An Ensemble and Iterative Recovery Strategy Based kGNN Method to Edit Data with Label Noise
Abstract
:1. Introduction
- We introduce the general nearest neighbor (GNN) [12] method to take the neighborhood information of all samples into account. For any query sample Q, its traditional k-general nearest neighbors and samples whose k nearest neighbors contain Q constitute Q’s k general nearest neighbors. In this way, the one-sided NN rule is substituted with the bilateral NN rule, improving the convincingness of the NN rule.
- We ensemble the prediction results of a finite set of ks to produce the inconsistent number as a vote to measure the noise levels for each sample using the kGNN classifier. A sample with a higher vote value is obviously much easier to be detected as label noise, as it has more predicted results contradictory to its given label.
- We flip the labels of easy-to-learn label noise to the expected ones and repeat the ensemble classifying process to detect more difficult-to-learn label noise, enhancing the precision of the detected label noise. Here, the samples selected to be flipped have different vote values. This happens one level each time, which is called cascade recovery.
- Two loops, i.e., the inner loop and the outer loop, are utilized to iteratively detect the suspected label noise. The inner loop will output the suspected label-noise samples for each fold, and the outer loop will produce the frequency of each sample being detected as label noise among the R folds. Frequency is later used as an indicator to categorize the detected label noise into two types, i.e., boundary noise and definite noise. Boundary noise is removed, and definite noise is relabeled to better recover the original distribution of the training set.
2. Related Work
3. An Ensemble and Iterative Recovery Strategy-Based kGNN Method
3.1. k Nearest Neighbors Algorithm
3.2. An Ensemble and Iterative Recovery Strategy-Based kGNN Method
3.2.1. kGNN Algorithm
- Given a data set , which contains n samples with d dimensions, for each sample , find its k nearest neighbors using the NN rule and generate an index matrix , where the first column refers to the index of each sample, the second column refers to the index of its first nearest neighbor, the third column refers to the index of its second nearest neighbor, and so on until all k nearest neighbors are found. Euclidean distance is used in this step. Then, the list in each row represents nearest neighbors of , where the nearest neighbor is itself.
