1. Introduction
Epileptic seizures occur due to repeated and sudden abnormal brain reactions as a response to the abnormal electrical discharges produced by neurons [
1,
2]. They result in the disorderliness of mood, sensation, movement, or mental function [
3,
4], attention delays, whole-body tremors, and unconsciousness. The accurate classification of epileptic seizure type plays an essential role in the treatment and management of epilepsy patients [
5]. Moreover, it can help neurosurgeons to understand brain cortical connectivity, obtain information about the potential triggers of seizures, and understand the risk of different potential comorbidities, such as learning difficulties, intellectual disability, and unexpected death [
6,
7].
Elliptic seizures are classified into focal, generalized, and unknown seizure types based on when and how the seizure starts in the patient’s brain. Focal seizures begin in a single area, or a set of cells located on the same side of the brain, and are categorized into simple and complex partial seizures. Generalized seizures begin simultaneously in sets of cells located on both sides of the brain and are categorized into myoclonic, clonic, tonic, and tonic–clonic seizures. In contrast, seizures that have unidentified beginnings are called unknown seizures [
8,
9]. The beginning of a seizure is detected from EEG recordings with the presence of abnormal discharges, which are called ictal epileptic discharges. At that time, neurologists start noticing the increase in frequency and the appearance of rapid continuous spikes and waves. Neurologists then capture the peak seizure activity when these waves progress to numerous spikes with buried waves. Finally, they detect the stop point when the seizure activity slows down, where waves reappear, and an obvious decrease in frequency is noticed until the stop [
10]. Neurologists defined two main events for the TUH EEG Corpus to record seizure segments from EEG recordings based on how these recordings are interpreted and the maximum amplitude of signals. The first event is the periodic lateralized epileptiform discharges (PLEDs), in which seizures are represented by a repetitive spike or sharp wave discharges that are lateralized or focal over one hemisphere and recur at fixed time intervals. The second event is the GPED, as described above, where tri-phasic waves (diffuse and bilaterally synchronous spikes with bifrontal predominance, with a rate of 1–2 Hz) are presented in recordings [
11].
The focus of this research is to identify the type of seizures. The most commonly used modality to identify types of epileptic seizures is the EEG; EEG signals include important information about the brain’s electrical activities and are widely used for epilepsy analysis. Machine learning, particularly deep learning, has been employed for various medical applications [
12,
13,
14,
15,
16,
17]. CNNs have been well suited for EEG signal representation learning among several learning models. However, most of the available CNN-based methods for classifying seizure types in the literature suffer from the overfitting problem due to the huge difference between the small number of EEG trials and the huge number of learnable parameters [
18,
19]. Moreover, 2D CNNs are the most commonly applied models, which result in high computational complexity. We propose a lightweight deep model based on residual learning and long short-term memory (LSTM) to overcome these issues.
Therefore, this paper aims to present the design of an efficient, lightweight, and expressive deep network model that adopts both the 1D ResNet module and LSTM module to classify epileptic seizure types, avoiding overfitting as it involves a relatively small number of learnable parameters. The main contributions of this paper are as follows:
This paper proposes a deep network model for identifying seizure types that adopts 1D ResNet module and LSTM. The model involves a small number of learnable parameters, so it prevents overfitting when trained on a small number of available EEG trials. ResNet learning helps to encounter vanishing gradient problems.
The deep network includes the LSTM module to encode and learn long-term dependencies over time.
To obtain a more promising performance and efficient solution to the overfitting problem, the SMOTE algorithm is applied to balance the training data by increasing the trials of minority classes.
The rest of this paper is organized as follows:
Section 2 reviews the state-of-the-art methods for the problem.
Section 3 presents the proposed deep network, details the model formulation, gives an overview of ResNet and LSTM, and discusses the proposed deep model architecture.
Section 4 discusses the evaluation procedure, introducing the benchmark dataset used for validation, the evaluation protocol and metrics adopted for evaluation, and the network training.
Section 5 describes the experiments conducted to study the impact of ResNet and LSTM modules and their hyper-parameters and data augmentation on the model performance. It then discusses the results obtained from the final network configuration.
Section 6 presents the analysis of the network performance by discussing the effect of frequency bands on the network performance, the use of both a confusion matrix and t-SNE plot to visualize the results, and the revealed space and time complexity of the network. It then gives a comparison between the performance of the proposed method and those of the state-of-the-art methods.
Section 7 discusses the model and obtained results.
Section 8 concludes the paper.
2. Related Work
Automatic seizure detection using multi-channel EEG trials has been considered an active area of research over the years. Hence, various research works have been conducted recently to identify different types of epileptic seizures from EEG trials using classical machine learning techniques. This section gives an overview of these studies describing the proposed methods on the one hand and determining their core research gaps, on the other hand, to be solved.
Raghu et al. [
20] presented a CNN-based framework that solves the eight-class classification problem and classifies different non-seizure and seizure types. Input trials fed into the AlexNet, VGG16, and VGG19 models. The presented method was tested on the TUH database using stochastic gradient descent with a momentum optimizer. Input trials were divided into a training set (70%) and a validation set (30%). The results revealed that AlexNet obtained the highest classification accuracy of 84.06%. The EEG trials were converted from 1D into 2D by the STFT using all 19 channels for inputs to be fed into CNNs.
Raghu et al. [
6] proposed a CNN-based framework that classifies seizure and non-seizure EEG data. It converts the 19 input channel EEG time series into a spectrogram stack and the EEG trials from 1D into 2D. The CNN model includes a Conv with ReLU activation function, pooling, and batch normalization. Moreover, its last layer comprises a fully connected layer, drop out, softmax, and classification output layers. The presented method was tested on the TUH database using ten pre-trained networks: AlexNet, VGG16, VGG19, SqueezeNet, GoogLeNet, Inception v3, DenseNet201, ResNet18, ResNet50, and ResNet101. Input data were divided into a training set (70%) and a testing set (30%). Results revealed that the highest recorded accuracy was 88.30% for the Inception v3 network.
Aristizaba et al. [
7] proposed an approach to classify seizure types using electrophysiological data. The proposed approach uses a temporal central parasagittal montage of 20 chosen pair channels as inputs. The architecture includes CNN and LSTM networks. The CNN is composed of two convolutional layers, followed by one max-pooling layer and a fully connected layer. The LSTM includes two LSTM layers (each one has 128 cells), followed by a densely connected layer and a softmax activation layer. The presented method was tested on the TUH database using 5-fold cross-validation, the Adam optimizer, and definite cross-entropy loss. Input data were classified into training (60%), validation (20%), and testing sets (20%). The results revealed that the proposed model recorded a score of 0.945.
Roy et al. [
5] conducted the first study on the classification of eight seizure types (simple partial, complex partial, focal non-specific, generalized non-specific, absence, tonic, tonic–clonic, and non-seizure) using machine learning algorithms. In this method, EEG trials are initially converted into overlapping segments of W1 second with an overlap of O seconds. All extracted features are then passed to a classifier to identify the type of seizure. The authors tested five classifiers: k-NN, SGD, XGBoost, AdaBoost, and ResNet50 models. They evaluated the method on the TUH EEG database using the 5-fold cross-validation in such a way that for each split (fold), the seizures of each type were divided into a training set (60%) of seizure type, a validation set (20%) of seizure type, and a test set (20%) of seizure type, considering the F1-score as a performance metric. The resulting F1-score of 0.907 was obtained with k-NN.
Liu et al. [
21] presented a hybrid bilinear deep learning network model to classify epileptic seizures using both a CNN and recurrent neural networks (RNNs). The CNN is composed of a feature extractor, including three convolutional blocks and a fully connected dense classifier. The RNNs, in contrast, include a feature extractor with two ConvLSTM layers and a fully connected dense classifier. The CNN extracts spatiotemporal patterns from inputs, whereas the RNNs discover the features of temporal dynamics related to longer intervals for the same inputs. Both the CNN and RNNs were initially trained using STFT of a one-second surface electroencephalogram (sEEG) and the Adam optimization algorithm with batch sizes of 32 over 200 epochs. The presented method was tested on the TUH database and EPILEPSIAE. The results revealed that the presented methods recorded 97% and 97.2% F1-scores for the TUH and EPILEPSIAE databases, respectively.
