Fully Automatic Segmentation, Identification and Preoperative Planning for Nasal Surgery of Sinuses Using Semi-Supervised Learning and Volumetric Reconstruction
Abstract
:1. Introduction
2. Methods and Experimental Data
- (1)
- Training dataset: For the labeled dataset, the number of images was doubled by the data augmentation method. The basis of the gradient descent backpropagation algorithm was calculated by the input image and actual labeling result.
- (2)
- Validation dataset: For the labeled dataset, the gradient descent backpropagation algorithm was not calculated. Only the difference between labeling and prediction was calculated in each iteration to evaluate the effect of the model in every iteration.
- (3)
- Semi-supervised learning dataset: For the unlabeled dataset, a dataset for generating pseudo-labels after a model was trained.
- (4)
- Testing dataset: The unlabeled dataset was a dataset for evaluating effectiveness after model training was completed. Doctors would give evaluation scores to confirm model accuracy.
2.1. Image Data Augmentation
2.1.1. Gaussian Blur
2.1.2. Gaussian Noise
2.1.3. Mixup
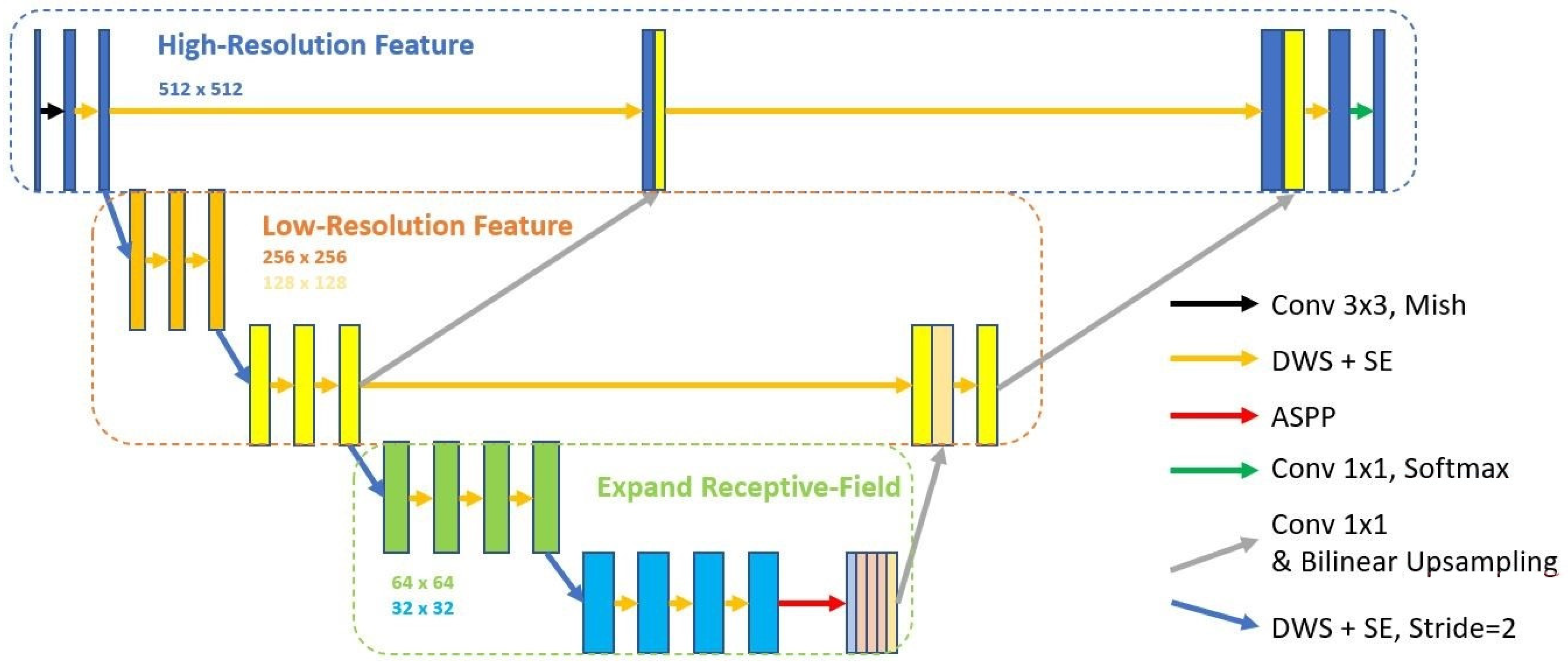
2.2. Semi-Supervised Learning
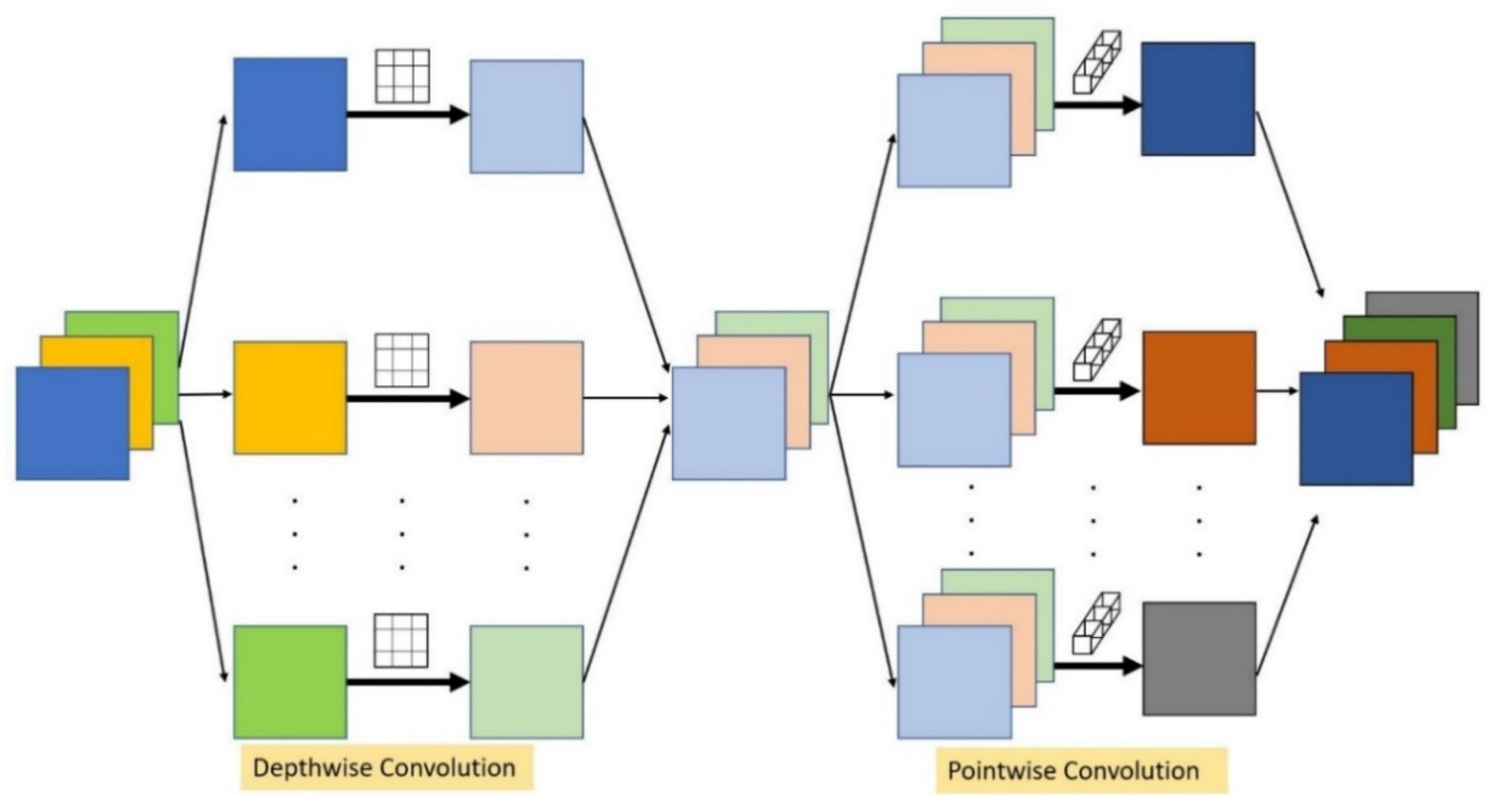
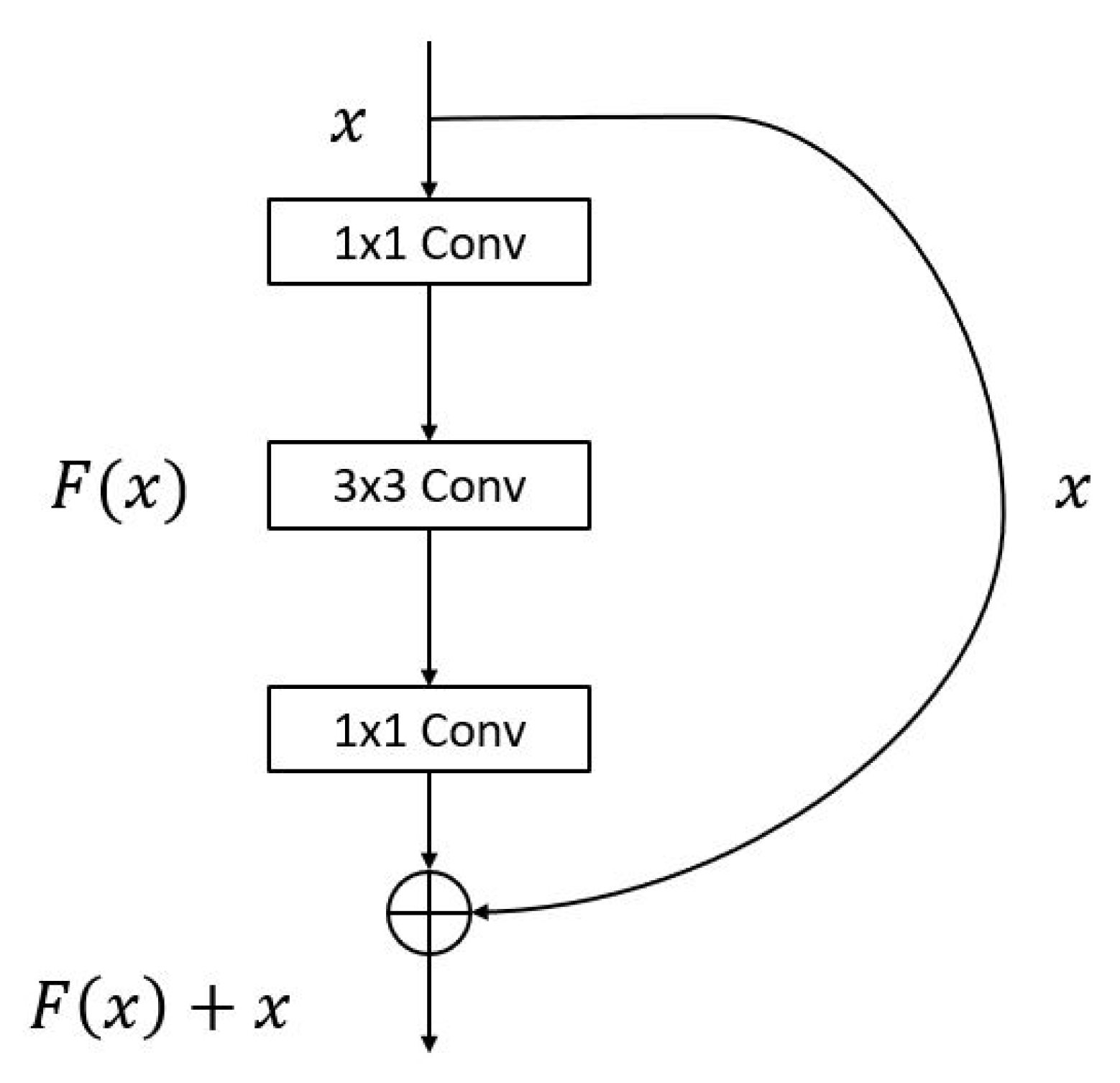
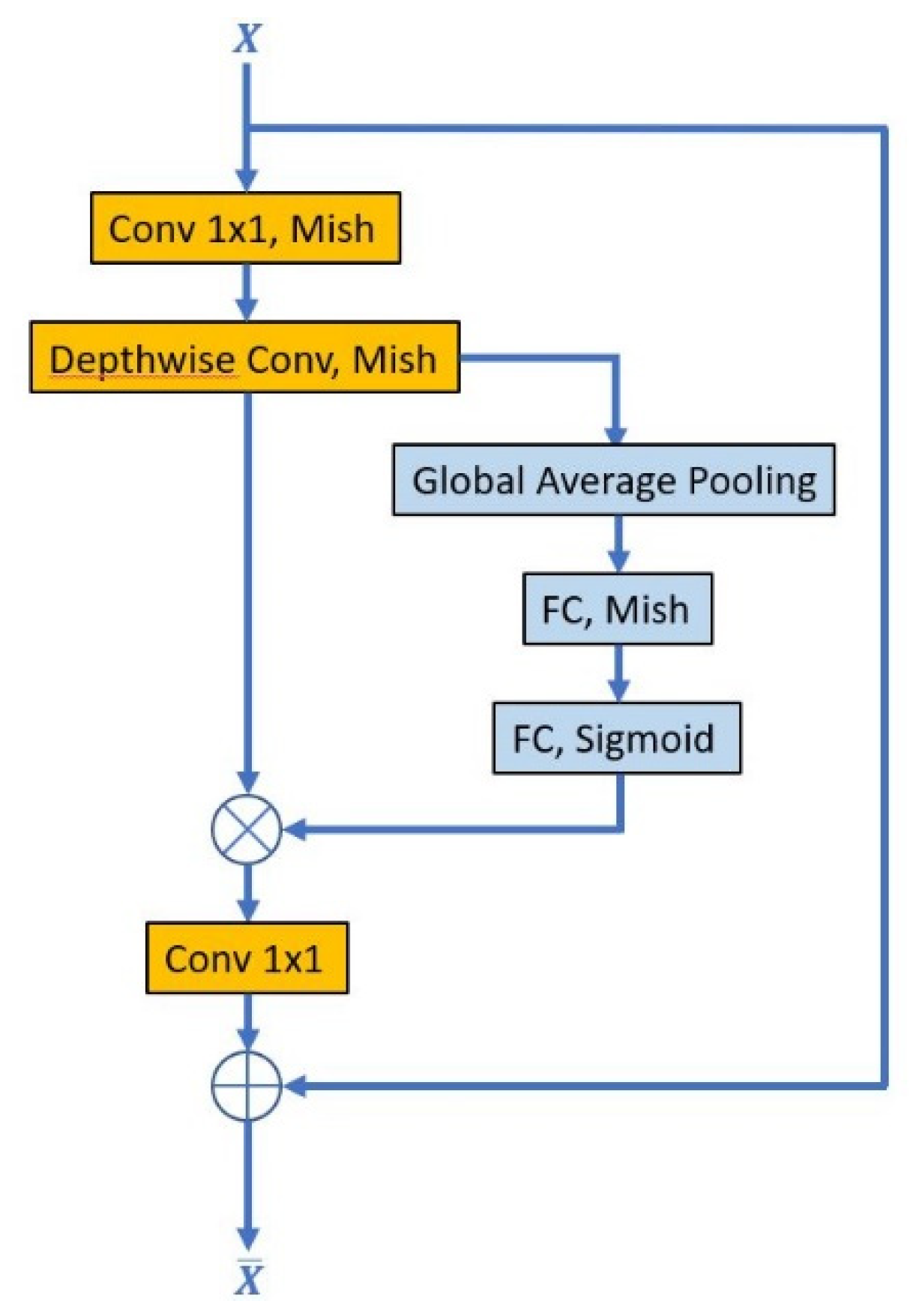
2.2.1. Convolution Layer
2.2.2. Activation Function
2.3. K-Fold Cross-Validation
2.4. Morphology
2.4.1. Erosion and Dilation
2.4.2. Opening
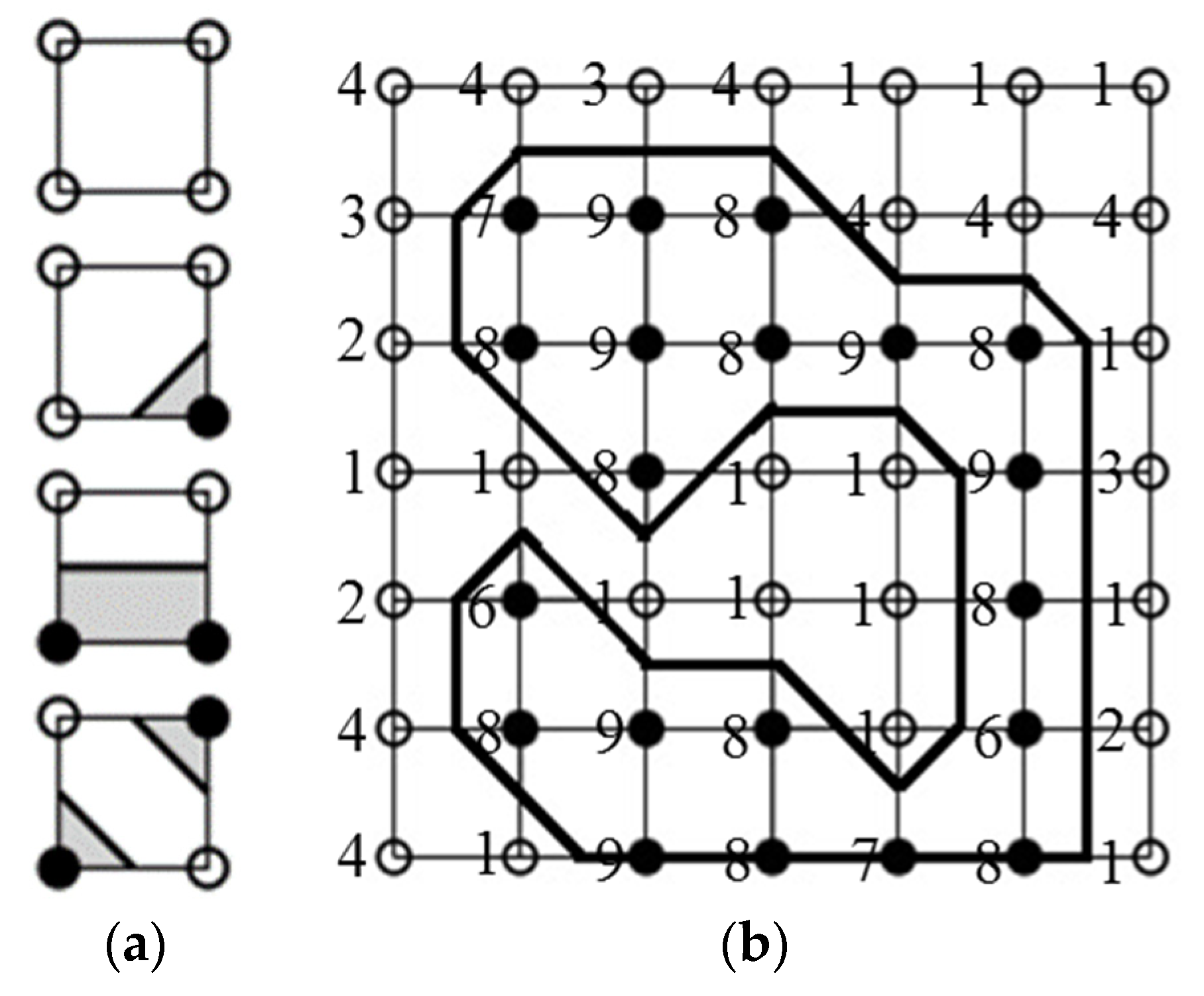
2.4.3. Labeling
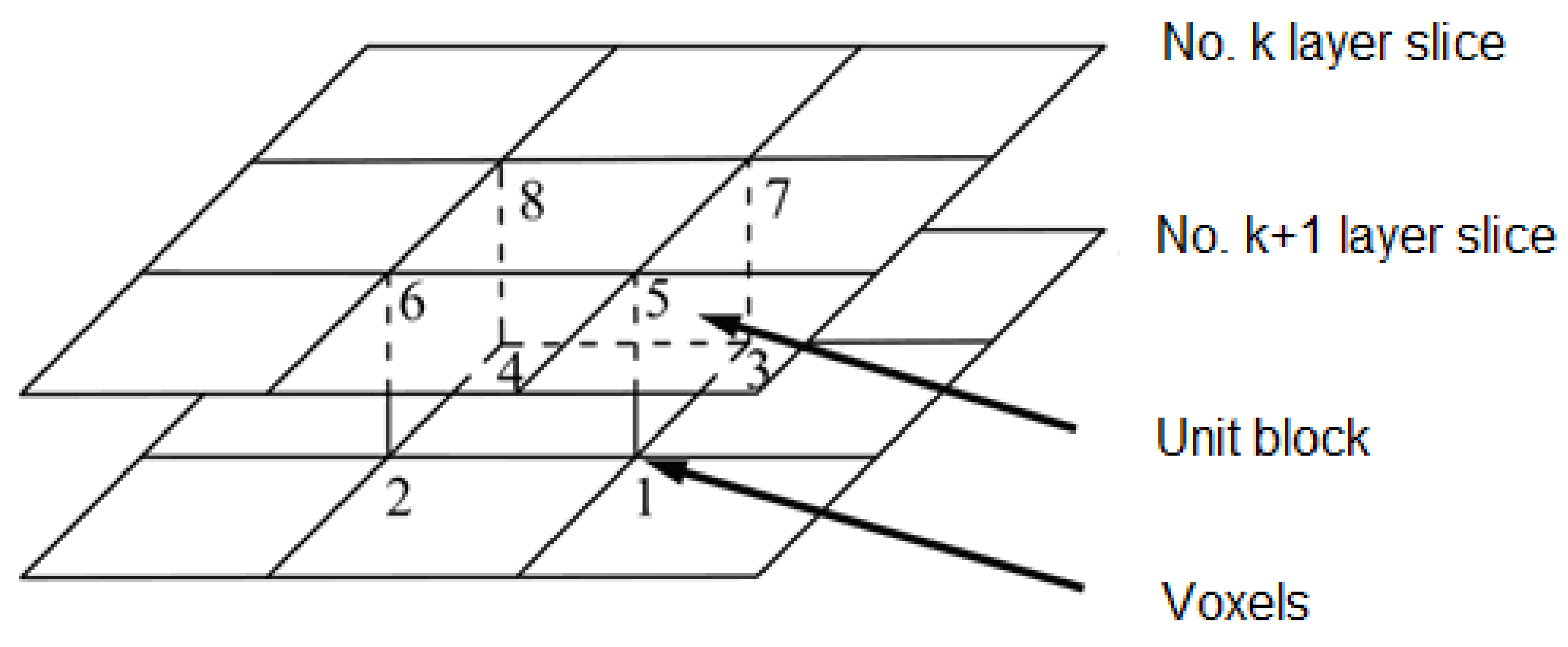
2.5. Marching Cube 3D Reconstruction
2.6. Implementation Challenges
3. Results
3.1. Image Data Augmentation
3.1.1. Gaussian Blur
3.1.2. Gaussian Noise
3.1.3. Mixup
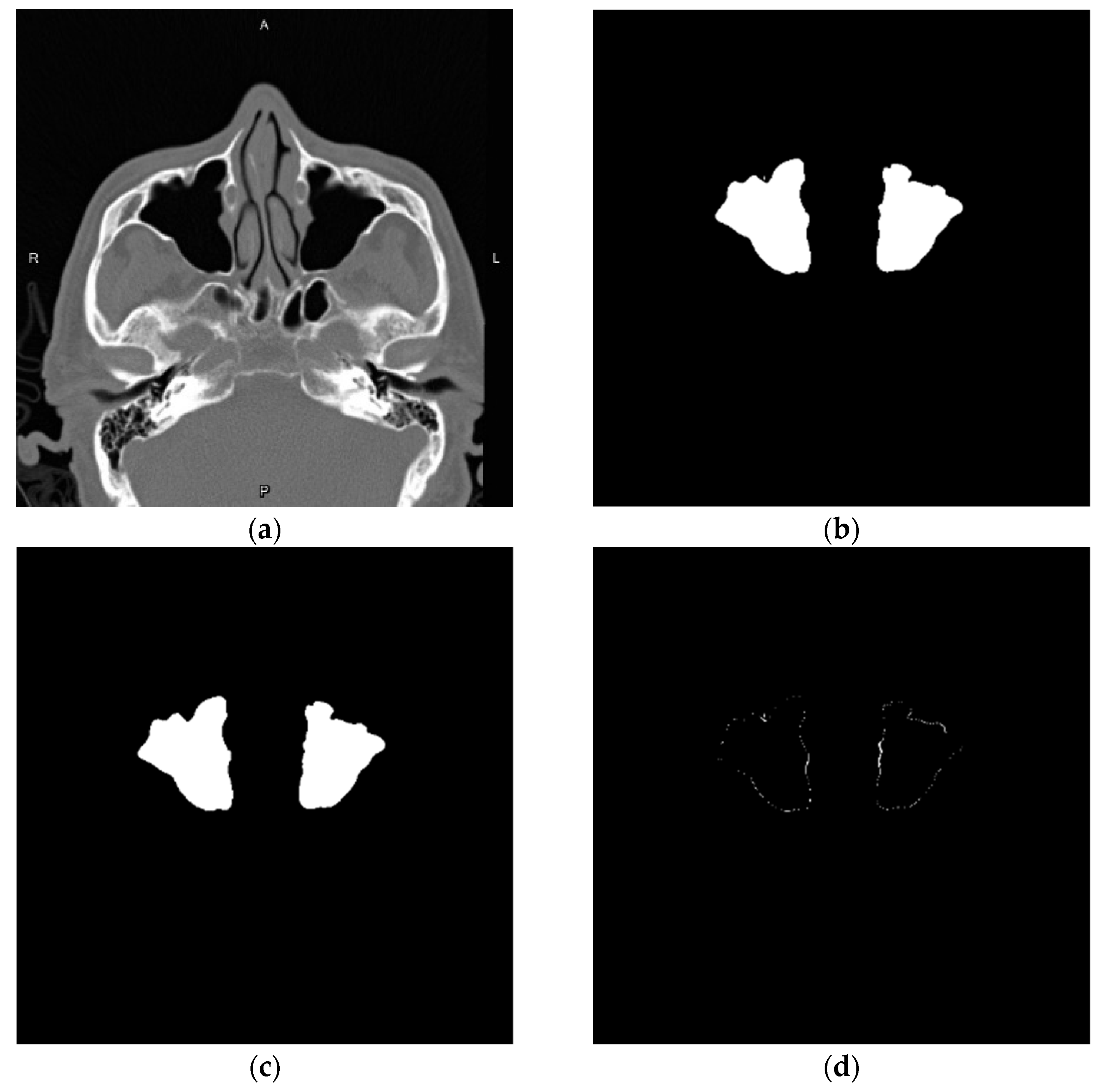
3.2. Nasal Sinus Region Segmentation
3.2.1. Loss Function Selection
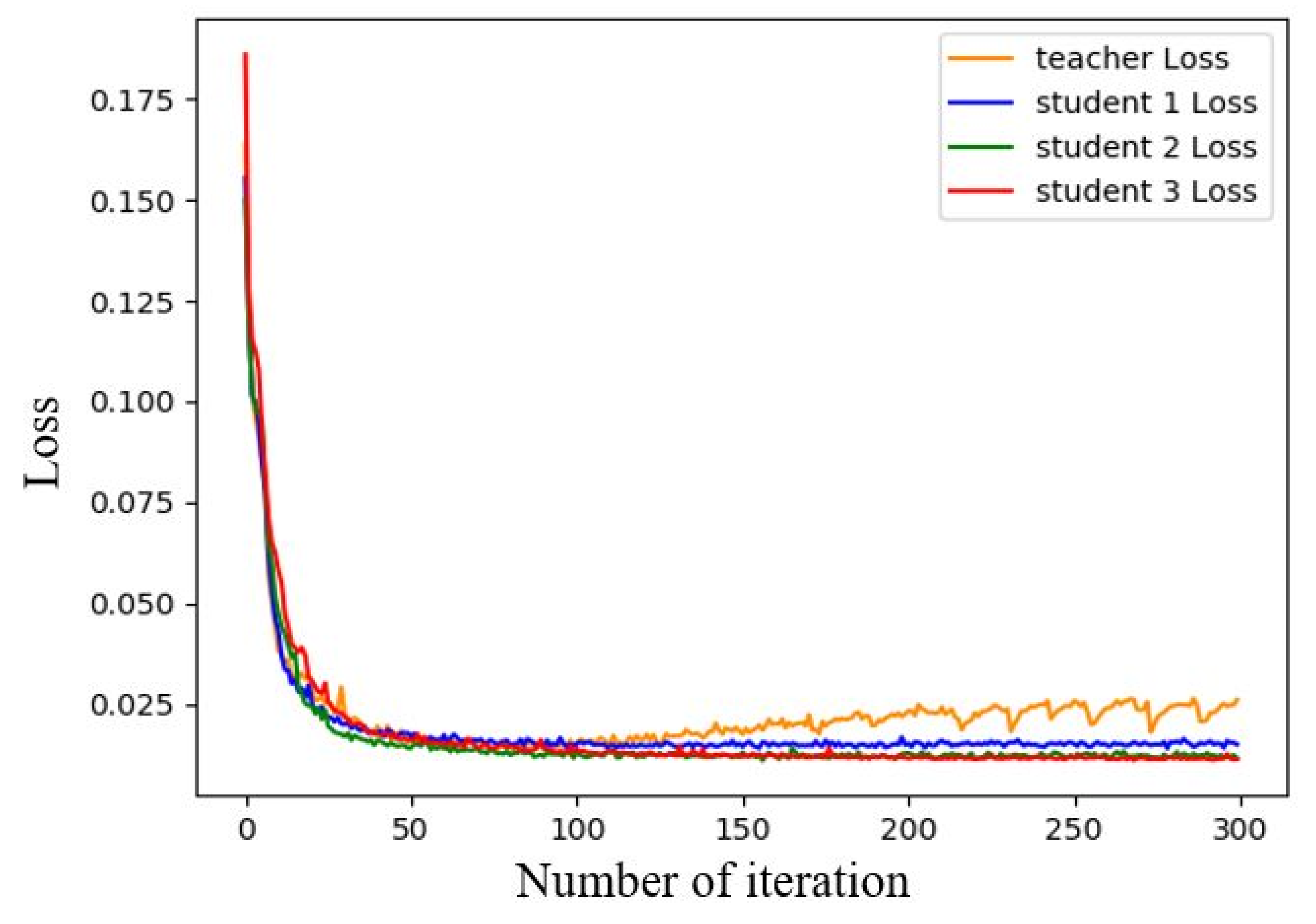
3.2.2. Semantic Segmentation Model Training
3.2.3. Effectiveness Evaluation Indexes
- (1)
- Pixel Accuracy (PA):
- (2)
- Mean Intersection-Over-Union (MIoU)
- (3)
- Dice Coefficient
3.3. Semi-Supervised Learning
3.3.1. Pseudo Label Generation
3.3.2. Semi-Supervised Learning Results
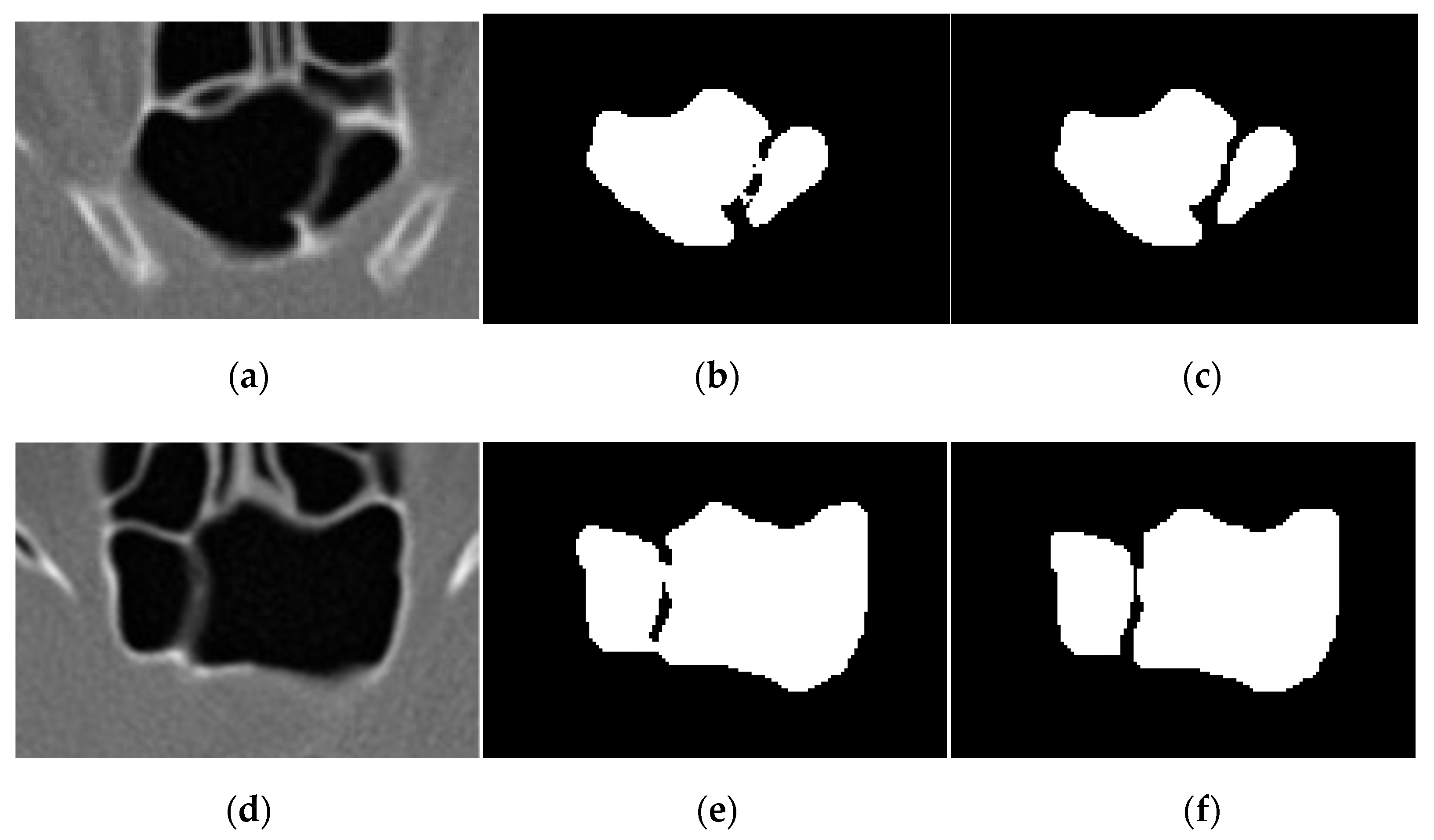
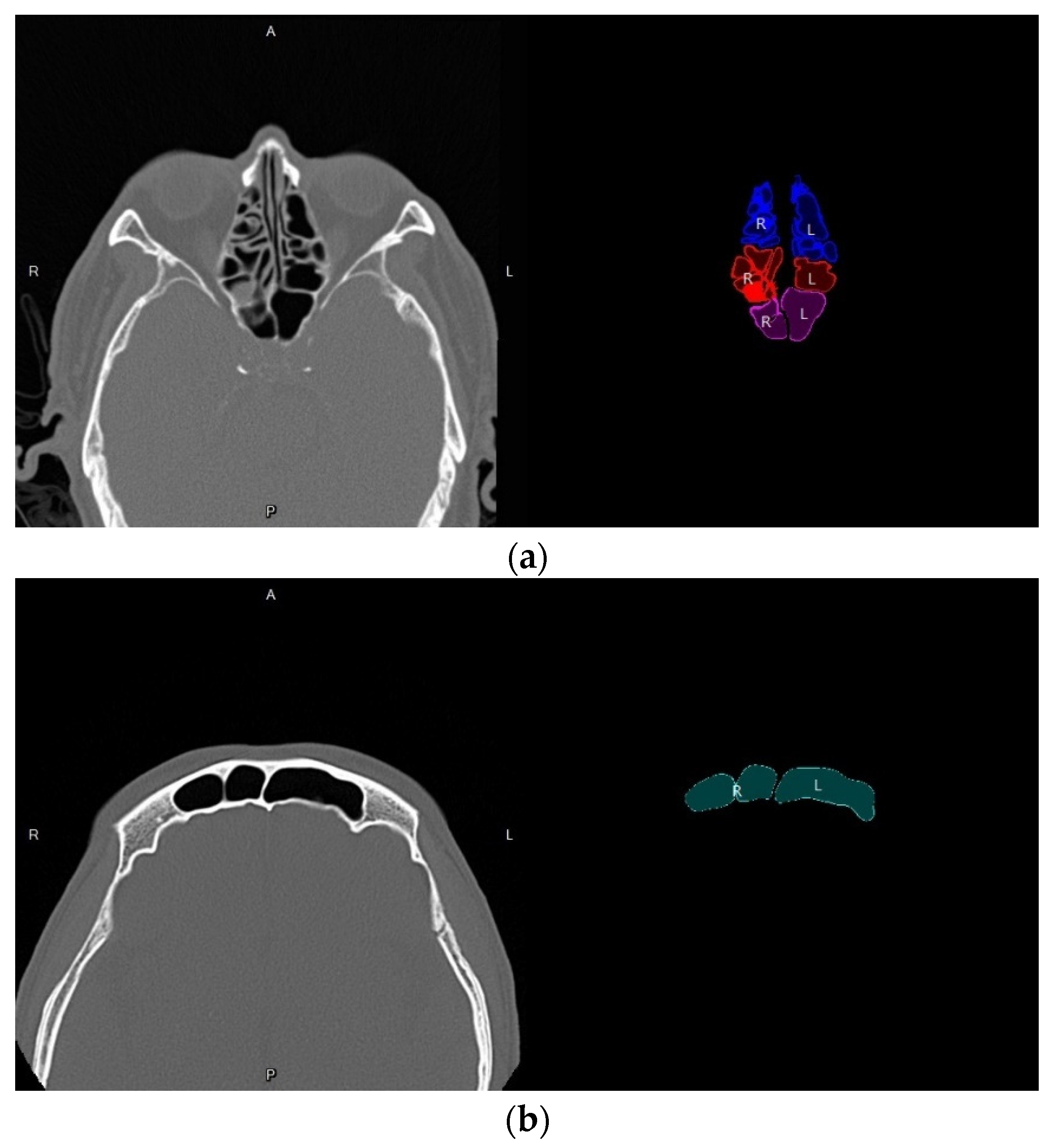
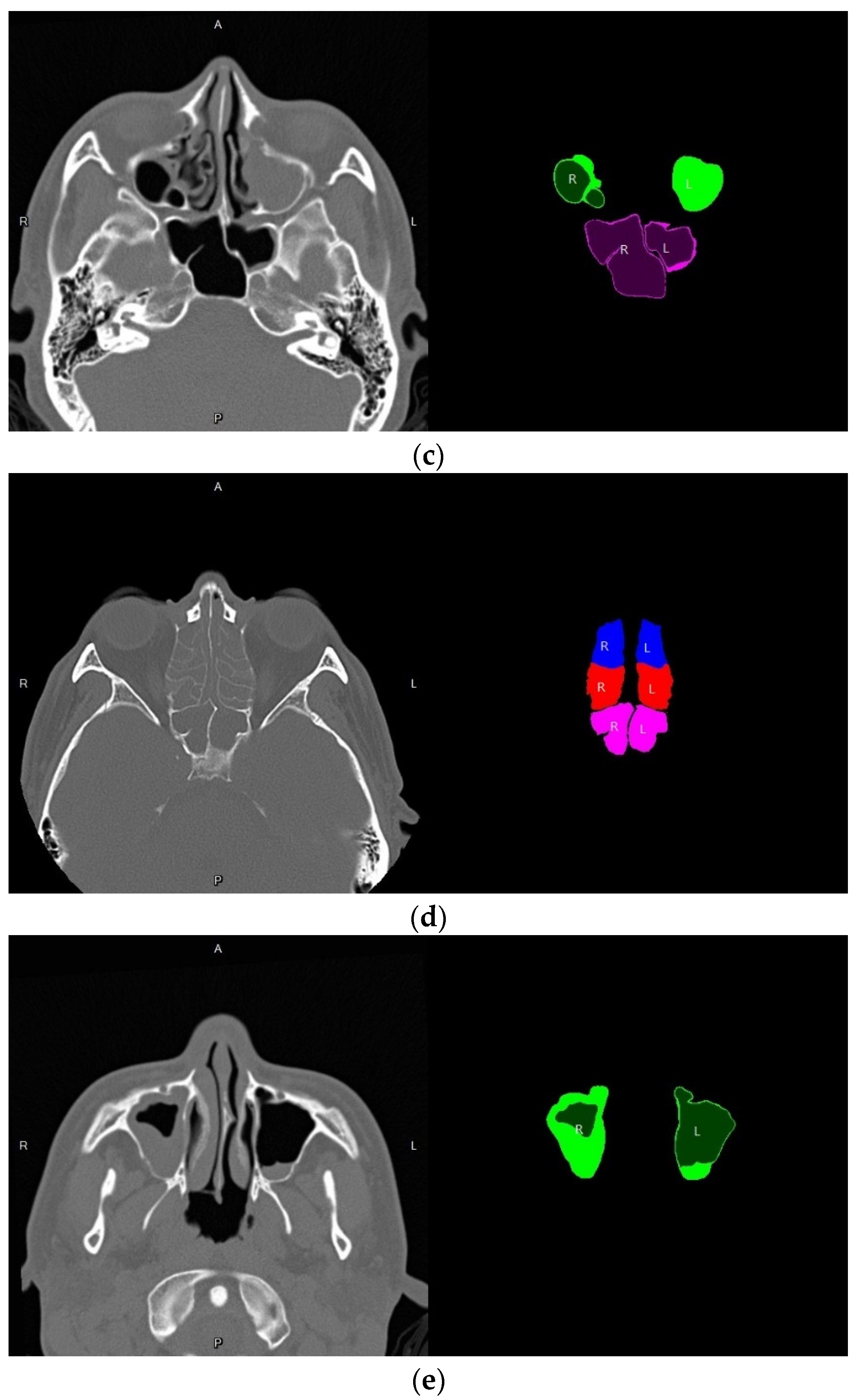
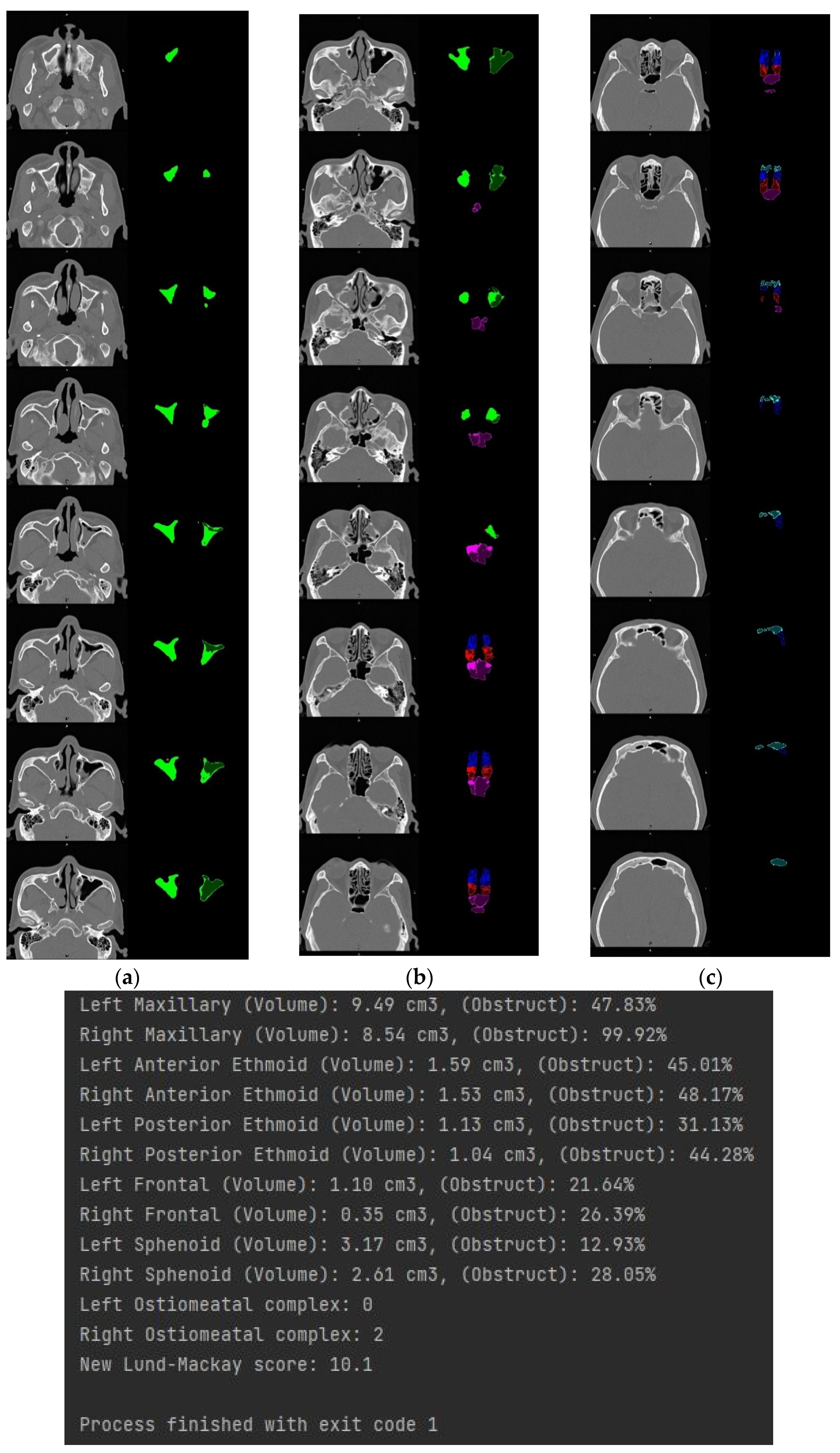
3.4. Volume Reconstruction
