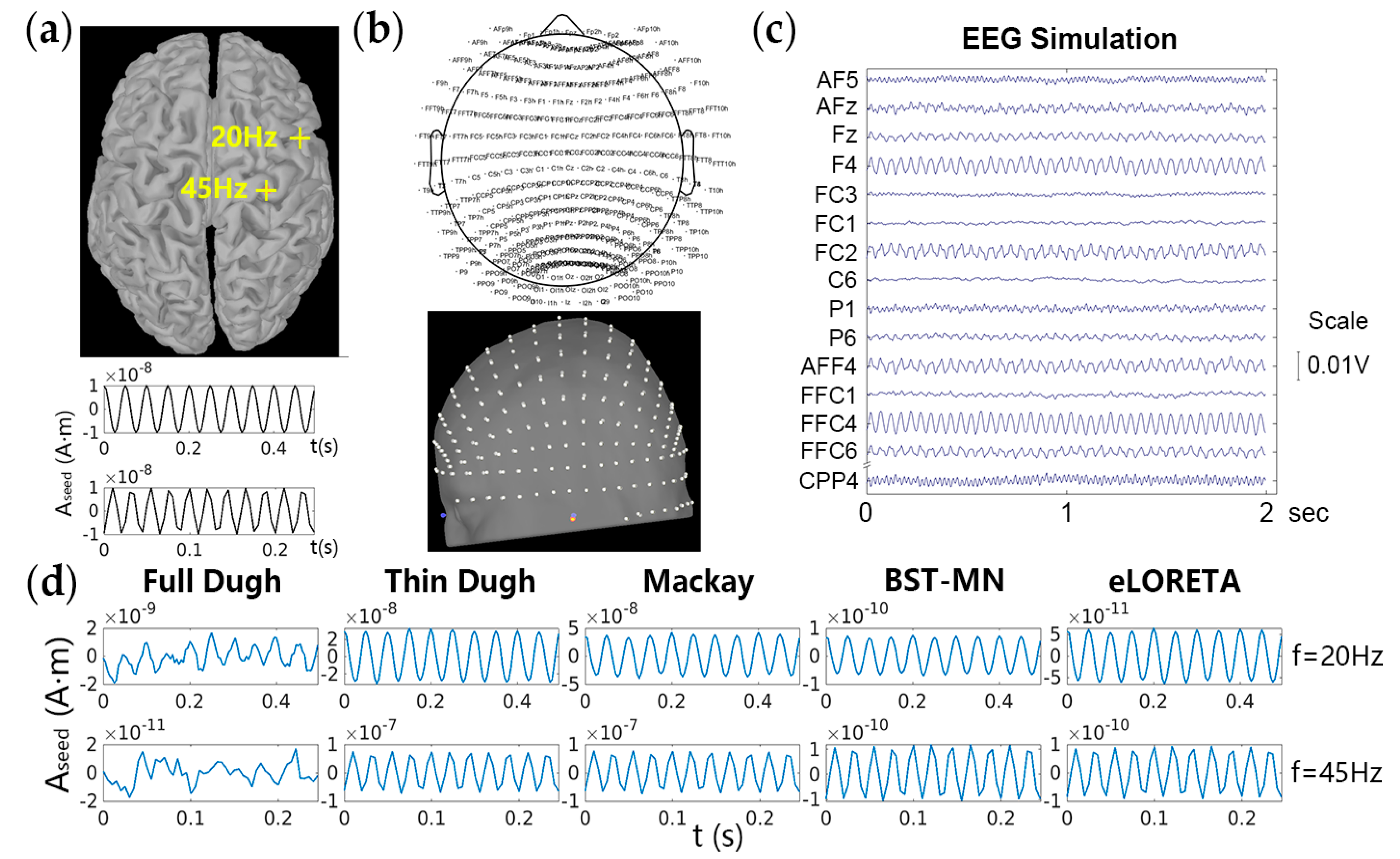
3.2. Electrical Propagation from EEG Sources to Scalp Electrodes
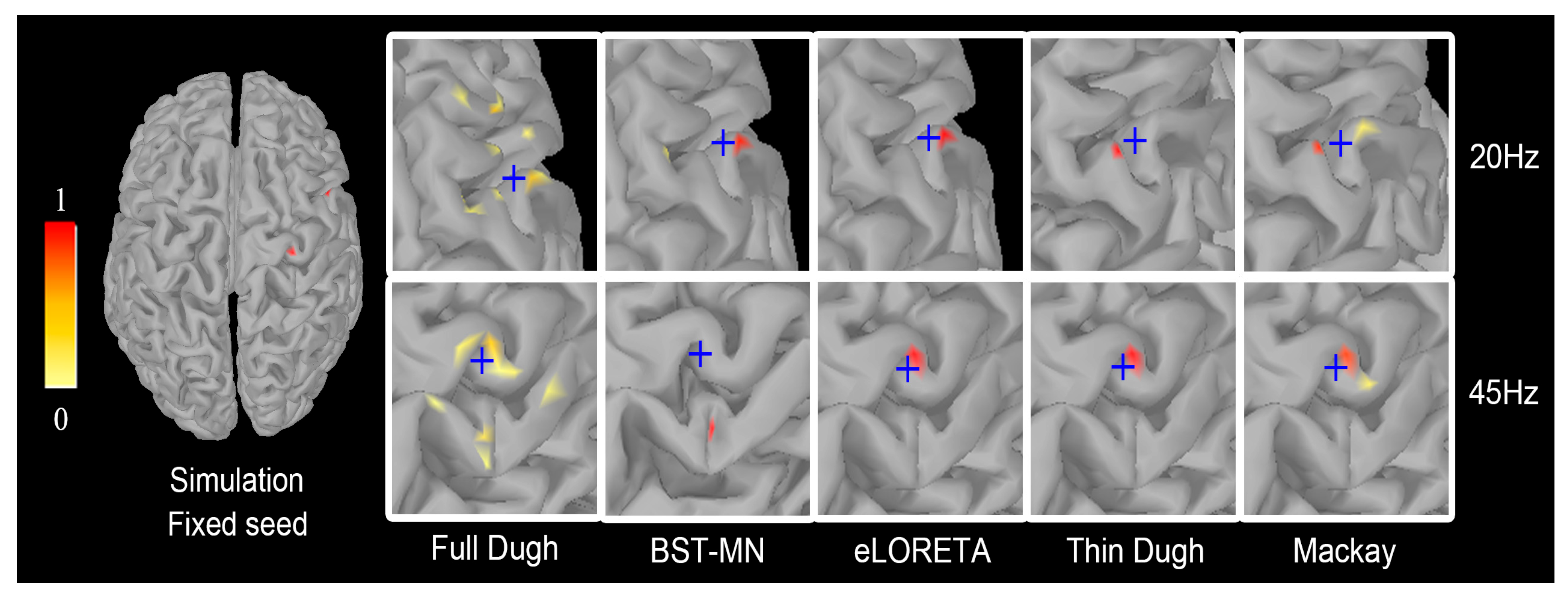
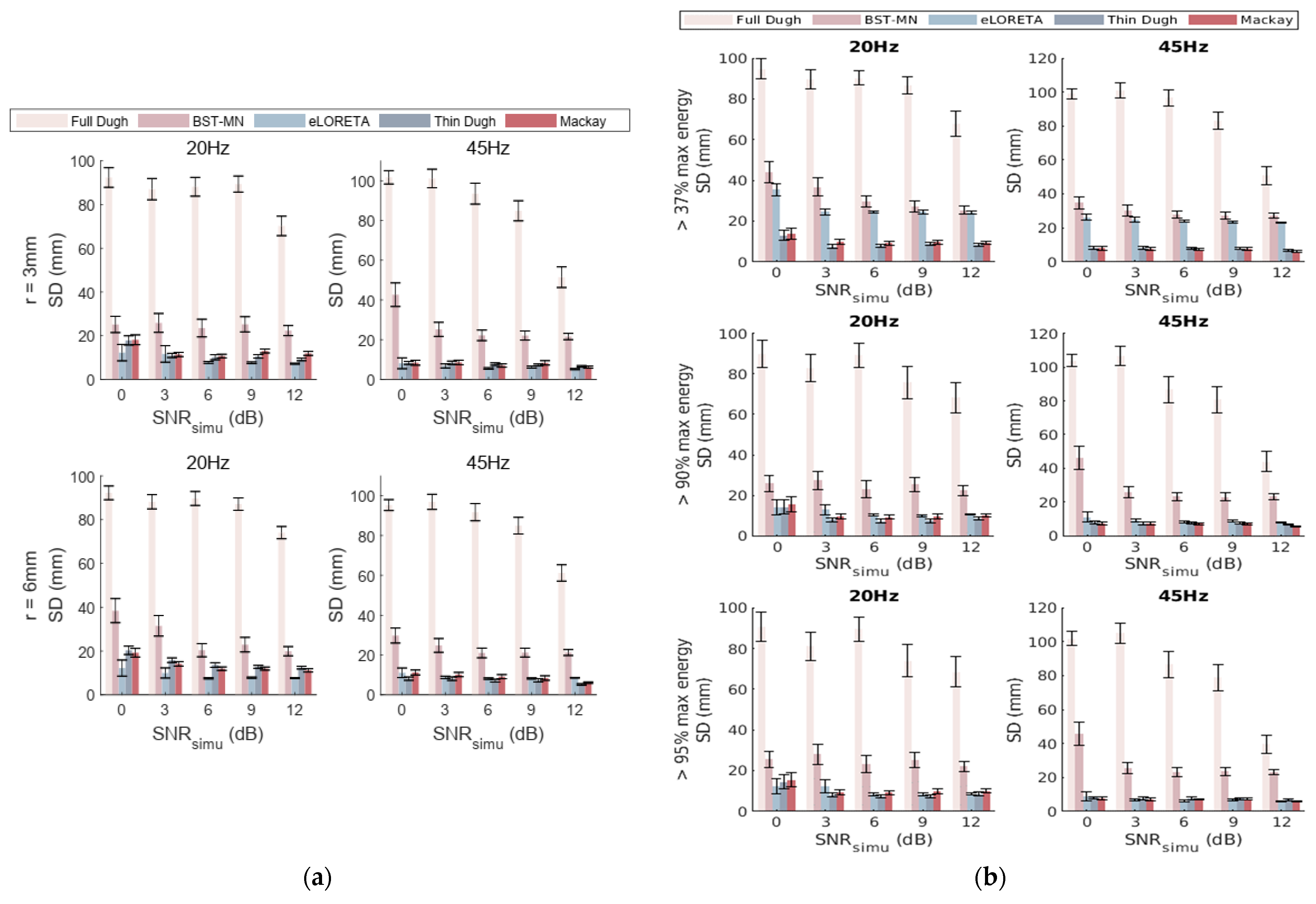
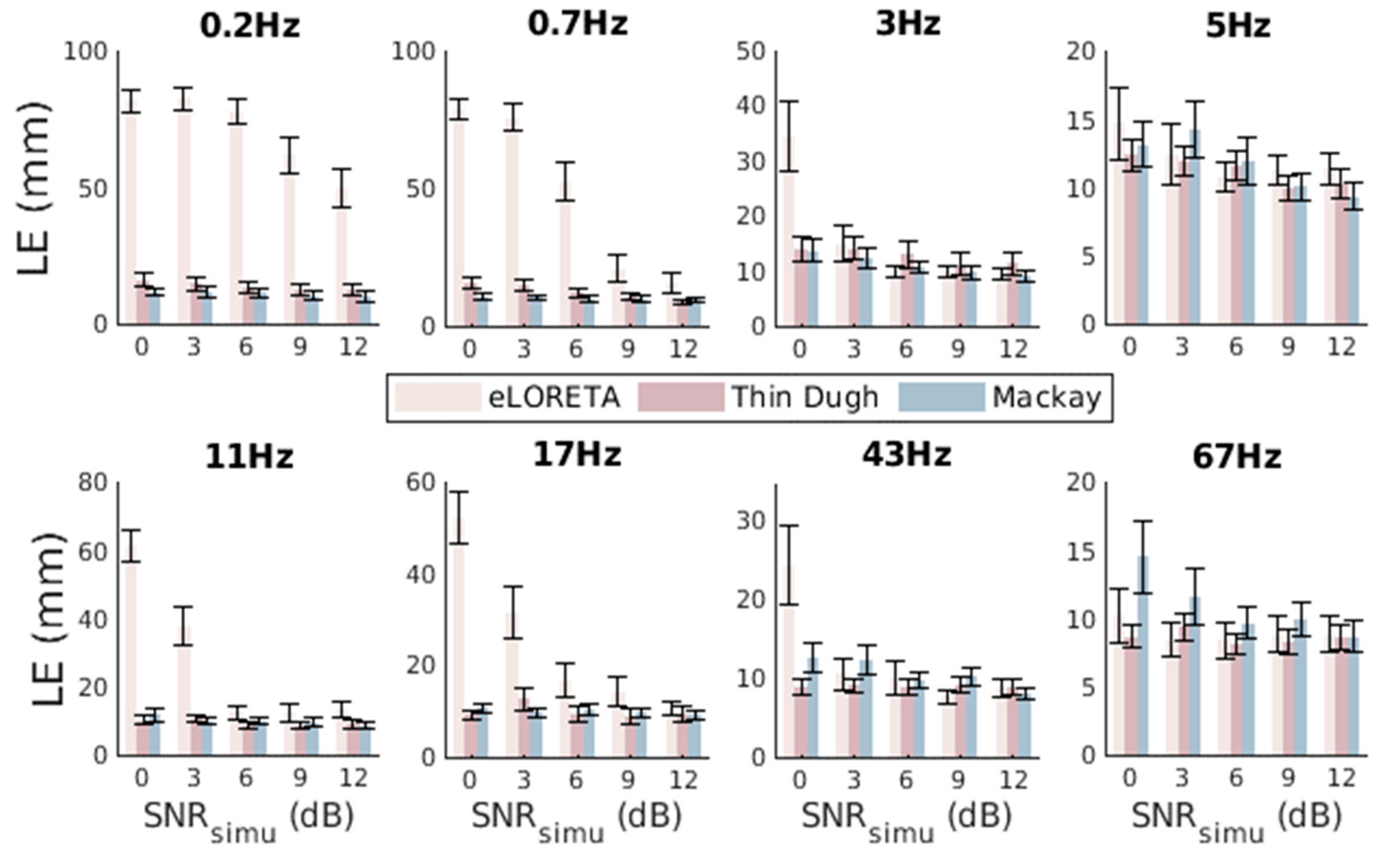
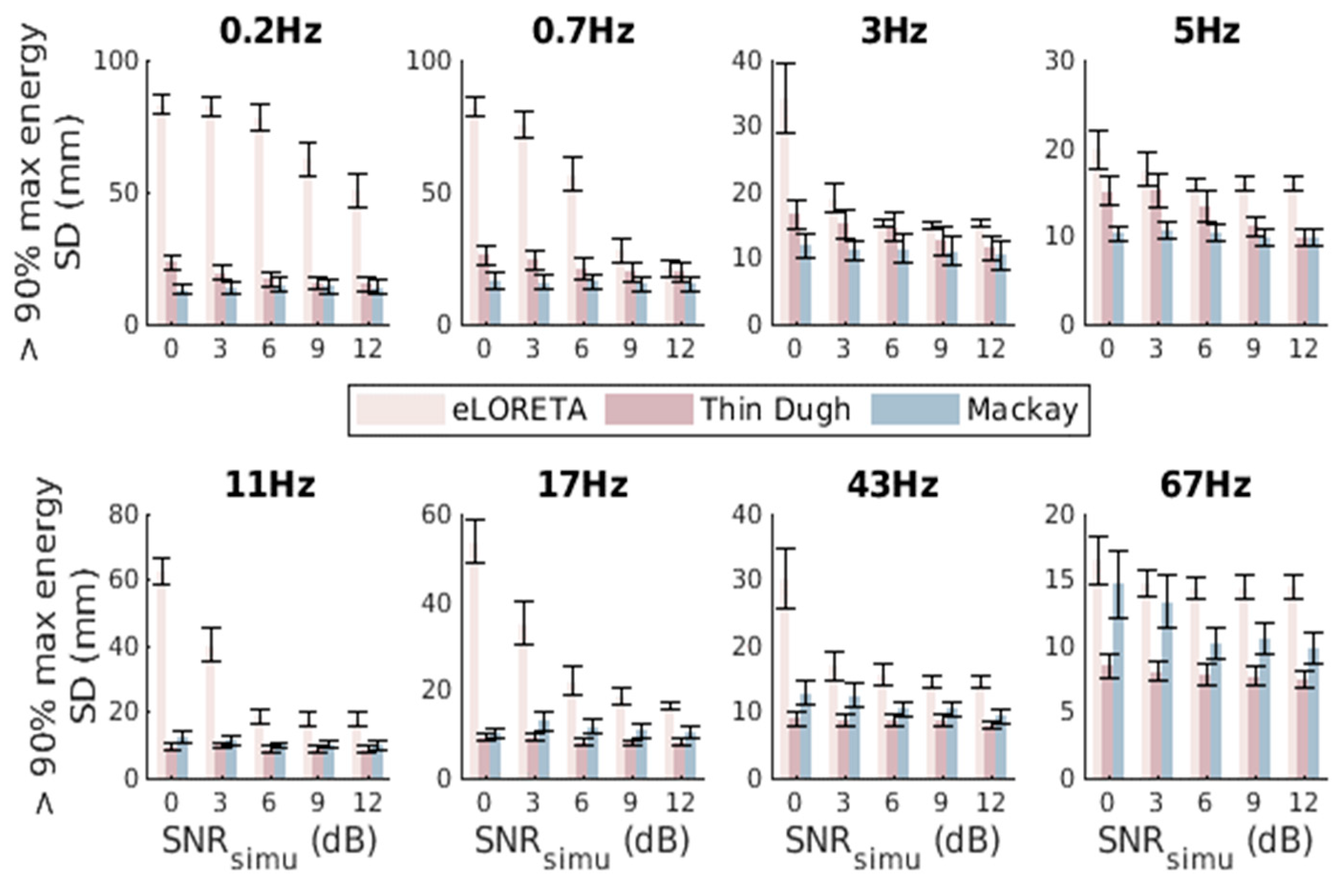
3.3. Comparison and Evaluation of Seed Activity Recovery
3.4. Fixed Dual-Seed Simulations for Algorithm Evaluations
3.5. Unfixed Dual-Seed Simulations for Algorithm Evaluations
3.6. Triple-Seed Simulations for Algorithm Evaluations
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cohen, M.X. Where Does EEG Come From and What Does It Mean? Trends Neurosci. 2017, 40, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Niedermeyer, E. The clinical relevance of EEG interpretation. Clin. Electroencephalogr. 2003, 34, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Lei, Y.; Li, Y.; Zhang, W.; Su, J.; Qi, X.; Chen, L.; Zhang, X.; Gu, Y.; Yu, Y.; et al. Changes in Brain Functional Network Connectivity in Adult Moyamoya Diseases. Cogn. Neurodyn. 2021, 15, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Li, Y.; Qi, X.; Zhang, W.; Yu, Y. Mental Calculation Drives Reliable and Weak Distant Connectivity While Music Listening Induces Dense Local Connectivity. Phenomics 2021, 1, 285–298. [Google Scholar] [CrossRef]
- Qi, X.; Wang, Y.; Lu, Y.; Zhao, Q.; Chen, Y.; Zhou, C.; Yu, Y. Enhanced brain network flexibility by physical exercise in female methamphetamine users. Cogn. Neurodyn. 2022, 26, 1–7. [Google Scholar] [CrossRef]