Ma et al. [
22] presented a Transformers for Seizure Detection (TSD) system to identify eight epilepsy seizures and justify the imbalance between false alarms and sensitivity to clinicians for large EEG datasets. A Vision Transformer (ViT) is used as a baseline model within the system. This research adopts 17-channel bipolar longitudinal montages with conventional 10–20 placements. The TSD architecture comprises three modules: input, encoders, and output. Input long-term EEG trials are initially preprocessed to remove the DC component by applying the STFT. In the input module, the processed input signals are divided into 200 patches, where each one is projected into a fixed-length vector to be embedded into the transformer. The encoder module uses a multi-head attention mechanism to recognize input embedding and add a specific token into the input sequence whose corresponding output predicts epileptic seizures. The output module extracts the specific token and obtains predictions. The presented method was tested on the TUH database which was divided into training, validation, and test sets. The model recognizes the hyper-parameters of the validation set to train and learn the characteristics of EEG signals of epileptic seizures. It then validates the model’s hyper-parameters on the validation set and finally assesses the model’s hyper-parameters on the validation set. Results revealed that the proposed system achieved an accuracy of 92.1%.
It is clear from the literature that all the state-of-the-art models offered promising performance for classifying different types of seizures with different CNN models. However, applying those methods to identify the types of seizure is a challenge for neurologists because of the complexity of 2D CNN models, which suffer from overfitting when trained using training data consisting of a small number of input trials compared to a large number of learnable parameters in the NNs. Therefore, the goal of this work is to overcome the challenge of state-of-the-art models by proposing an efficient, lightweight, and expressive deep network model. It is based on ResNet theory and LSTM for classifying seizure types from EEG trials from the TUH database.
3. Proposed Method
The problem is to identify the types of epileptic seizures using EEG trials. To design the method for this problem, first, we formulate the problem. As the proposed method is based on deep learning techniques, we give an overview of ResNet theory and LSTM. Finally, we present the details of the proposed deep learning-based method.
3.1. Problem Formulation
A trial of an EEG trial is used to identify the type of an epileptic seizure. A trial is represented as a matrix
where
C is the number of channels and
T is the number of time stamps:
where:
C1,
C2, ……
Cc are 1D signals (channels of EEG trials) captured at different locations on the scalp and
ci (
tj) is the potential captured at time
tj.
is the space of EEG trials. Let Y = {AB, CP, FN, GN, SP, TN, TC, and MY} be the set of class labels, where AB, CP, FN, GN, SP, TN, TC, and MY represent the epileptic seizure types absence, complex partial, focal non-specific, generalized non-specific, simple partial, tonic, tonic–clonic, and myoclonic seizure, respectively. The problem is to identify the seizure class using EEG trial
x. It means that we need to design a mapping
that maps an EEG trial
to a label
, i.e.,
Motivated by the success of deep learning in various applications using EEG signals, such as in emotion recognition [
23], epilepsy detection [
20], and drowsiness detection [
24], we model
using a deep network.
3.2. ResNet
ResNet is a widely used efficient residual learning-based deep CNN model [
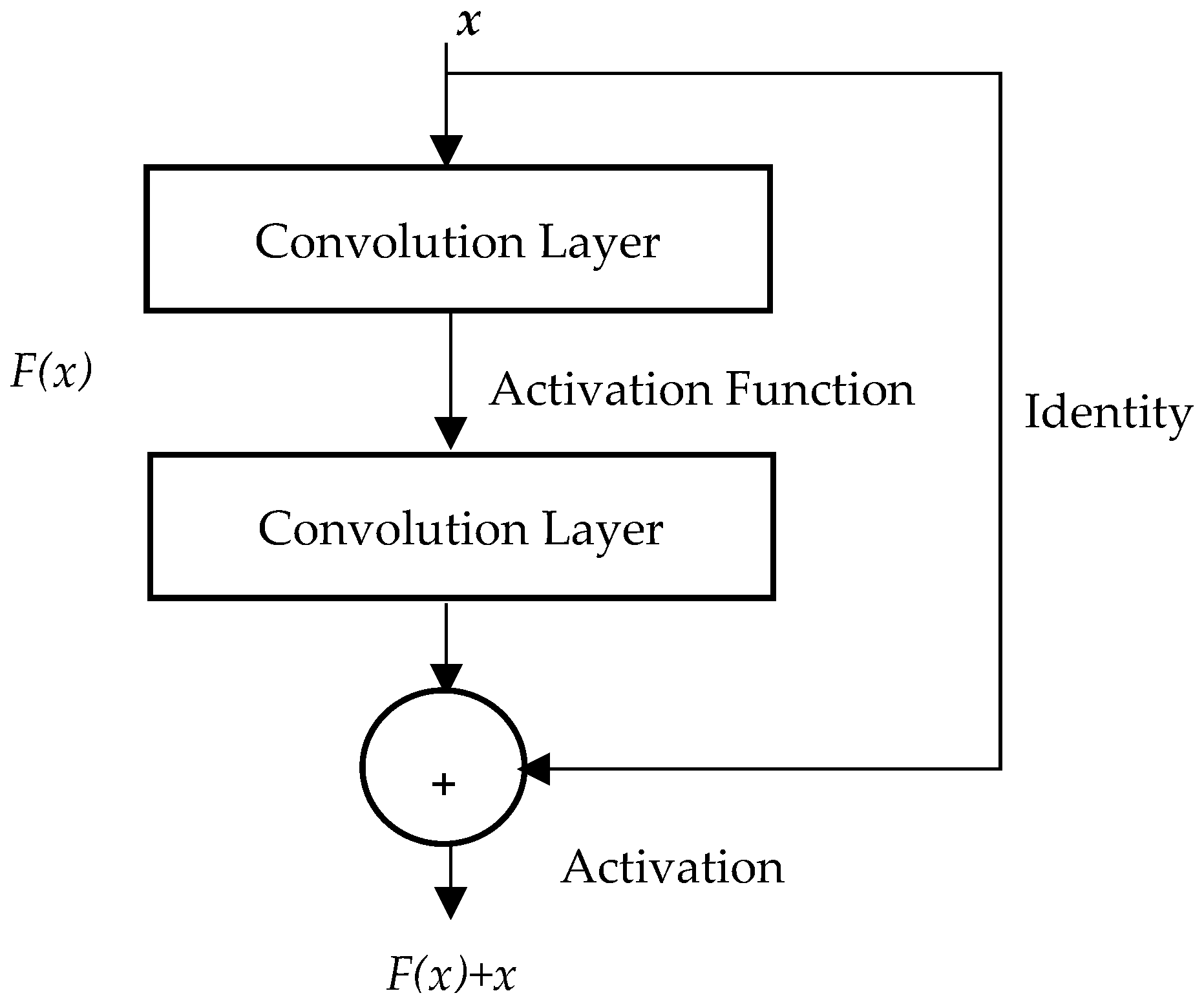
25]. It allows a network to learn the identity mapping of residual blocks instead of the underlying mapping. The residual block is shown in
Figure 1; it consists of a residual function
F(
x), an identity mapping, and the desired underlying mapping
H(
x), which is expressed as
H(
x) =
F(
x) +
x. The residual function
F(
x) is modeled with two convolutional layers. One activation function is added after the first convolutional layer and another after the summation of the outputs of the second convolutional layer and the identity layer. This block prevents the vanishing gradient problem by shortening the path between the input and output of the neural network.
The residual block learns a residual mapping
F(
x) instead of fitting a desired underlying mapping
H(
x) =
F(
x) +
x. As shown in
Figure 1, the output from the block is then expressed as follows:
It is demonstrated that optimizing the residual mapping rather than optimizing the underlying mapping
H(x) is easier. In other words, pushing the residual to zero is simpler than fitting an identity mapping via a stack of non-linear layers for optimal identity mapping. The block output y can be realized via feedforward neural networks with “shortcut connections,” as shown in
Figure 1. Those shortcuts skip one or more layers and carry out identity mapping, where their outputs are added to the output of stacked layers. Practically, the identity shortcut connections do not add any computational complexity or extra parameter.
The ResNet architecture consists of a series of residual blocks, each containing multiple convolution layers. The convolution layers enable the network to learn complex features. Additionally, they allow the network to capture patterns at different levels of abstraction better. The shortcut connection is added to each residual block, and the input of each block is added to its output. The output is then fed into a non-linearity ReLU activation function to avoid the vanishing gradient problem, which can occur in deep neural networks when the gradients become very small, and the networks become difficult to train. The next network layer is the average pooling that reduces the overfitting and dimensionality of the feature map. It is finally followed by a fully connected layer with softmax activation.
Deep networks based on adopting residual modules can be learned without encountering a vanishing gradient problem and can also avoid the overfitting problem of CNN models when trained using training data consisting of a small number of input trials compared to a large number of learnable parameters. This suits our problem because we have limited data for training the network.
3.3. LSTM
The RNN is a useful network for analyzing time series such as EEG trials, but it has a problem with learning [
26]. The most commonly used learning algorithms for training RNNs store information over long intervals and take too long due to the deficient, decaying error backflow. Hence, LSTM was proposed to handle the vanishing gradient problem in traditional RNNs. It is capable of learning long-term dependencies in sequential data.