3.4.1. Morphology
3.4.2. 3D Reconstruction
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bhattacharyya, N. Clinical and Symptom Criteria for the Accurate Diagnosis of Chronic Rhinosinusitis. Laryngoscope 2006, 116, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Lund, V.J.; Mackay, I.S. Staging in rhinosinusitis. Rhinology 1993, 31, 183. [Google Scholar] [CrossRef]
- Garneau, J.; Bs, M.R.; Armato, S.G.; Sensakovic, W.; Ford, M.K.; Poon, C.S.; Ginat, D.T.; Starkey, A.; Baroody, F.M.; Pinto, J.M. Computer-assisted staging of chronic rhinosinusitis correlates with symptoms. Int. Forum Allergy Rhinol. 2015, 5, 637–642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, S.; Ramirez, M.V.; Garneau, J.C.; Ford, M.K.; Bs, K.M.; Ginat, D.T.; Baroody, F.M.; Armato, S.G.; Pinto, J.M. Three-dimensional image analysis for staging chronic rhinosinusitis. Int. Forum Allergy Rhinol. 2017, 7, 1052–1057. [Google Scholar] [CrossRef]
- Younis, R.T.; Anand, V.K.; Childress, C. Sinusitis Complicated by Meningitis: Current Management. Laryngoscope 2001, 111, 1338–1342. [Google Scholar] [CrossRef]
- Younis, R.T.; Lazar, R.H.; Bustillo, A.; Anand, V.K. Orbital Infection as a Complication of Sinusitis: Are Diagnostic and Treatment Trends Changing? Ear Nose Throat J. 2002, 81, 771–775. [Google Scholar] [CrossRef]
- Gulec, M.; Tassoker, M.; Magat, G.; Lale, B.; Ozcan, S.; Orhan, K. Three-dimensional volumetric analysis of the maxillary sinus: A cone-beam computed tomography study. Folia Morphol. 2020, 79, 557–562. [Google Scholar] [CrossRef]
- Saccucci, M.; Cipriani, F.; Carderi, S.; Di Carlo, G.; D’Attilio, M.; Rodolfino, D.; Festa, F.; Polimeni, A. Gender assessment through three-dimensional analysis of maxillary sinuses by means of cone beam computed tomography. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 185–193. [Google Scholar]
- Likness, M.M.; Pallanch, J.F.; Sherris, D.A.; Kita, H.; Mashtare, J.T.L.; Ponikau, J.U. Computed Tomography Scans as an Objective Measure of Disease Severity in Chronic Rhinosinusitis. Otolaryngol. Neck Surg. 2013, 150, 305–311. [Google Scholar] [CrossRef]
- Bui, N.L.; Ong, S.H.; Foong, K.W.C. Automatic segmentation of the nasal cavity and paranasal sinuses from cone-beam CT images. Int. J. Comput. Assist. Radiol. Surg. 2014, 10, 1269–1277. [Google Scholar] [CrossRef]