- Lei, Y.; Li, Y.; Yu, L.; Xu, L.; Zhang, X.; Zheng, G.; Chen, L.; Zhang, W.; Qi, X.; Gu, Y.; et al. Faded Critical Dynamics in Adult Moyamoya Disease Revealed by EEG and fMRI. Oxid. Med. Cell. Longev. 2021, 2021, 6640108. [Google Scholar] [CrossRef]
- Michel, C.M.; Murray, M.M.; Lantz, G.; Gonzalez, S.; Spinelli, L.; Grave de Peralta, R. EEG source imaging. Clin. Neurophysiol. 2004, 115, 2195–2222. [Google Scholar] [CrossRef]
- Berger, H. Electroencephalogram in humans. Arch. Psychiatr. Nervenkrankh. 1929, 87, 527–570. [Google Scholar] [CrossRef]
- He, B.; Sohrabpour, A.; Brown, E.; Liu, Z. Electrophysiological Source Imaging: A Noninvasive Window to Brain Dynamics. Annu. Rev. Biomed. Eng. 2018, 20, 171–196. [Google Scholar] [CrossRef]
- Cai, C.; Kang, H.; Hashemi, A.; Chen, D.; Diwakar, M.; Haufe, S.; Sekihara, K.; Wu, W.; Nagarajan, S.S. Bayesian algorithms for joint estimation of brain activity and noise in electromagnetic imaging. IEEE Trans. Med. Imaging 2022, 42, 762–773. [Google Scholar] [CrossRef]
- Hashemi, A.; Gao, Y.; Cai, C.; Ghosh, S.; Müller, K.-R.; Nagarajan, S.; Haufe, S. Efficient hierarchical Bayesian inference for spatio-temporal regression models in neuroimaging. In Proceedings of the Advances in Neural Information Processing Systems, Virtual, 6–14 December 2021; pp. 24855–24870. [Google Scholar]
- Sohrabpour, A.; Lu, Y.; Worrell, G.; He, B. Imaging brain source extent from EEG/MEG by means of an iteratively reweighted edge sparsity minimization (IRES) strategy. NeuroImage 2016, 142, 27–42. [Google Scholar] [CrossRef]