The LSTM architecture comprises three layers; input, hidden, and output. The input layer consists of biased, multiplicative gate units and memory cells that receive incoming signals. The hidden layer (fully connected or semi-connected) receives signals from the input layer. The output layer, in turn, receives signals only from memory cells. The LSTM applies a constant error backpropagation flow within its memory cells, and it can learn bridging time intervals of more than 1000 time steps.
Moreover, using multiplicative gate units ensures protection of the error flow from unwanted perturbations by learning when to open/close the access to the constant error flow.
Figure 2 illustrates the LSTM building block: forget, input, and output gates (from left to right). This block allows the processing of sequence information through the cumulative linear form to prevent the gradient disappearance problem and learn long-term information. Therefore, it is practical for learning long-term sequence information [
27,
28].
The equation for the forget gate, as shown in
Figure 2, is:
where
is the output value of the forget gate,
is the output value of the last moment,
is the input value of the current moment,
is the weight matrix,
is the bias vector, and the sigmoid function
is the activation function of the gate. In this equation,
represents the concatenation of
and
.
The equations for the input gate, as shown in
Figure 2, are:
where
and
are the output values of the input gate,
and
are the weight matrix and bias in the
of the input gate, respectively,
and
are the weight matrix and bias vector in the input gate
tanh function, respectively. The equations for the output gate, as shown in
Figure 2, are:
where
represents the output value of the output gate,
and
represent the weight matrix and bias in the
of the output gate, respectively, and
represents the output value of the current moment. The updated cell state is expressed as follows:
where
represents the cell state of the current mode, and
represents the cell state of the previous moment.
As an EEG trial is a time series, LSTM is helpful for analyzing it. In our method, we use the LSTM layer as one building block to encode the time series characteristics of each EEG trial as well as to learn long-term dependencies and avoid the vanishing gradient problem.
3.4. Proposed Deep Network
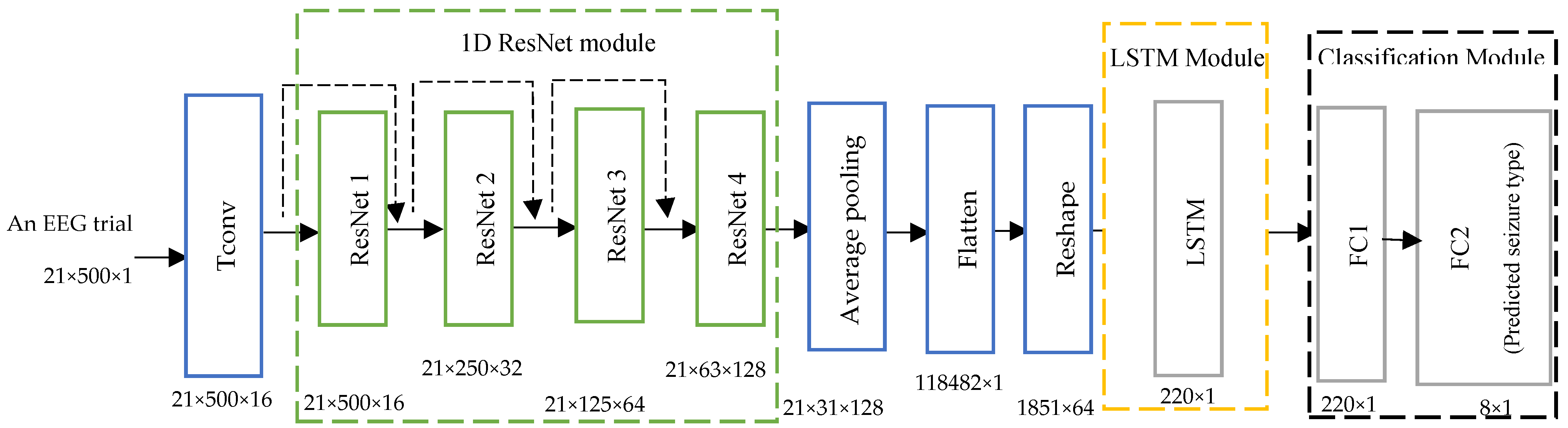
As mentioned, the CNN is one of the most successful deep learning-based models for analyzing EEG trials. However, a sequential CNN model suffers from overfitting and vanishing gradient problems because it involves a huge number of learnable parameters and uses conventional learning. Based on the residual learning theory and LSTM, we propose a deep network with fewer parameters to overcome these problems. The network consists of three main modules: 1D ResNet, LSTM, and the classification module, as shown in
Figure 3. A description of the architecture of each module is presented in the following paragraphs.
The architecture shows that the input trials are first processed by a temporal convolution (Tconv) layer to extract temporal features. This layer transforms EEG trials into a set of temporal feature maps that are then passed to the ResNet module.
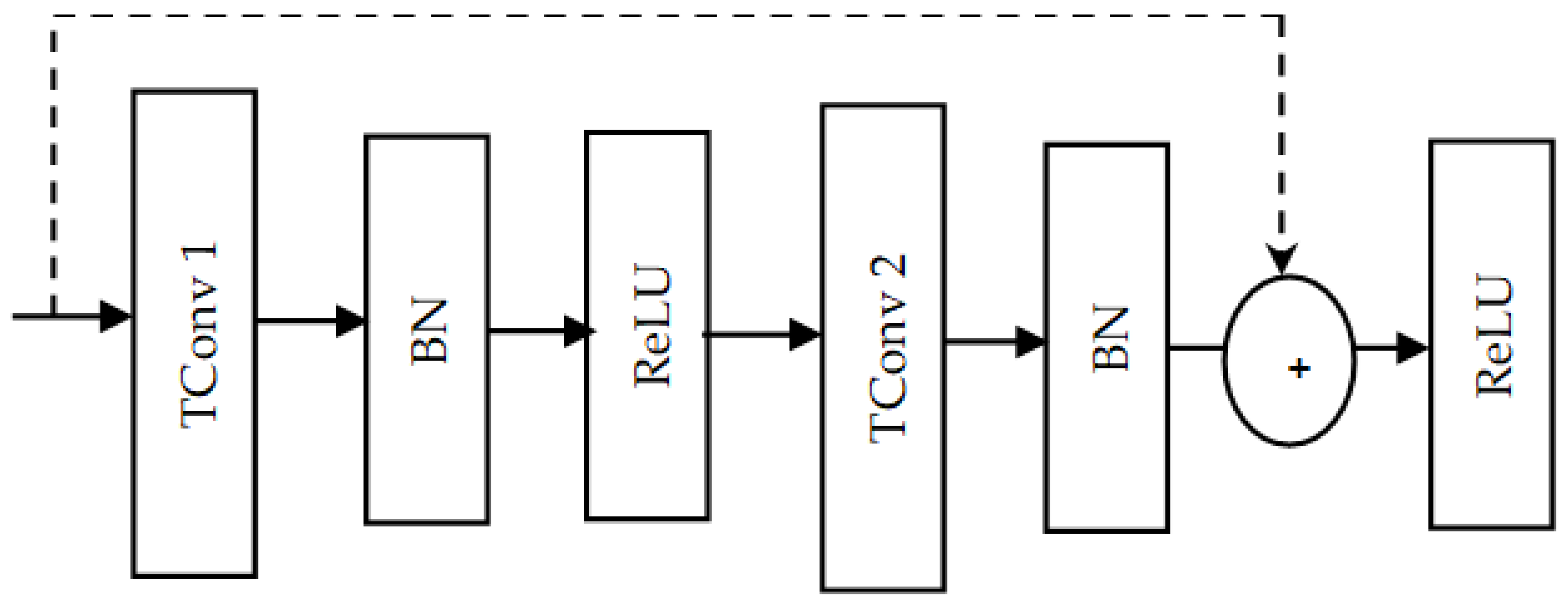
The architecture of the ResNet module, as shown in
Figure 3, consists of four 1D residual blocks. Each ResNet block has two temporal convolution (Tconv) layers, as shown in
Figure 4. The temporal convolution layers allow the network to learn and capture temporal features. Each temporal convolution layer is followed by a batch normalization layer that helps to improve the stability and speed of the training process by normalizing the activation. Two rectified linear unit (ReLU) activation functions are included in the ResNet module. The first ReLU is added after the first batch normalization layer, whereas the second ReLU is added after the summation step. The purpose of adding ReLU layers is to address the vanishing gradient problem. This in turn helps to reduce the computational complexity of the network.