- Okushi, T.; Nakayama, T.; Morimoto, S.; Arai, C.; Omura, K.; Asaka, D.; Matsuwaki, Y.; Yoshikawa, M.; Moriyama, H.; Otori, N. A modified Lund–Mackay system for radiological evaluation of chronic rhinosinusitis. Auris Nasus Larynx 2013, 40, 548–553. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.F.; Gamba, T.D.O.; Yamasaki, M.C.; Groppo, F.C.; Neto, F.H.; Possobon, R.D.F. Development and validation of a formula based on maxillary sinus measurements as a tool for sex estimation: A cone beam computed tomography study. Int. J. Leg. Med. 2018, 133, 1241–1249. [Google Scholar] [CrossRef] [PubMed]
- de Souza, L.A.; Marana, A.N.; Weber, S.A.T. Automatic frontal sinus recognition in computed tomography images for person identification. Forensic Sci. Int. 2018, 286, 252–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodacre, B.J.; Swamidass, R.S.; Lozada, J.; Al-Ardah, A.; Sahl, E. A 3D-printed guide for lateral approach sinus grafting: A dental technique. J. Prosthet. Dent. 2018, 119, 897–901. [Google Scholar] [CrossRef]
- Giacomini, G.; Pavan, A.L.M.; Altemani, J.M.C.; Duarte, S.B.; Fortaleza, C.M.; Miranda, J.R.D.A.; De Pina, D.R. Computed tomography-based volumetric tool for standardized measurement of the maxillary sinus. PLoS ONE 2018, 13, e0190770. [Google Scholar] [CrossRef] [Green Version]
- Souadih, K.; Belaid, A.; BEN Salem, D.; Conze, P.-H. Automatic forensic identification using 3D sphenoid sinus segmentation and deep characterization. Med. Biol. Eng. Comput. 2019, 58, 291–306. [Google Scholar] [CrossRef]
- Humphries, S.M.; Centeno, J.P.; Notary, A.M.; Gerow, J.; Cicchetti, G.; Katial, R.K.; Lynch, D.A. Volumetric assessment of paranasal sinus opacification on computed tomography can be automated using a convolutional neural network. Int. Forum Allergy Rhinol. 2020, 10, 1218–1225. [Google Scholar] [CrossRef]
- Jung, S.-K.; Lim, H.-K.; Lee, S.; Cho, Y.; Song, I.-S. Deep Active Learning for Automatic Segmentation of Maxillary Sinus Lesions Using a Convolutional Neural Network. Diagnostics 2021, 11, 688. [Google Scholar] [CrossRef]
- Kim, H.-G.; Lee, K.M.; Kim, E.J.; Lee, J.S. Improvement diagnostic accuracy of sinusitis recognition in paranasal sinus X-ray using multiple deep learning models. Quant. Imaging Med. Surg. 2019, 9, 942–951. [Google Scholar] [CrossRef]