- Hallez, H.; Vanrumste, B.; Grech, R.; Muscat, J.; De Clercq, W.; Vergult, A.; D’Asseler, Y.; Camilleri, K.P.; Fabri, S.G.; Van Huffel, S.; et al. Review on solving the forward problem in EEG source analysis. J. Neuroeng. Rehabil. 2007, 4, 46. [Google Scholar] [CrossRef]
- Chen, Y.; Akutagawa, M.; Emoto, T.; Kinouchi, Y. The removal of EMG in EEG by neural networks. Physiol. Meas. 2010, 31, 1567–1584. [Google Scholar] [CrossRef]
- Gurve, D.; Delisle-Rodriguez, D.; Bastos-Filho, T.; Krishnan, S. Trends in Compressive Sensing for EEG Signal Processing Applications. Sensors 2020, 20, 3703. [Google Scholar] [CrossRef]
- Zorzos, I.; Kakkos, I.; Ventouras, E.M.; Matsopoulos, G.K. Advances in Electrical Source Imaging: A Review of the Current Approaches, Applications and Challenges. Signals 2021, 2, 378–391. [Google Scholar] [CrossRef]
- Baillet, S.; Mosher, J.C.; Leahy, R.M. Electromagnetic brain mapping. IEEE Signal Process. Mag. 2001, 18, 14–30. [Google Scholar] [CrossRef]
- Asadzadeh, S.; Yousefi Rezaii, T.; Beheshti, S.; Delpak, A.; Meshgini, S. A systematic review of EEG source localization techniques and their applications on diagnosis of brain abnormalities. J. Neurosci. Methods 2020, 339, 108740. [Google Scholar] [CrossRef]
- Tadel, F.; Baillet, S.; Mosher, J.C.; Pantazis, D.; Leahy, R.M. Brainstorm: A user-friendly application for MEG/EEG analysis. Comput. Intell. Neurosci. 2011, 2011, 879716. [Google Scholar] [CrossRef]