As shown in the ResNet block architecture, skip (shortcut) connections are applied. The purpose of those connections is to help in solving the vanishing gradient problem. In a 1D ResNet block, the linear projection residual shortcut connection can be expressed as follows:
where:
x is the input of the residual block,
F(
x) is the output of the residual block without shortcut, and
represents the learnable weights (parameters) of
F(
x). In this formula, the linear projection
is used to match the dimension of both the input x and output
F(x). This is necessary when the dimensions of the output
F(
x) and the input x are different; in case
F(
x) and
x have the same dimensions,
is identity. This shortcut helps to solve the vanishing gradient problem by directly backpropagating the trials. Each residual block down-samples the EEG channels along the temporal dimension, so the ResNet module extracts the temporal features of the input signals at different resolution levels.
The output of the ResNet module is then fed into the average pooling layer. The average pooling layer with stride two is applied on the output from the 1D ResNet module to reduce the temporal dimension by half and hence down-sample the input. The purpose of using stride two is to avoid the loss of temporal features, which occurs when using a stride bigger than two. This, in turn, helps to reduce the number of parameters, prevent overfitting, and reduce the computational complexity. The output of the average pooling layer is then fed into the flatten layer.
The flatten layer is mainly applied within the deep neural network in transitioning from a convolution layer into a fully connected layer to convert the multi-dimensional input into a one-dimensional vector for passing it to the next block. The purpose of flattening the output is to fuse the temporal feature along each channel independently and then create a single long feature vector. Our method applies the flatten layer to convert the multi-dimensional output from the average pooling layer into a vector that concatenates the EEG channels. A reshape layer is used to transform the input vector into a time series shape suitable for the LSTM module.
This module consists of just a single LSTM layer. The architecture of the LSTM layer comprises a memory cell, input gate, output gate, and forget gate, as described in
Section 3.3. The input gate’s weights and biases control the new value’s flow into the memory cell. The weights and biases to the forget gate control the remaining value within the cell. In contrast, the weights and biases to the output gate control the use of the value in the computation of the LSTM output activation.
The LSTM layer is applied to solve the problem of the limited capacity of the ResNet module to learn longer-term dependencies by using additional gates to control what information in the hidden cell is to be transferred as output to the next hidden state. Hence, the LSTM permits the network to effectively learn and fuse long-term dependencies in the data and important temporal features. Moreover, the LSTM has low sensitivity to the time gap, so it efficiently analyzes temporal features learned by the ResNet module. The output of the LSTM module is then fed into the classification module.
This module consists of two fully connected layers, as shown in
Figure 3. The first fully connected layer is a time-distributed dense layer that is used to reduce the dimension of the output from the LSTM module. The output from this layer is then fed into the second fully connected layer that consists of eight neurons, according to the eight seizure classes, and uses a softmax function. It yields the probability of each class that is used to predict the class of the input EEG trials. In summary, two important modules are added to the proposed deep neural network model to obtain more efficient and accurate classification outcomes: ResNet and LSTM. The 1D ResNet block extracts temporal features at different resolution levels to increase the receptive field of each neuron. It helps to avoid the overfitting problem of CNN models when trained using training data consisting of a small number of input trials compared to a large number of learnable parameters. The LSTM, in turn, is used to learn, process, and classify sequential data to learn long-term dependencies between time steps of data.
4. Evaluation Procedure
For validating the effectiveness and performance of the proposed model to classify the type of epileptic seizure using EEG trials, it is trained and tested using the TUH database. Hence, this section initially introduces the TUH database. Next, the applied evaluation protocol and the evaluation metrics, which are applied to assess the trained deep network model performance, are described.
4.1. Dataset
The TUH corpus was produced to support research about interpreting EEGs using machine learning approaches [
29,
30]. It comprises clinical data collected at Temple University Hospital (TUH) from several sectors, including the epilepsy monitoring unit (EMU), the intensive care unit (ICU), the emergency room (ER), and outpatient services, from 2002 until the present. The used arrangement of electrodes in EEG recordings is according to the international 10/20 system. It includes 21 regularly distributed electrodes on the scalp, where the distance between two electrodes is either 10% or 20% of the complete distance between the nasion (front) and the inion (back) [
31].
The TUH database version 1.5.2, published in 2022, was adopted, which is the largest open-source, available dataset of seizure type and non-seizure. For the experiments, 21 channels (FP1, FP2, F3, F4, C3, C4, P3, P4, F7, F8, T3, T4, T5, T6, O1, O2, A1, A2, FZ, CZ, and PZ) are considered, where the total number of EEG files is 5612; 3050 of them are seizure files and the rest are non-seizure (NS) ones. Practically, the non-seizure events were deleted due to the focus on epileptic seizures only. Data were collected from 642 subjects, 242 of them suffering from seizures. The database includes eight seizure types (classes), which are: absence (AB), complex partial (CP), focal non-specific (FN), generalized non-specific (GN), simple partial (SP), tonic (TN), tonic–clonic (TC), and myoclonic (MY) seizures.
Table 1 shows the number of files in each class. The range of sampling rate varies from 250 Hz to 512 Hz.
The TUH database was used for experiments to learn and validate the model. All EEG trials in the database were initially preprocessed to standardize the data. This was based on resampling the EEG trials to 250 Hz because 87% of the EEG signals recorded were 250 Hz. Each EEG trial was then segmented into sub-trials using a fixed window of 2 s and a stride of 0.5 s so that there were enough trials to train the model, avoiding overfitting. Next, the SMOTE algorithm was applied to balance the minority classes, such as AB, TN, TC, and MY seizures, based on producing and increasing the trials to 500,000 [
32,
33].
4.2. Evaluation Protocol and Metrics
For the evaluation process, the TUH dataset was initially divided into a 90% training set and a 10% testing set based on the subjects. Next, 90% of the data were divided into 80% training and 10% validation sets. The data augmentation (SMOTE) was applied to 80% of the data to train the data to oversample the training set and solve the imbalanced data problem. The TUH database was divided into three core sets: 80% training set, 10% validation set, and 10% testing set.
The performance evaluation of the deep learning model for prediction and classification can be conducted using different evaluation metrics. One is the F1-score, a significant machine learning evaluation metric that measures the model’s accuracy. It depends on combining the precision and recall scores of the model. In other words, it balances the model precision and recall on the positive class, unlike the accuracy that looks at the correctly classified positive and negative observations. The F1-score can be computed after calculating four basic measures: true positives (TP), false positives (FP), true negatives (TN), and false negatives (FN). The TP represents the number of seizure trials that are correctly classified. The FN represents the number of seizure trials incorrectly classified as non-seizures. The TN represents the number of correctly classified non-seizure trials. Finally, the FP represents the number of non-seizure trials that are incorrectly classified as seizure trials. Using these measures, the accuracy, precision, recall, and F1-score evaluation metrics are computed as follows:
4.3. Training the Network
The EEG trials obtained from the TUH dataset were divided into 80% training set, 10% testing set, and 10% validation set based on subjects, i.e., data from 80% of the subjects were selected for training, 10% of the subjects for testing, and the remaining 10% of the subjects were used for validation. The three sets are disjointed, i.e., no subject is common in the three sets. The training set is used to learn the learnable parameters of the model, and the validation set is used to control the training process. The training process was conducted using the Google Cloud Platform. The learning rate used for training is based on the number of epochs adopted as follows:
For epochs > 40, the learning rate is multiplied by 1 × 10−1;
For epochs > 80, the learning rate is multiplied by 0.5 × 10−1;
For epochs > 130, the learning rate is multiplied by 0.25 × 10−2;
For epochs > 150, the learning rate is multiplied by 0.5 × 10−3.
The total number of epochs considered in the model is 190, with a batch size of 128. The Adam optimizer was adapted to fit the model by updating its weights iteratively based on the training data. The model classification outputs were then assessed using the testing set. The L2 regularization technique was used to prevent overfitting. In addition, the SMOTE algorithm was applied only to the training data to increase the number of training trials.
After training the model using training and validation sets, it was tested on the stand-alone test set.
5. Experiments and Results
The challenge was to determine the efficacy and performance of the proposed model in classifying the type of epileptic seizure using EEG signal trials. We performed several experiments to validate the suitability and effectiveness of the proposed model. The first experiment was choosing the most appropriate architecture configuration for the proposed model. Then, several experiments were conducted to show the effectiveness of the LSTM module and the hyper-parameters of the ResNet module and validate the effectiveness of data augmentation using the SMOTE algorithm. Then, other experiments were performed to reveal the most effective brain area and frequency band for identifying the seizure class.
5.1. Configuration of the Network
The first experiment was conducted to assess the architecture configuration of the deep network model (described in
Section 3) to classify seizure types automatically. The first layer of the proposed model is the input layer, which takes EEG trials represented as tensors of size C × T × 1, where C is the number of channels, T is the number of time stamps, and 1 is the depth. As mentioned in the Dataset section, the number of channels is 21, and the number of time stamps is 500 since the window size is 2 s and the sampling rate is 250 Hz.
Following the input layer, a temporal convolution layer processes the EEG trials to extract temporal features. This convolution layer converts the EEG trials from the input layer into 16 temporal feature maps, which are then fed into the ResNet module.