- Ahmad, M.; Ai, D.; Xie, G.; Qadri, S.F.; Song, H.; Huang, Y.; Wang, Y.; Yang, J. Deep Belief Network Modeling for Automatic Liver Segmentation. IEEE Access 2019, 7, 20585–20595. [Google Scholar] [CrossRef]
- Qadri, S.F.; Shen, L.; Ahmad, M.; Qadri, S.; Zareen, S.S.; Akbar, M.A. SVseg: Stacked Sparse Autoencoder-Based Patch Classification Modeling for Vertebrae Segmentation. Mathematics 2022, 10, 796. [Google Scholar] [CrossRef]
- Zhang, X.-D.; Li, Z.-H.; Wu, Z.-S.; Lin, W.; Lin, J.-C.; Zhuang, L.-M. A novel three-dimensional-printed paranasal sinus–skull base anatomical model. Eur. Arch. Oto-Rhino-Laryngol. 2018, 275, 2045–2049. [Google Scholar] [CrossRef] [PubMed]
- Valtonen, O.; Ormiskangas, J.; Kivekäs, I.; Rantanen, V.; Dean, M.; Poe, D.; Järnstedt, J.; Lekkala, J.; Saarenrinne, P.; Rautiainen, M. Three-Dimensional Printing of the Nasal Cavities for Clinical Experiments. Sci. Rep. 2020, 10, 502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Perez, L. The effectiveness of data augmentation in image classification using deep learning. Convolutional Neural Netw. Vis 2017, 11, 1–8. [Google Scholar]
- Hussain, Z.; Gimenez, F.; Yi, D.; Rubin, D. Differential Data Augmentation Techniques for Medical Imaging Classification Tasks. AMIA Annu. Symp. Proc. 2018, 2017, 979–984. [Google Scholar] [PubMed]
- Zhang, H.; Cisse, M.; Dauphin, Y.N.; Lopez-Paz, D. Mixup: Beyond empirical risk minimization. arXiv 2017, arXiv:1710.09412. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4510–4520. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Gu, R.; Wang, G.; Song, T.; Huang, R.; Aertsen, M.; Deprest, J.; Ourselin, S.; Vercauteren, T.; Zhang, S. CA-Net: Comprehensive Attention Convolutional Neural Networks for Explainable Medical Image Segmentation. IEEE Trans. Med. Imaging 2020, 40, 699–711. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Wan, S.; Liang, Y.; Zhang, Y. Deep convolutional neural networks for diabetic retinopathy detection by image classification. Comput. Electr. Eng. 2018, 72, 274–282. [Google Scholar] [CrossRef]
- Jung, H.; Choi, M.K.; Jung, J.; Lee, J.H.; Kwon, S.; Young, J.W. ResNet-based vehicle classification and localization in traffic surveillance systems. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Honolulu, HI, USA, 21–26 July 2017; pp. 61–67. [Google Scholar]