- Pascual-Marqui, R.D. Discrete, 3D distributed, linear imaging methods of electric neuronal activity. Part 1: Exact, zero error localization. arXiv 2007, arXiv:0710.3341. [Google Scholar]
- Hyder, F.; Herman, P.; Bailey, C.J.; Møller, A.; Globinsky, R.; Fulbright, R.K.; Rothman, D.L.; Gjedde, A. Uniform distributions of glucose oxidation and oxygen extraction in gray matter of normal human brain: No evidence of regional differences of aerobic glycolysis. J. Cereb. Blood Flow Metab. 2016, 36, 903–916. [Google Scholar] [CrossRef]
- Schrader, S.; Westhoff, A.; Piastra, M.C.; Miinalainen, T.; Pursiainen, S.; Vorwerk, J.; Brinck, H.; Wolters, C.H.; Engwer, C. DUNEuro-A software toolbox for forward modeling in bioelectromagnetism. PLoS ONE 2021, 16, e0252431. [Google Scholar] [CrossRef] [PubMed]
- Medani, T.; Garcia-Prieto, J.; Tadel, F.; Schrader, S.; Antonakakis, M.; Joshi, A.; Engwer, C.; Wolters, C.; Mosher, J.; Leahy, R. Realistic head modeling of electromagnetic brain activity: An integrated Brainstorm-DUNEuro pipeline from MRI data to the FEM solutions. In Proceedings of the SPIE Medical Imaging, San Diego, CA, USA, 14–18 February 2021. [Google Scholar]
- Thielscher, A.; Antunes, A.; Saturnino, G.B. Field modeling for transcranial magnetic stimulation: A useful tool to understand the physiological effects of TMS? In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 222–225. [Google Scholar]
- Yao, D. A method to standardize a reference of scalp EEG recordings to a point at infinity. Physiol. Meas. 2001, 22, 693–711. [Google Scholar] [CrossRef] [PubMed]