The number of filters (F) in 1D ResNet blocks starts at 16, and it is increased by a multiple of 2 in subsequent ResNet blocks, whereas the temporal kernel size is 1 × 7, as represented in
Table 2. The temporal kernel determines the number of time steps in the input sequence that the convolutional filter considers at a time. The purpose of the convolution layer is to extract the temporal feature from each channel. As described in the ResNet module in
Figure 1, there are four 1D ResNet blocks stacked on each other to extract the most informative and relevant temporal features at different scales from EEG trials. The time dimension is reduced by a factor of two from residual block two to the last block.
An average pooling layer with stride 2 further reduces the temporal dimension to half and effectively down-samples the input. It reduces the number of parameters to prevent the overfitting problem and avoid computational complexity. The LSTM layer in the LSTM module is designed to capture long-term dependencies in sequential data, which is common in EEG trials. The number of nodes used in the LSTM is 220, represented by (L) in
Table 2. The LSTM obtains input from the flatten layer that concatenates the temporal features along the time dimension.
Finally, FC1 and FC2 are the fully connected layers, where the number of neurons in the first fully connected layer is 110, while the second layer has 8 neurons with softmax activation to classify the trials into the 8 classes.
Table 2 shows the specification of each layer and the total number of parameters. The pseudocodes of the main modules are presented in
Appendix A.
This model achieved an F1-score of 97.4%. It can be noticed from
Table 2 that the total number of learnable parameters is equal to 516,652. This reveals the model’s ability to balance the difference between the small number of input trials used for model training and the total learnable parameters in the proposed model, unlike the millions of learnable parameters used in the state-of-the-art models [
6,
7], where 53 and 23 million learnable parameters were involved. Further, the ResNet and LSTM modules help solve the vanishing gradient problem based on learning long-term dependencies.
5.2. The Impact of LSTM Layer
This experiment focuses on examining the effect of using the LSTM module in the deep network model performance. The LSTM is a valuable addition to the model. It effectively learns, processes, and classifies sequential data due to its ability to learn long-term dependencies between time steps of data. The results in
Table 3 show that higher performance is achieved with the architecture containing the LSTM module. In other words, it reveals that the LSTM layers can effectively model the temporal dependencies between the EEG trial samples, leading to improved classification performance.
The results of experiments 6, 7, and 8 in
Table 3, when the LSTM module is used with four stacks in the ResNet module, indicate that the model performance is enhanced by learning more temporal long-term dependencies. On the other hand, considering the results of experiments 7 and 8 for the same model hyper-parameters and different numbers of LSTM nodes, an F1-score of 97.4% is achieved for experiment 8 with 220 LSTM neurons, while it is 96% for experiment 7 with 250 neurons. This indicates that increasing the number of nodes leads to overfitting the network and reduces the model performance.
5.3. The Selection of Hyper-Parameters
Five experiments were conducted to tune different hyper-parameters of the deep network architecture, including the number of ResNet stacks in the ResNet block, window size, and kernel size; the results are shown in
Table 4. The first experiment reveals that for the model with three blocks in the residual module when EEG trials are segmented by a window size of one second and a stride of a quarter second, an F1-score of 80% was achieved. From the second experiment onwards, the window size was kept at two seconds, while the stride was kept at half a second. Increasing the window size and overlapping led to higher performance. It is because longer windows can contain more information and capture more data per interval.
The window size used to capture the EEG trials significantly impacts the model performance, as demonstrated by the higher performance values in cases where larger window sizes are used, as shown in
Table 3 and
Table 4. It is because capturing more EEG trials over a longer period can help improve classification accuracy.
It can be noticed from
Table 3 and
Table 4 that a larger temporal kernel size results in a higher performance due to its ability to capture longer-term dependencies in EEG trials. Practically, a temporal kernel size of 7 allows for smoothing and filtering of the EEG trials, which suffer from noise.
It can also be noticed from
Table 3 and
Table 4 that stacking four layers in 1D ResNet helps to efficiently learn the complex temporal features of the EEG trials and create more accurate results than stacking three layers. Furthermore, stacking four layers can enhance the richness of features since the deeper layers can capture more temporal features. It can help to reduce the overfitting problem and improve the performance, as we can see from experiments three onwards.
5.4. The Impact of Data Augmentation
This experiment investigates the effect of applying the data augmentation method, SMOTE, on the model performance. The SMOTE algorithm is one of the efficient oversampling methods used to solve the data imbalance problem. It focuses on balancing the class distribution based on randomly increasing the minority class examples by replicating them and synthesizing new minority instances between existing minority instances.
It is evident from the outcomes of experiments in
Table 3 that the highest achieved performance F1-score is 97.4% in the case when all the classes are balanced. It is due to the use of the SMOTE algorithm to avoid the overfitting problem and balance the minority classes, AB, TN, TC, and MY seizures. This algorithm increases the number of trials by taking the difference between the trial and its nearest neighbor. In this configuration, the three nearest neighbors used SMOTE.
Based on comparing the results in
Table 3 and
Table 4, using the data augmentation SMOTE algorithm significantly impacts deep network model performance. By artificially increasing the size of the training dataset, the SMOTE algorithm can help to prevent the overfitting problem and improve the generalization capabilities of the network. Moreover, comparing the results of experiments 1 and 2 in
Table 3 and experiments 4 and 5 in
Table 4 reveals that although the same values of hyper-parameters are used in each experiment, the application of the SMOTE algorithm in experiments 1 and 2, shown in
Table 3, slightly enhances the performance.
5.5. The Final Network and the Results
This work presents the design of an efficient, lightweight, and expressive deep network model to classify epileptic seizure types. It was revealed that combining the model with the ResNet module, LSTM module, and the SMOTE algorithm achieved the highest F1-score results. The ResNet module is designed and added to the model to train a much deeper network without encountering vanishing gradient problems and avoid the overfitting problem of CNN models. The LSTM module, in turn, can handle learning long-term dependencies and prevent the vanishing gradient problem. The SMOTE algorithm can avoid the overfitting problem and provide enhanced performance based on balancing the data by increasing the minority classes.
The proposed model was applied to the TUH database, where the Adam optimizer was used for model fitting. The F1-score was adopted as the main evaluation indicator for seizure type classification.
Table 5 below summarizes the seizure type classification results obtained on the TUH database. It can be noticed that the proposed deep learning model achieved a 97.4% F1-score in classifying EEG signals into the eight main seizure classes.
6. Analysis of the Performance of the Network
The effect of frequency bands on the performance of the proposed deep network is discussed to analyze the performance of the proposed deep network. Moreover, a confusion matrix and t-SNE plot were adopted to analyze the proposed model’s decision-making mechanism. The performance analysis was also based on illustrating and discussing the number of learnable parameters. Finally, a comparison was then conducted between the proposed network and state-of-the-art works.
6.1. The Effect of Frequency Bands
EEG trials was decomposed into five core component frequency bands (delta 1–3 Hz, theta ( 4–7 Hz, alpha 8–12 Hz, beta 13–30 Hz, and gamma 30–100 Hz). Therefore, this section focuses on comparing the impact of these bands on the deep model performance.
The delta band is associated with slow wave activity and is important for monitoring long-term changes in EEG trials. The high delta achieved an F1-score of 82%, showing that the deep network model performs well in detecting slow wave activity in EEG trials, as shown in
Table 6.
Theta and alpha F1-scores are 12% and 18%, respectively. Theta waves are slower than alpha but faster than delta waves. However, they demonstrate that the deep network model is not performing well. A good beta F1-score of 40% suggests that the deep network model may perform well with the beta band. Finally, an F1-score of 33% was recorded for the gamma frequency band, indicating that the model is inaccurate in classifying the fastest brain wave of the gamma band.
6.2. ROC Curve
The receiver operating characteristic (ROC) curve is a graphical representation of a binary classification model’s diagnostic ability and performance at all classification thresholds. It plots the true positive rate of positive samples in the
Y-coordinate and the false positive rate along the
X-coordinate at different classification thresholds. The true positive rate is a synonym for recall, where it equals the TP divided by the summation of TP and FN. In contrast, the false negative rate equals the FP divided by the summation of FP and TN. Lowering the classification threshold results in classifying more items as positive, thus increasing both false positives and true positives. Moreover, the closer the area under the curve within the plot to 1, the better the overall model performance, where a test with an area under the curve (true positive rate) value of 1 is perfectly accurate. The practical lower limit for the ROC is 0.5, which is represented by the dotted line segment from (0, 0) to (1, 1) in the curve. The proposed model was evaluated using the ROC curve for all eight classes and both the training and test ROC curves obtained using the one-against-all strategy for each class are shown in
Figure 5.