- Jiang, Y.; Li, Y.; Zhang, H. Hyperspectral Image Classification Based on 3-D Separable ResNet and Transfer Learning. IEEE Geosci. Remote Sens. Lett. 2019, 16, 1949–1953. [Google Scholar] [CrossRef]
- Zhang, Q.; Bai, C.; Liu, Z.; Yang, L.T.; Yu, H.; Zhao, J.; Yuan, H. A GPU-based residual network for medical image classification in smart medicine. Inf. Sci. 2020, 536, 91–100. [Google Scholar] [CrossRef]
- Misra, D. Mish: A self regularized non-monotonic neural activation function. arXiv 2019, arXiv:1908.08681. [Google Scholar]
- Kim, D.; MacKinnon, T. Artificial intelligence in fracture detection: Transfer learning from deep convolutional neural networks. Clin. Radiol. 2018, 73, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Haralick, R.M.; Sternberg, S.R.; Zhuang, X. Image analysis using mathematical morphology. IEEE Trans. Pattern Anal. Mach. Intell. 1987, 9, 532–550. [Google Scholar] [CrossRef] [PubMed]
- Chang, F.; Chen, C.-J.; Lu, C.-J. A linear-time component-labeling algorithm using contour tracing technique. Comput. Vis. Image Underst. 2004, 93, 206–220. [Google Scholar] [CrossRef] [Green Version]
- Lorensen, W.E.; Cline, H.E. Marching cubes: A high resolution 3D surface construction algorithm. In Proceedings of the 14th Annual Conference on Computer Graphics and Interactive Techniques, Anaheim, CA, USA, 27–31 July 1987; Volume 21, pp. 163–169. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Wang, M.; Lu, S.; Zhu, D.; Lin, J.; Wang, Z. A high-speed and low-complexity architecture for softmax function in deep learning. In Proceedings of the 2018 IEEE Asia Pacific Conference on Circuits and Systems (APCCAS), Chengdu, China, 26–30 October 2018; pp. 223–226. [Google Scholar]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Li, F.-F. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2881–2890. [Google Scholar]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision, Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- Wang, J.; Sun, K.; Cheng, T.; Jiang, B.; Deng, C.; Zhao, Y.; Liu, D.; Mu, Y.; Tan, M.; Wang, X.; et al. Deep High-Resolution Representation Learning for Visual Recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 43, 3349–3364. [Google Scholar] [CrossRef] [Green Version]
- Bochkovskiy, A.; Wang, C.Y.; Liao, H.Y.M. Yolov4: Optimal speed and accuracy of object detection. arXiv 2020, arXiv:2004.10934. [Google Scholar]