- Mandija, S.; Petrov, P.I.; Vink, J.J.T.; Neggers, S.F.W.; van den Berg, C.A.T. Brain Tissue Conductivity Measurements with MR-Electrical Properties Tomography: An In Vivo Study. Brain Topogr. 2021, 34, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Marino, M.; Cordero-Grande, L.; Mantini, D.; Ferrazzi, G. Conductivity Tensor Imaging of the Human Brain Using Water Mapping Techniques. Front. Neurosci. 2021, 15, 694645. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Corazza, M.; Turovets, S.; Luu, P.; Price, N.; Muravchik, C.H.; Tucker, D. Skull Modeling Effects in Conductivity Estimates Using Parametric Electrical Impedance Tomography. IEEE Trans. Bio-Med. Eng. 2018, 65, 1785–1797. [Google Scholar] [CrossRef]
- Wolters, C.H.; Anwander, A.; Tricoche, X.; Weinstein, D.; Koch, M.A.; MacLeod, R.S. Influence of tissue conductivity anisotropy on EEG/MEG field and return current computation in a realistic head model: A simulation and visualization study using high-resolution finite element modeling. NeuroImage 2006, 30, 813–826. [Google Scholar] [CrossRef]
- Murakami, S.; Okada, Y. Contributions of principal neocortical neurons to magnetoencephalography and electroencephalography signals. J. Physiol. 2006, 575, 925–936. [Google Scholar] [CrossRef]
- Lamus, C.; Hämäläinen, M.S.; Temereanca, S.; Brown, E.N.; Purdon, P.L. A spatiotemporal dynamic distributed solution to the MEG inverse problem. NeuroImage 2012, 63, 894–909. [Google Scholar] [CrossRef]