For the training ROC curve shown in
Figure 5a, the model recorded an average ROC true positive rate of 0.99. Moreover, a perfect performance was revealed for all classes, as illustrated in the upper left corner of the graph. A true positive rate of 1 is recorded for classes MY and AB, indicating a perfectly accurate classification. A true positive rate of 0.99 is recorded for classes TC, SP, and TN, with a false positive rate of 0.01. Additionally, the recorded true positive rate for classes GN, FN, and CP is 0.98, with a false positive rate of 0.02. Hence, the model revealed a negligible false positive rate for classes TC, SP, TN, GN, FN, and CP and a zero false positive rate for classes MY and AB. Therefore, the model is promising in distinguishing between positive and negative samples. In contrast, the test ROC curve revealed a positive rate of 0.99 for class MY and 0.95 for classes GN, FN, and CP, while the false positive rate is between 0 and 0.03 for those classes at that point. Hence, the proposed model achieved a promising performance in distinguishing between positive and negative samples with an average ROC true positive rate of 98%.
6.3. Confusion Matrix
Generally, the confusion matrix is an efficient performance measurement for machine learning classification. It allows visualizing and summarizing a classification algorithm’s performance and permits measuring recall, precision, and accuracy. The purpose of applying the confusion matrix for classification algorithms is to show how much confusion there is between positive (correctly classified) classifications and negative (misclassified) classifications. Therefore, the results of classifying the eight seizure type classes are validated using the confusion matrix shown in
Figure 6.
It can be noticed from
Figure 7 that 1973 GN seizure trials are correctly classified as GN, while around 65 are misclassified as FN. Around 3999 FN seizure trials are correctly classified as FN, while 79 are misclassified as GN. It indicates that there is some confusion between the classes FN and GN.
Moreover, it can also be noticed that there are also misclassifications between the CP and FN classes. These misclassifications between these three classes are mainly due to the overlapping or similarity of their characteristics. It means that there are similar patterns or features between these classes that are difficult for the model to differentiate. On the other hand, the other classes, TC, SP, MY, AB, and TN, are mostly correctly classified with few or no misclassified data.
6.4. Analysis of Features Learned by the Network
The t-distributed stochastic neighbor embedding (t-SNE) is a non-linear, powerful, statistical visualization technique that assists in visualizing high-dimensional data and finding patterns by projecting them in lower dimensions. Thus, a 2D t-SNE model was applied in this work to visualize the distribution of classified data over the eight classes, as shown in
Figure 7. It can be noticed that the samples of eight classes are well separated and clearly clustered, as we can see in the plot. There is a clear separation between classes, especially AB and TC. However, there are some outliers from classes FN and GN moving to other classes, as described in the confusion matrix. In addition, FN and GN seizure classes are close to each other. It indicates an overlap in their respective features.
6.5. Visualization of the Decision-Making Process of the Model
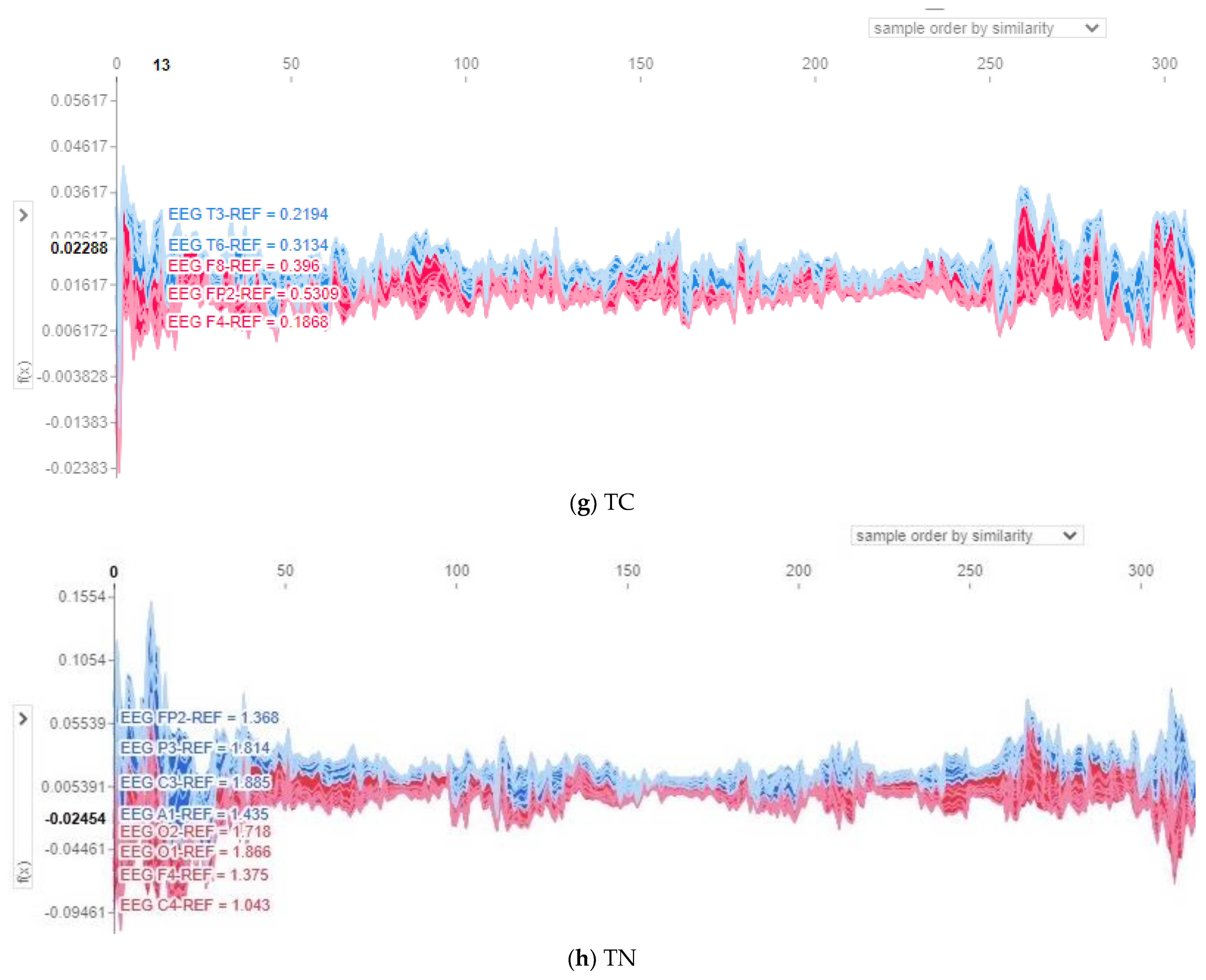
The proposed deep network model is like a black box, in which it is difficult to recognize how it classifies input data. Therefore, Shapley additive explanations (SHAP), which is an individualized, interpretable, model-agnostic explainer, is applied in this paper to explain the proposed model by assuming it as a black box without knowing how it internally works. SHAP is based on computing the contribution of each input channel of the input trials to the output and finding out the most vital channels and their impact on the model output. SHAP is based on finding the Shapley values of channels. In other words, after obtaining the model output, SHAP values are computed for each channel per input trial. Next, each channel with its related SHAP values contributes to pushing the model output from that predicted output up and down.
SHAP force plots are used to visualize the SHAP values and show how features explain the model output and which features had the most impact on the model output for a single observation. In this paper, multi-output force plots are presented for all data points. Force plots are flipped to be vertical and then stacked against each other for the eight classes, as shown in
Figure 8, to illustrate how channels positively or negatively affect the model output. In all presented multi-output force plots, red bars illustrate channels that increase the predicted value above the baseline, whereas blue bars represent channels that decrease the predicted value. Moreover, the
X-axis represents the input instances sorted by similarity, while the
Y-axis shows the output prediction values of the model before being converted into probabilities.