- Xie, Q.; Luong, M.T.; Hovy, E.; Le, Q.V. Self-training with noisy student improves imagenet classification. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 10687–10698. [Google Scholar]
- Hahn, S.; Choi, H. Understanding dropout as an optimization trick. Neurocomputing 2020, 398, 64–70. [Google Scholar] [CrossRef] [Green Version]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Nowak, R.; Mehls, G. X-ray film analysis of the sinus paranasales from cleft patients (in comparison with a healthy group) (author’s transl). Anat. Anzeiger. 1977, 142, 451–470. [Google Scholar]
- Hopkins, C.; Browne, J.P.; Slack, R.; Lund, V.; Brown, P. The Lund-Mackay staging system for chronic rhinosinusitis: How is it used and what does it predict? Otolaryngol. Neck Surg. 2007, 137, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Girshick, R.; Dollár, P.; Tu, Z.; He, K. Aggregated residual transformations for deep neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1492–1500. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, inception-resnet and the impact of residual connections on learning. In Proceedings of the AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017; Volume 31, pp. 4278–4284. [Google Scholar]
- Sahlstrand-Johnson, P.; Jannert, M.; Strömbeck, A.; Abul-Kasim, K. Computed tomography measurements of different dimensions of maxillary and frontal sinuses. BMC Med. Imaging 2011, 11, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwamoto, Y.; Xiong, K.; Kitamura, T.; Han, X.-H.; Matsushiro, N.; Nishimura, H.; Chen, Y.-W. Automatic Segmentation of the Paranasal Sinus from Computer Tomography Images Using a Probabilistic Atlas and a Fully Convolutional Network. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 2789–2792. [Google Scholar] [CrossRef]






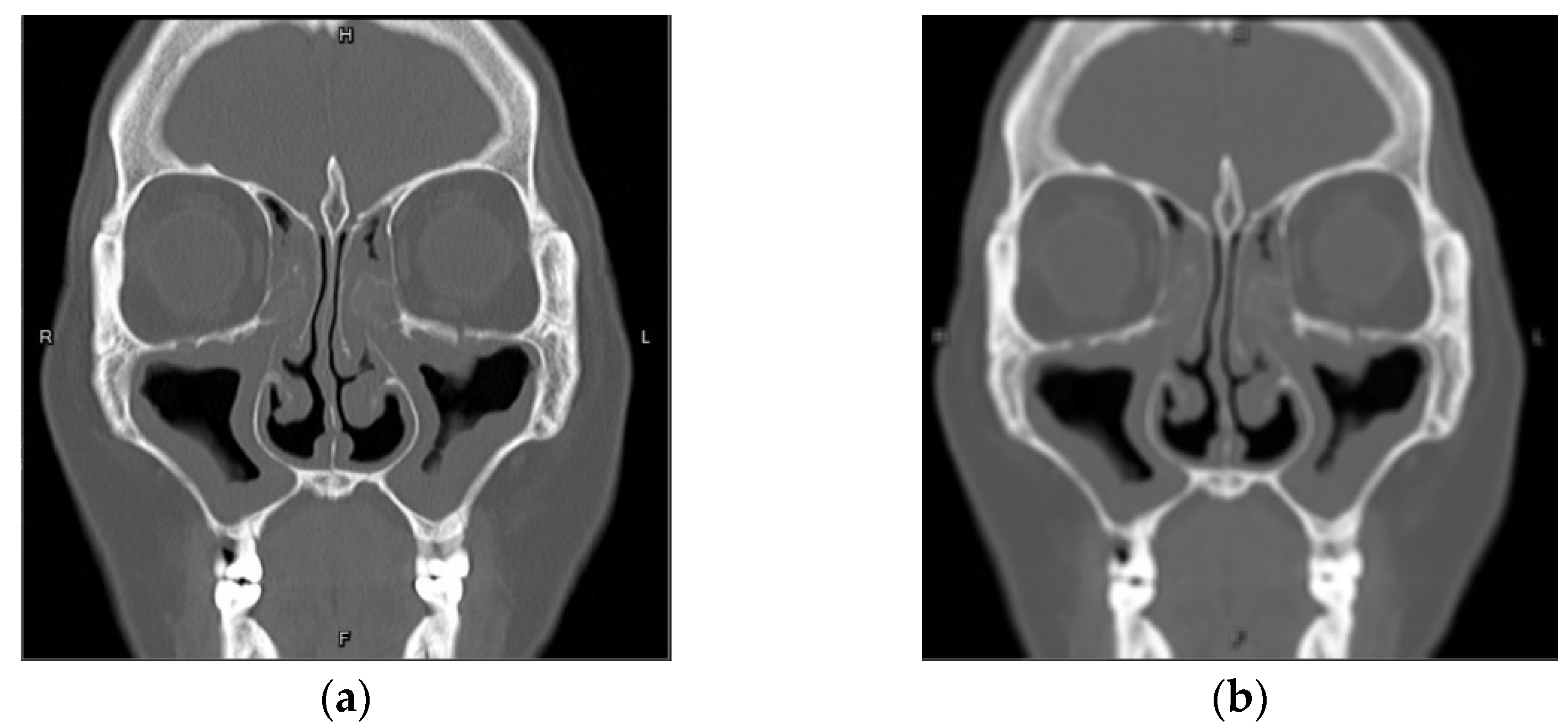
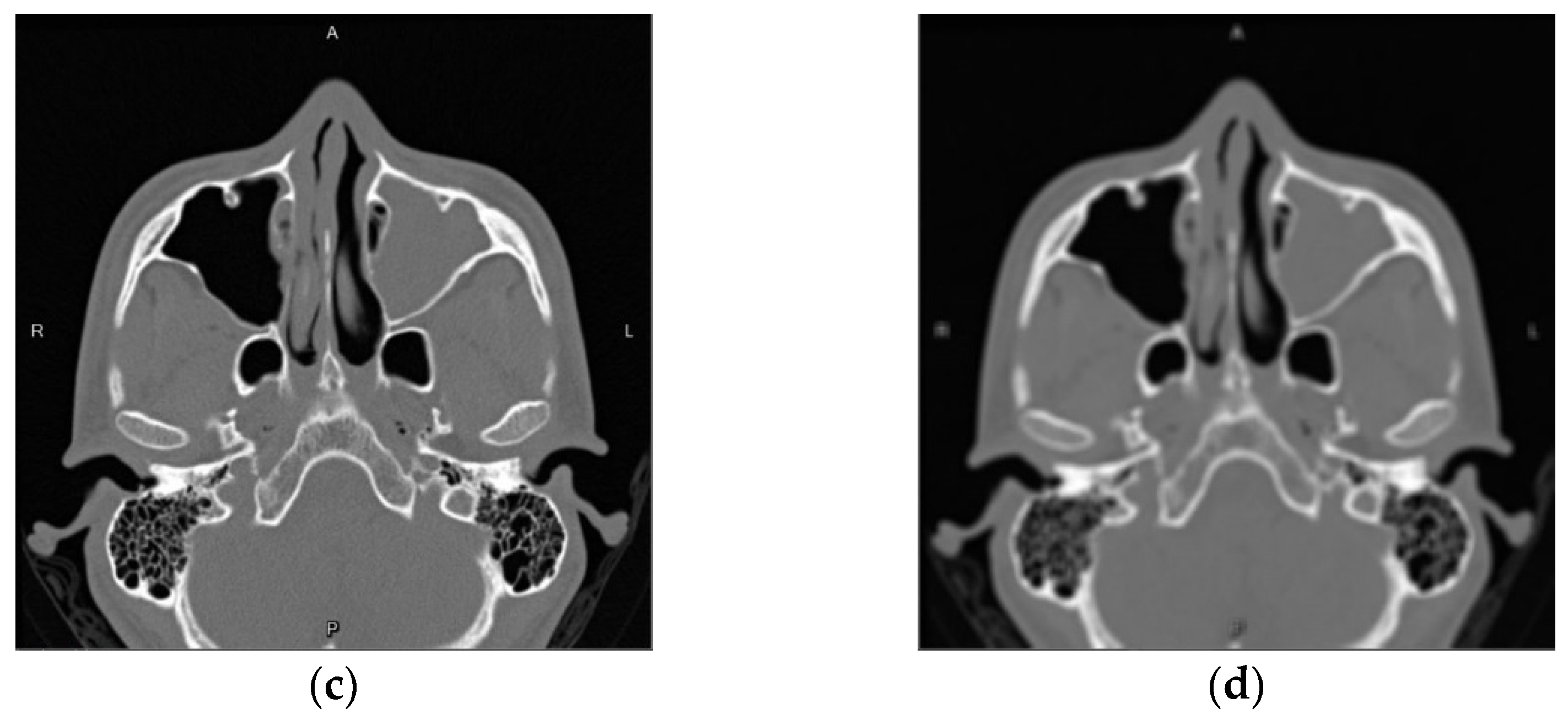
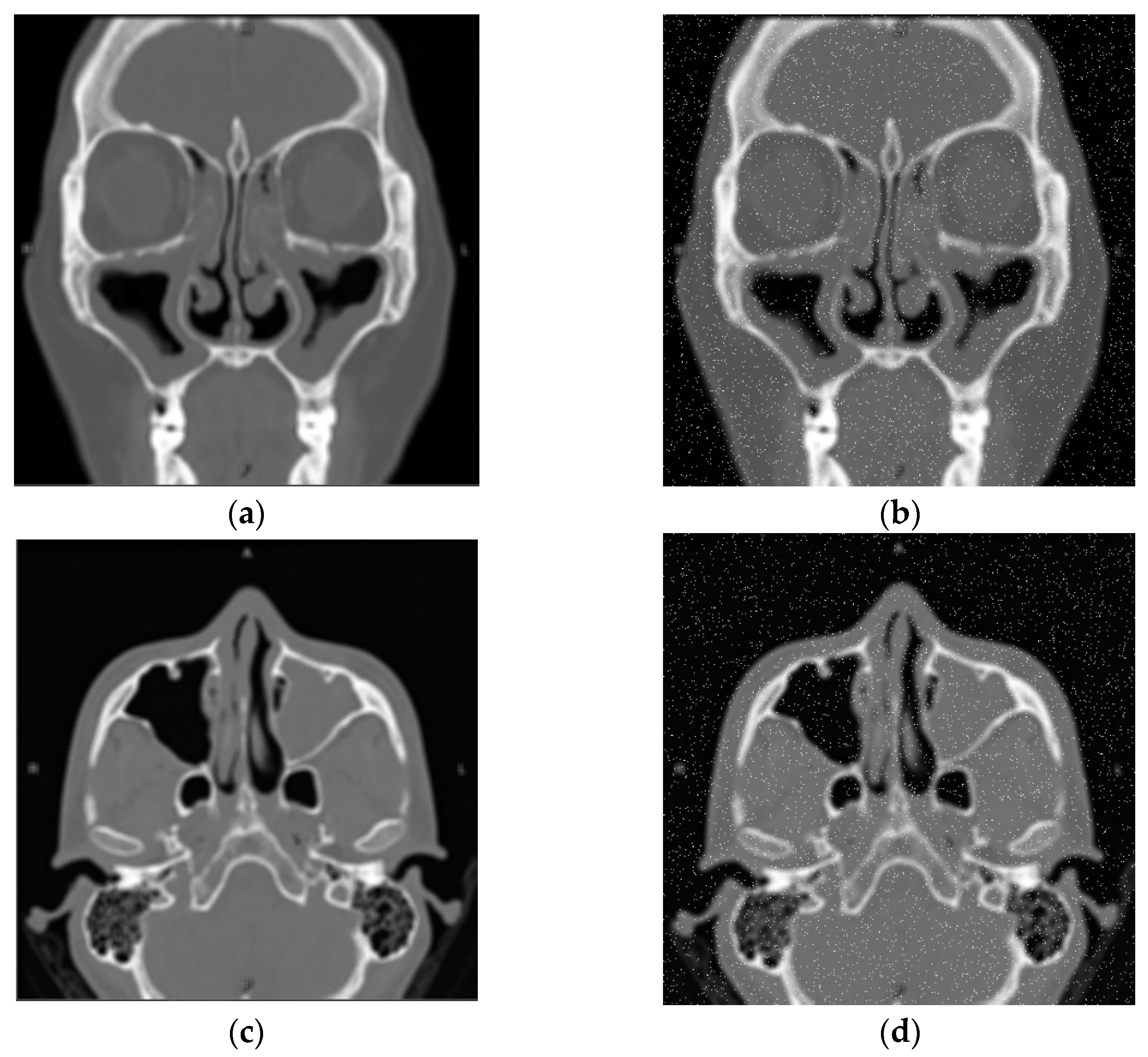



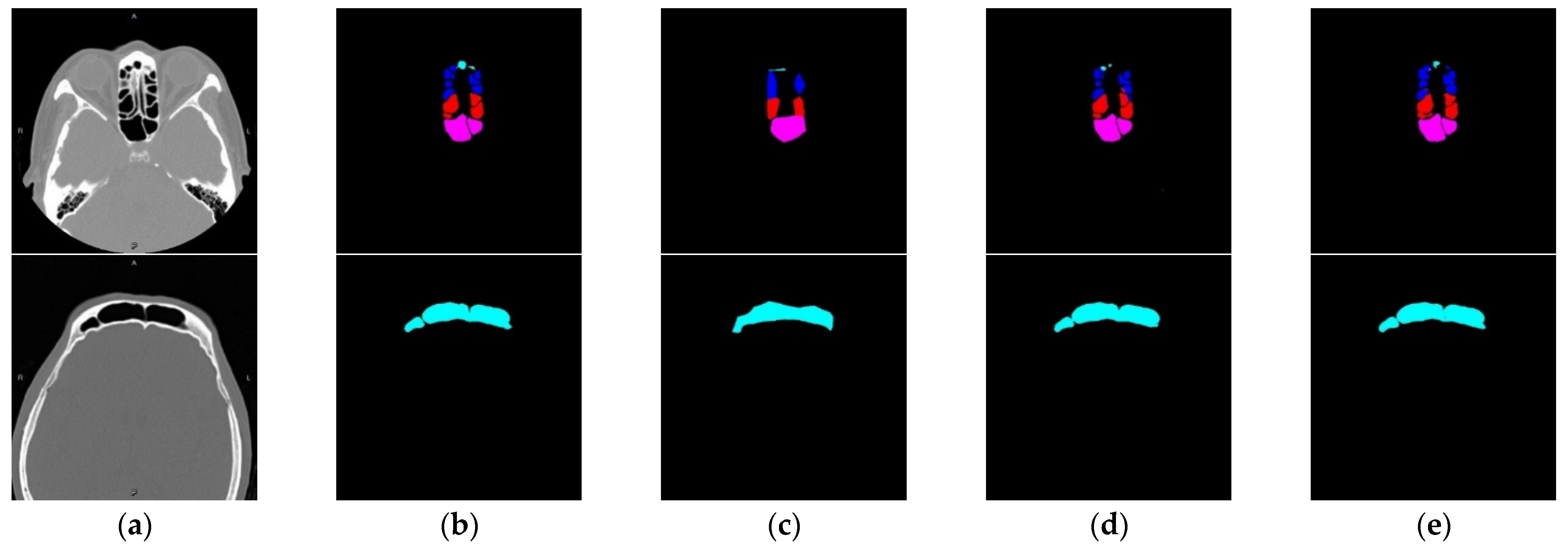




| Self-Confidence Threshold | Dice |
|---|---|
| 0.3 | 90.79% |
| 0.4 | 90.79% |
| 0.5 | 90.79% |
| 0.6 | 90.81% |
| 0.7 | 90.83% |
| 0.8 | 90.67% |
| 0.9 | 90.16% |
| Frequency of Repetitive Training | Minimum Loss |
|---|---|
| 0 | 0.0211 |
| 1 | 0.0192 |
| 2 | 0.0175 |
| 3 | 0.0172 |
| Method | PA | Dice | mIOU |
|---|---|---|---|
| UNet | 99.56% | 89.68% | 87.83% |
| PSPNet | 99.31% | 87.52% | 85.78% |
| HRNet | 99.12% | 86.68% | 83.02% |