- Huang, G.; Liu, K.; Liang, J.; Cai, C.; Gu, Z.H.; Qi, F.; Li, Y.; Yu, Z.L.; Wu, W. Electromagnetic Source Imaging via a Data-Synthesis-Based Convolutional Encoder–Decoder Network. IEEE Trans. Neural Netw. Learn. Syst. 2022, 2022, 1–15. [Google Scholar] [CrossRef]
- Teplan, M. Fundamentals of EEG measurement. Meas. Sci. Rev. 2002, 2, 1–11. [Google Scholar]
- Kumar, J.S.; Bhuvaneswari, P. Analysis of Electroencephalography (EEG) signals and its categorization—A study. Procedia Eng. 2012, 38, 2525–2536. [Google Scholar] [CrossRef]
- Chávez, C.E.; Alonzo-Atienza, F.; Álvarez, D. Avoiding the inverse crime in the Inverse Problem of electrocardiography: Estimating the shape and location of cardiac ischemia. In Proceedings of the Computing in Cardiology 2013, Zaragoza, Spain, 22–25 September 2013; pp. 687–690. [Google Scholar]
- Munck, J.C.D.; Huizenga, H.M.; Waldorp, L.J.; Heethaar, R.A. Estimating stationary dipoles from MEG/EEG data contaminated with spatially and temporally correlated background noise. IEEE Trans. Signal Process. 2002, 50, 1565–1572. [Google Scholar] [CrossRef]
- Voytek, B.; Kramer, M.A.; Case, J.; Lepage, K.Q.; Tempesta, Z.R.; Knight, R.T.; Gazzaley, A. Age-Related Changes in 1/f Neural Electrophysiological Noise. J. Neurosci. 2015, 35, 13257–13265. [Google Scholar] [CrossRef]
- Dave, S.; Brothers, T.A.; Swaab, T.Y. 1/f neural noise and electrophysiological indices of contextual prediction in aging. Brain. Res. 2018, 1691, 34–43. [Google Scholar] [CrossRef]