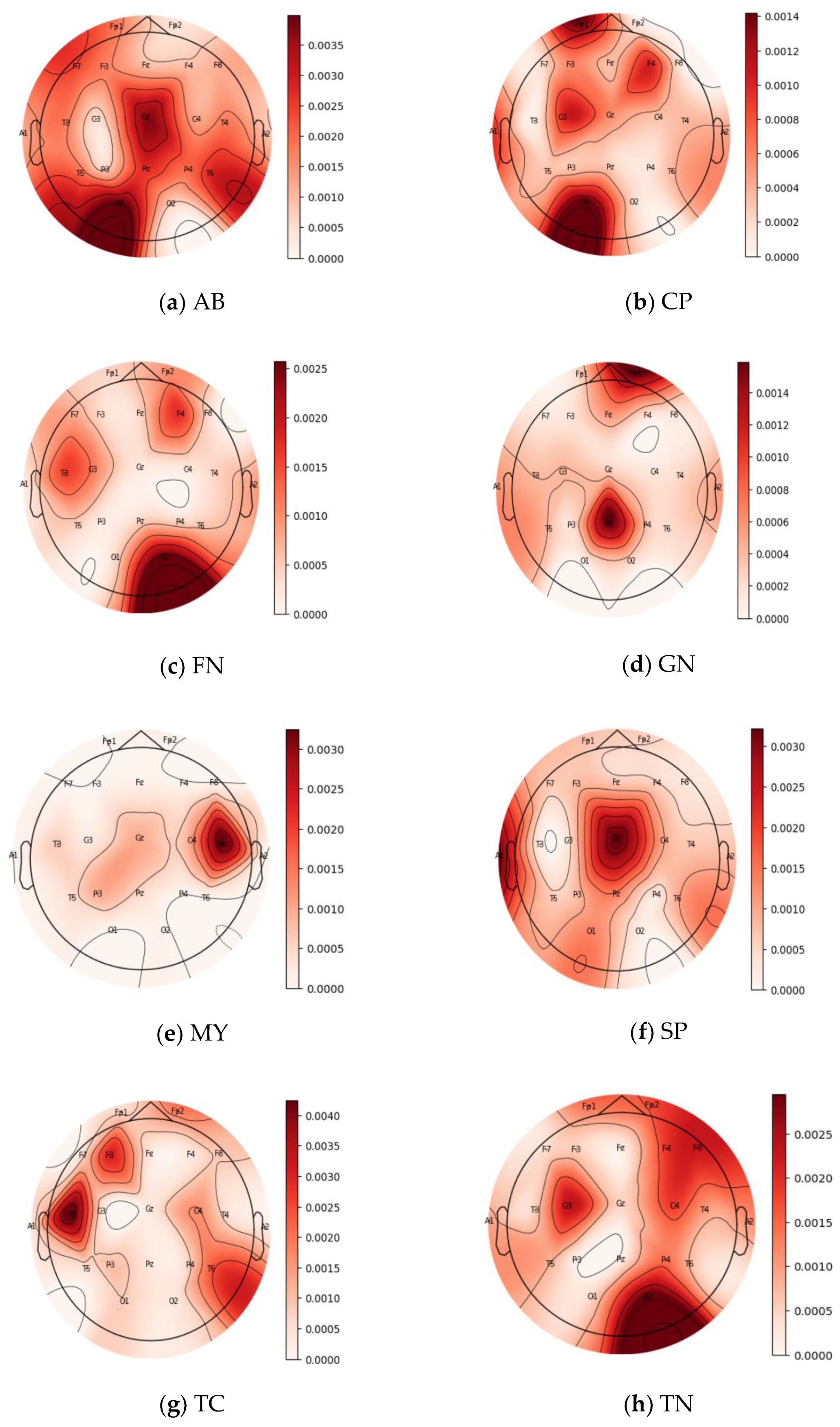
In this section, both the force plots and topo maps are adopted to represent the contribution and impact of the 21 channels (FP1, FP2, F3, F4, C3, C4, P3, P4, F7, F8, T3, T4, T5, T6, O1, O2, A1, A2, FZ, CZ, and PZ) of EEG trials for each one of the eight seizure classes; AB, CP, FN, GN, SP, TN, TC, and MY. In the 21 channels, F stands for frontal, C for central, T for temporal, P for parietal, and O for occipital. Moreover, the odd numbers represent the brain’s left hemisphere, the even numbers represent the right hemisphere, and Z stands for center channels.
Figure 8 and
Figure 9 show the force plots and topo maps of the eight seizure classes, respectively. For the topo maps, we used the activations of the ResNet4 block of the ResNet module to plot topo maps for visualizing the model’s decision-making process. Topographic (topo) mapping is an efficient, non-invasive technique to display the spatial distribution of the brain’s electrical activation. Topo maps, also called contribution maps, are mainly visualized as color maps. In these maps, values (contribution amplitudes) are mapped as colors after normalization, where the darker the color, the higher the contribution is. Those values are measured by placing a cap of 21 sensors on the patient’s head, including small electrodes that record the electrical patterns coming from the brain.
For class AB, as shown in
Figure 8a, contributions spread over almost all channels with varying contribution values. This is also clear in the topo map of class AB, shown in
Figure 9a, represented in dark colors. It is obvious that channels O1, T6, F7, and T5, represented in
Figure 8a and in the darkest color in
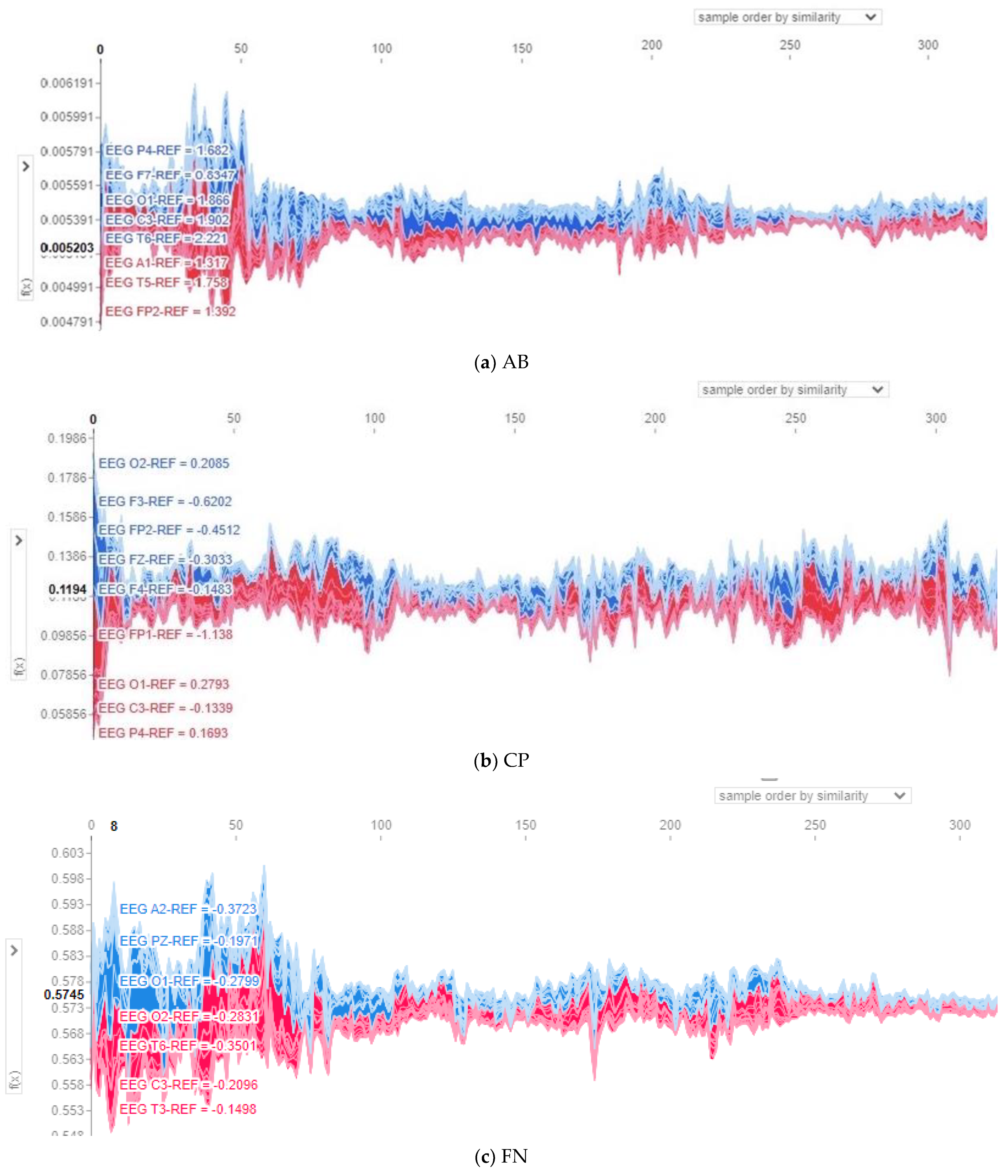
Figure 9a, have a positive impact on the model output and recorded magnitude of contribution in the range of 0.004 to 0.006 to the predicted label. For class CP shown in
Figure 8b, channels O1, C3, FP1, and F4 positively impact the model output, which is also proved in the topo map of class CP shown in
Figure 9b. The revealed contribution magnitude for those channels ranges from −0.13 to 0.17. For class FN, as illustrated in
Figure 8c, channels O2, C3, and T3 positively impact the model output and reveal contribution magnitude in the range from −0.2 to 0.3. Those channels are also revealed in a darker color in the topo map of class FN shown in
Figure 9c. It is obvious from the force plot of class GN shown in
Figure 8d and the topo map shown in
Figure 9d that contributions concentrate in two channels only, FZ and FP1, where those channels have a positive impact on the model output and reveal contribution magnitude in the range from 0.04 to 0.06.