| DeepLab v3+ | 99.55% | 89.64% | 87.69% |
| Our + MobilenNet [53] | 99.62% | 90.41% | 88.53% |
| Our + ResNet50 [31] | 99.63% | 90.47% | 88.57% |
| Our + ResNet101 [31] | 99.63% | 90.51% | 88.58% |
| Our + ResNext50 [31] | 99.63% | 90.49% | 88.58% |
| Our + ResNext101 [31] | 99.64% | 90.55% | 88.59% |
| Our + Inception [56] | 99.65% | 90.57% | 88.59% |
| Our + DWS + SE | 99.67% | 90.79% | 88.75% |
| Method | PA | Dice | mIOU |
|---|---|---|---|
| UNet | 99.56% | 89.68% | 87.83% |
| Our | 99.67% | 90.79% | 88.75% |
| Our + Semi-supervised learning | 99.75% | 91.57% | 89.43% |
| Computer Equipment 1 | Computer Equipment 2 | |
|---|---|---|
| CPU | Intel®Core™ i5-7300 | Intel®Xeon® Silver-4110 |
| Memory | 16GB DDR4 | 64GB DDR4 |
| Graphic card | NVIDIA GTX1050 4 GB | NVIDIA Quadro GP100 16 GB |
| UNet prediction time | 0.076 s | 0.057 s |
| Prediction time of this model | 0.082 s | 0.062 s |
| Maxillary Sinus | Anterior Ethmoid Sinus | Posterior Ethmoid Sinus | Frontal Sinus | Sphenoid Sinus | |
|---|---|---|---|---|---|
| Left | 98% | 88.5% | 92.5% | 95% | 98.5% |
| Right | 98.5% | 88.5% | 91% | 96% | 99.5% |
| Males’ Average | Males’ Standard Deviation | Females’ Average | Females’ Standard Deviation | |
|---|---|---|---|---|
| Age | 46.19 | 16.35 | 51.16 | 14.31 |
| Height (cm) | 171.06 | 7.64 | 159.32 | 5.54 |
| Weight (kg) | 71.16 | 11.10 | 58.06 | 10.71 |
| BMI | 24.30 | 3.35 | 22.87 | 3.98 |
| Left maxillary sinus (cm3) | 15.39 | 6.27 | 11.05 | 5.60 |
| Right maxillary sinus (cm3) | 15.50 | 6.45 | 11.27 | 5.60 |
| Left anterior ethmoid sinus (cm3) | 1.60 | 0.72 | 1.28 | 0.52 |
| Right anterior ethmoid sinus (cm3) | 1.58 | 0.57 | 1.34 | 0.50 |
| Left posterior ethmoid sinus (cm3) | 1.50 | 0.61 | 1.21 | 0.57 |
| Right posterior ethmoid sinus (cm3) | 1.47 | 0.57 | 1.13 | 0.54 |
| Left frontal sinus (cm3) | 1.68 | 1.30 | 0.91 | 0.92 |
| Right frontal sinus (cm3) | 1.64 | 1.33 | 0.77 | 0.69 |
| Left sphenoid sinus (cm3) | 3.64 | 2.04 | 2.70 | 1.77 |
| Right sphenoid sinus (cm3) | 3.81 | 2.47 | 2.70 | 1.90 |
| Overall Average | Males’ Average | Females’ Average | Overall Average (Absolute Value) | Males’ Average (Absolute Value) | Females’ Average (Absolute Value) | |
|---|---|---|---|---|---|---|
| Maxillary sinus | −0.014 | −0.022 | 0.003 | 0.145 | 0.143 | 0.148 |
| Anterior ethmoid sinus | 0.005 | −0.015 | 0.045 | 0.144 | 0.138 | 0.153 |