- Yu, Y.; Romero, R.; Lee, T.S. Preference of sensory neural coding for 1/f signals. Phys. Rev. Lett. 2005, 94, 108103. [Google Scholar] [CrossRef]
- Qu, G.; Fan, B.; Fu, X.; Yu, Y. The Impact of Frequency Scale on the Response Sensitivity and Reliability of Cortical Neurons to 1/fβ Input Signals. Front. Cell. Neurosci. 2019, 13, 311. [Google Scholar] [CrossRef]
- Grova, C.; Daunizeau, J.; Lina, J.M.; Bénar, C.G.; Benali, H.; Gotman, J. Evaluation of EEG localization methods using realistic simulations of interictal spikes. NeuroImage 2006, 29, 734–753. [Google Scholar] [CrossRef]
- Molins, A.; Stufflebeam, S.M.; Brown, E.N.; Hämäläinen, M.S. Quantification of the benefit from integrating MEG and EEG data in minimum ℓ2-norm estimation. NeuroImage 2008, 42, 1069–1077. [Google Scholar] [CrossRef]
- Chang, W.-T.; Nummenmaa, A.; Hsieh, J.-C.; Lin, F.-H. Spatially sparse source cluster modeling by compressive neuromagnetic tomography. NeuroImage 2010, 53, 146–160. [Google Scholar] [CrossRef]
- Gaubert, S.; Raimondo, F.; Houot, M.; Corsi, M.C.; Naccache, L.; Diego Sitt, J.; Hermann, B.; Oudiette, D.; Gagliardi, G.; Habert, M.O.; et al. EEG evidence of compensatory mechanisms in preclinical Alzheimer’s disease. Brain A J. Neurol. 2019, 142, 2096–2112. [Google Scholar] [CrossRef]












| Seeds | Seed Positions | Seed Frequencies and Amplitudes |
|---|---|---|
| 1 | Random point on the cortex from either the left or the right hemisphere. | Low-frequency bands (0.2 Hz, 0.7 Hz, 3 Hz, and 5 Hz) with amplitudes of 20 nA·m and random initial phases for each frequency. |