The contribution in the MY class concentrates on channel T4 only with −0.47 magnitude, as shown in the force plot in
Figure 8e and the topo map in
Figure 9e. The PZ channel is the most effective for the SP class, as shown in
Figure 8f, with a −0.08 contribution magnitude. This is also clear in the topo map of the class shown in
Figure 9f. The force plot of class TC shown in
Figure 8g revealed that channels T3, T6, and FP2 have a positive impact on the model output and reveal contribution magnitude in the range from 0.2 to 0.5. This is also clearly indicated in the topo map shown in
Figure 9g. Finally, the most effective channels for class TN, as shown in
Figure 8h and
Figure 9h, are O2, C3, FP2, F4, and C4, with contribution magnitudes in the range from 1.05 to 1.7. Hence, it is obvious from these results that all channels in EEG trials have good contributions in each seizure class. These findings are in accordance with clinical findings [
8]. Moreover, EEG trials are able to classify different types of seizures.
6.6. Comparison with the State-of-the-Art Methods
Table 7 shows the performance of the proposed deep learning model and a comparison with the state-of-the-art models on the same public dataset, the TUH dataset. Our model and those of Raghu et al. [
20], 2019, Raghu et al. [
6], 2020, Roy et al. [
5], 2019, and Ma et al. [
22], 2023, classified the input EEG trials into eight classes. Our model considered the highest number of channels (21 channels) compared with all other state-of-the-art models.
Moreover, all previous models used 2D CNNs, which resulted in an overfitting problem due to the huge number of learnable parameters (in millions) compared to the small number of input trials. In comparison, our model applied both 1D ResNet and LSTM modules and the SMOTE algorithm to avoid the computational complexity and the overfitting problem by balancing the number of input trials and the number of learnable parameters, which is 516,652 parameters. The work by Ma et al. [
22] considered using transformers for detecting seizure types. Although it recorded a high accuracy outcome, our method still outperforms.
Considering the revealed F1-score and accuracy results, our method recorded the highest F1-score compared with the state-of-the-art models, indicating that our method has the best performance and is the most precise model in classifying seizure types. Although the work of Liu et al. [
21] recorded a similar F1-score, it still suffers from complexity and overfitting problems because of the model complexity.
It can be observed that our method overall outperforms the state-of-the-art models. It uses the 1D ResNet module and SMOTE algorithm to avoid the overfitting problem and ensure data balance. It uses the LSTM module to learn long-term dependencies, avoid the vanishing gradient problem, and record the highest F1-score value. It is an efficient and accurate model that allows neurologists to effectively identify the types of seizures and decide on the most applicable treatment and therapy for each patient, improving patients’ quality of life.
7. Discussion
Based on the results, it is clear that the proposed deep network model efficiently classifies the eight types of epileptic seizures using two fully connected layers with an F1-score of 97.4%. It is a useful method for neurologists to decide the most appropriate treatment and therapy for each patient and improve their quality of life. Results revealed a slight misclassification among three classes, i.e., GN, FN, and CP, as noticed in both the confusion matrix and t-SNE plots. It is due to the overlapping or similarity of their characteristics. In other words, similar patterns or features between these classes are difficult for the model to differentiate.
Most of the proposed models use 2D CNNs for feature learning. Although CNNs can automatically learn features by passing the training data through multiple convolutions, it is a time-consuming process that suffers from a vanishing gradient problem that makes it unable to learn long-term dependencies. Moreover, using 2D CNNs results in overfitting problems due to the huge number of learnable parameters (in millions) compared to the small number of input trials. To avoid these problems, our model comprises two core modules, 1D ResNet and LSTM modules, along with the SMOTE algorithm. The 1D ResNet module is adopted to train deep networks and avoid both the vanishing gradient problems and the overfitting problem of CNN models when trained using training data consisting of a small number of input trials. The ResNet module is combined with the LSTM to encode long-term dependencies to solve the vanishing gradient problem successfully.
Further, to solve the overfitting problem, the SMOTE algorithm was adopted to enhance the model performance by balancing the data by increasing the minority classes and samples by using an overlapping window of 2 s and a stride of 0.5 s. It decreased the number of learnable parameters from millions to only 516,652 parameters. The proposed model was fitted using the Adam optimizer, an efficient replacement optimization algorithm for stochastic gradient descent for training deep learning models.
The performance of the proposed model was greatly enhanced by combining the ResNet module with the LSTM layers that effectively model the long-term temporal dependencies between EEG trials over time. The number of used LSTM nodes has a noticeable impact on the model performance, where the F1-score value increased from 96% to 97.4% when the number of LSTM nodes decreased from 250 to 220. This indicates that increasing the number of nodes led to overfitting the network, degrading the model performance.
The appropriate selection of hyper-parameters, such as the number of ResNet stacks in the ResNet block, window size, stride, and kernel size, also revealed a major impact on the model performance. Results show that stacking four layers in 1D ResNet instead of three layers helps learn the complex temporal features of the EEG trials and create more accurate results. Stacking four layers enhances their usage since deeper layers can capture discriminative temporal features and reduce the overfitting problem. Moreover, a positive relation was revealed between the model performance and the stride and window size. Another positive relation was shown between the model performance and the temporal kernel size due to its ability to capture longer-term dependencies in EEG trials. A temporal kernel size of 7 was the best choice since it allows smoothing and filtering of the EEG trials that suffer from noise.
One important addition that improved the proposed model performance is the SMOTE algorithm, where for two conducted experiments with the same values of used hyper-parameters, the F1-score was the highest for the experiment that adopted the SMOTE algorithm. It is due to its ability to prevent the overfitting problem and improve the generalization capabilities of the network. In summary, the highest achieved model performance was with an F1-score of 97.4% when both the LSTM module and the SMOTE algorithm were combined with the 1D ResNet model stacking four 1D ResNet layers and choosing a kernel size of 7 with half stride and a window size of 2.
The analysis of the model performance showed that among five core EEG component frequency bands (delta 1–3 Hz, theta ( 4–7 Hz, alpha 8–12 Hz, beta 13–30 Hz, and gamma 30–100 Hz), the delta band is associated with slow wave activity, and it is important for monitoring long-term changes in EEG signal. The highest F1-score was revealed for the delta band, which shows that the deep network model performs well in detecting slow wave activity in EEG signals.
Table 8 summarizes a comparison between the proposed and state-of-the-art models and both the advantages and disadvantages of each model.
During a seizure, there is abnormal electrical activity in the brain that can be detected by EEG signals. The specific EEG findings can vary depending on the type and location of the seizure types. The results of employing deep learning algorithms to classify seizure types from EEG recordings proved their clinical acceptability, adaptability, and effective help to neurologists in their decisions in various relative state-of-the-art models. Liu et al. [
21] showed that their seizure type classification system could assist clinical specialists in diagnosing the disease, decreasing time taken, and enhancing accuracy and reliability. The proposed system combines CNN and RNN modules trained using one-second EEG. When ResNet and LSTM modules are combined, they lead to a robust classification system for seizure type classification. This can offer valuable clinical assistance to neurologists in their analysis to determine the correct treatment plans and precise drug prescriptions for patients based on the type of seizure they suffer from.
8. Conclusions
In conclusion, the design of an efficient, lightweight, and expressive deep network model to classify epileptic seizure types is presented in this paper. The model solves the problems of previous state-of-the-art models by adopting three essential components that complement each other with their strengths to produce an efficient model with improved performance: 1D ResNet module, LSTM module, and SMOTE algorithm. The 1D ResNet module is used to train a deep network without encountering vanishing gradient problems and avoiding the overfitting problem of CNN models when trained using training data consisting of a small number of input trials compared to a large number of learnable parameters in the NNs. The LSTM module encodes long-term dependencies and avoids the vanishing gradient problem. The SMOTE algorithm is applied to avoid the overfitting problem and obtain enhanced performance based on balancing the data by increasing the minority classes and samples by using an overlapping window of 2 s and a stride of 0.5 s.
Moreover, among the different frequency bands, the model performance outcomes show that the highest F1-score (82%) is due to the delta of 1–3 Hz. The model was evaluated using the TUH database, accuracy, and F1-score with 4 stacks, kernel size of 7, and 220 LSTM nodes. The proposed method showed the highest F1-score of 97.4%. This lightweight model can help neurologists to detect seizure types effectively, improve their clinical decisions, and determine the best treatment for each patient based on the seizure type. Future work will incorporate attention mechanisms in the proposed model to further enhance its effectiveness. In addition, we will address which frequency bands play a key role in the discrimination of different seizure types.