| Posterior ethmoid sinus | −0.011 | −0.006 | 0.044 | 0.161 | 0.163 | 0.158 |
| Frontal sinus | −0.207 | −0.153 | −0.315 | 0.452 | 0.398 | 0.549 |
| Sphenoid sinus | −0.143 | −0.137 | −0.155 | 0.446 | 0.455 | 0.432 |
| Item | Maxillary Sinus | Anterior Ethmoid Sinus | |||
| Left | Right | Left | Right | ||
| Age | Pearson Correlation p-value | −0.169 0.042 | −0.155 0.063 | −0.006 0.944 | −0.006 0.944 |
| Height (cm) | Pearson Correlation p-value | 0.412 0.001 | 0.417 0.001 | 0.305 0.001 | 0.299 0.001 |
| Weight (kg) | Pearson Correlation p-value | 0.151 0.071 | 0.149 0.074 | 0.241 0.004 | 0.199 0.017 |
| BMI | Pearson Correlation p-value | −0.098 0.241 | −0.106 0.207 | 0.079 0.348 | 0.032 0.705 |
| Item | Posterior Ethmoid Sinus | Frontal Sinus | |||
| Left | Right | Left | Right | ||
| Age | Pearson Correlation p-value | −0.110 0.189 | −0.107 0.203 | −0.068 0.418 | −0.088 0.294 |
| Height (cm) | Pearson Correlation p-value | 0.370 0.001 | 0.387 0.001 | 0.367 0.001 | 0.343 0.001 |
| Weight (kg) | Pearson Correlation p-value | 0.196 0.019 | 0.208 0.012 | 0.129 0.122 | 0.094 0.260 |
| BMI | Pearson Correlation p-value | −0.028 0.735 | −0.019 0.818 | −0.093 0.266 | −0.117 0.163 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuo, C.-F.J.; Liu, S.-C. Fully Automatic Segmentation, Identification and Preoperative Planning for Nasal Surgery of Sinuses Using Semi-Supervised Learning and Volumetric Reconstruction. Mathematics 2022, 10, 1189. https://doi.org/10.3390/math10071189
Kuo C-FJ, Liu S-C. Fully Automatic Segmentation, Identification and Preoperative Planning for Nasal Surgery of Sinuses Using Semi-Supervised Learning and Volumetric Reconstruction. Mathematics. 2022; 10(7):1189. https://doi.org/10.3390/math10071189
Chicago/Turabian StyleKuo, Chung-Feng Jeffrey, and Shao-Cheng Liu. 2022. "Fully Automatic Segmentation, Identification and Preoperative Planning for Nasal Surgery of Sinuses Using Semi-Supervised Learning and Volumetric Reconstruction" Mathematics 10, no. 7: 1189. https://doi.org/10.3390/math10071189
APA StyleKuo, C.-F. J., & Liu, S.-C. (2022). Fully Automatic Segmentation, Identification and Preoperative Planning for Nasal Surgery of Sinuses Using Semi-Supervised Learning and Volumetric Reconstruction. Mathematics, 10(7), 1189. https://doi.org/10.3390/math10071189