| 2 | Random point on the cortex from the opposite hemisphere of Seed 1. | High-frequency bands (11 Hz, 17 Hz, 43 Hz, and 67 Hz) with amplitudes of 10 nA·m and random initial phases for each frequency. |
| 3 | Random point on the cortex from either the left or the right hemisphere. | Randomly choose two frequencies from the low-frequency bands with amplitude of 20 nA·m. Randomly choose two frequencies from the high-frequency bands with amplitude of 10 nA·m. Set random initial phases for each frequency. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, H.; Yu, Y. Robust Evaluation and Comparison of EEG Source Localization Algorithms for Accurate Reconstruction of Deep Cortical Activity. Mathematics 2023, 11, 2450. https://doi.org/10.3390/math11112450
Shen H, Yu Y. Robust Evaluation and Comparison of EEG Source Localization Algorithms for Accurate Reconstruction of Deep Cortical Activity. Mathematics. 2023; 11(11):2450. https://doi.org/10.3390/math11112450
Chicago/Turabian StyleShen, Hao, and Yuguo Yu. 2023. "Robust Evaluation and Comparison of EEG Source Localization Algorithms for Accurate Reconstruction of Deep Cortical Activity" Mathematics 11, no. 11: 2450. https://doi.org/10.3390/math11112450
APA StyleShen, H., & Yu, Y. (2023). Robust Evaluation and Comparison of EEG Source Localization Algorithms for Accurate Reconstruction of Deep Cortical Activity. Mathematics, 11(11), 2450. https://doi.org/10.3390/math11112450