- For each sample , let us check which rows in contain its index i, and append the index of this row j to to form ’s general nearest neighbors. Additionally, a difference operation between sets and i is conducted, as the index of each sample i in is redundant. Finally, as the length of each sample’s GNN may be different, we use a dictionary to store the indices of each sample’s general nearest neighbors.
3.2.2. An Ensemble and Iterative Recovery Strategy
| Algorithm 1 EIRS-kGNN. |
|
- Vote values of all samples 2, where , represents the rounding down function, and 5 is the size of ks;
- .
3.3. Time Complexity Analysis
4. Experimental Results, Analyses, and Discussions
- ENN: , sampling_strategy = all
- RENN: , sampling_strategy = all
- AllkNN: , sampling_strategy = all
- NCR: , sampling_strategy = all
- RD:
- IPF: , , ,
- DynamicCF: ,
- EIRS-kGNN: ,
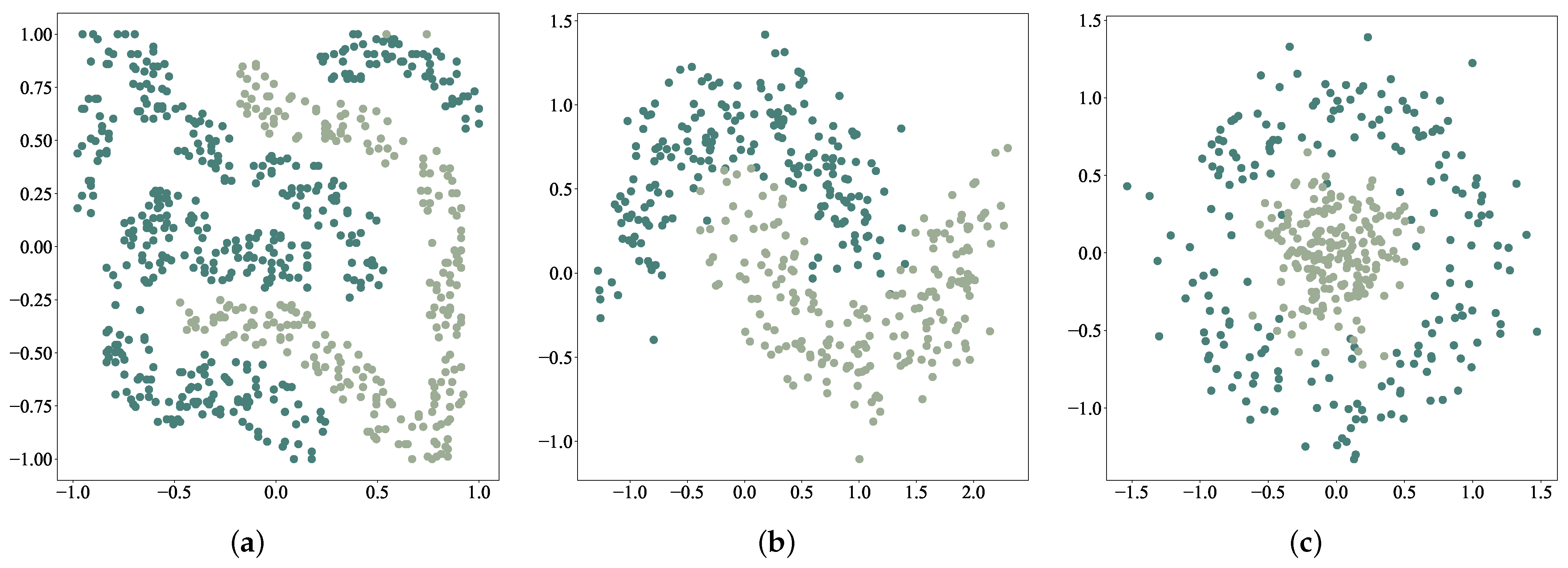
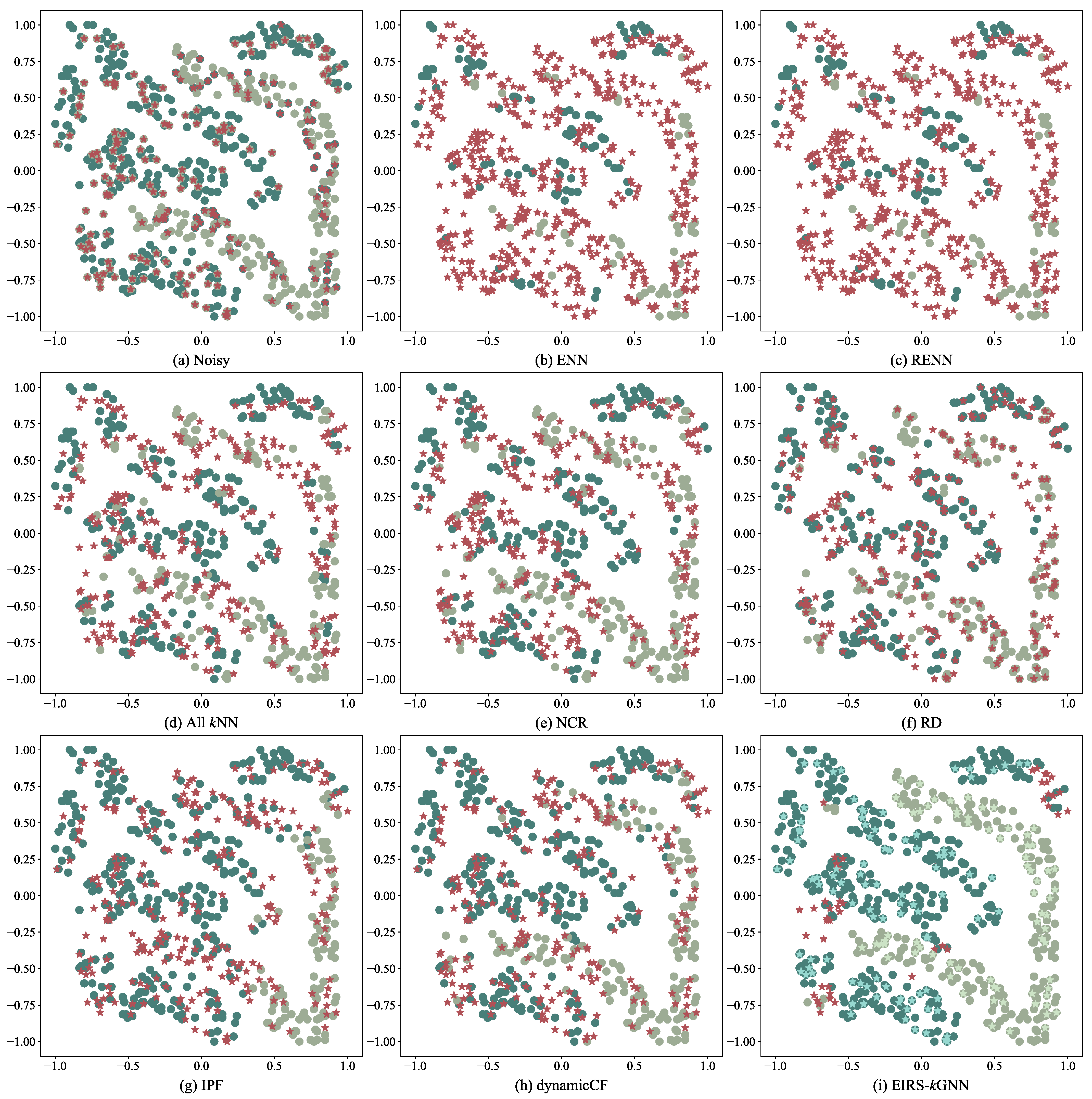
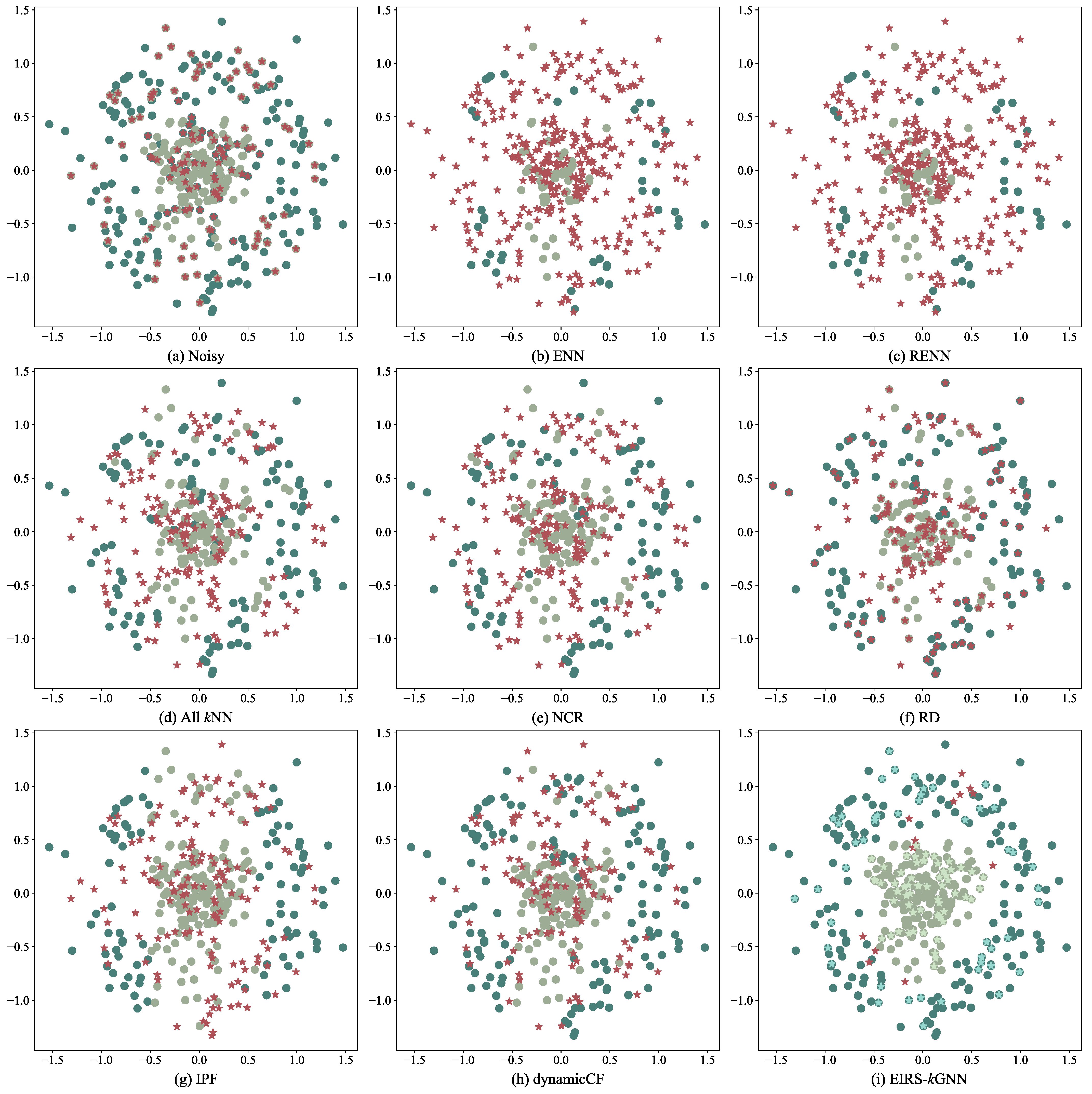
4.1. Experimental Results on Two-Dimensional Data Sets
4.2. Experimental Results on Binary UCI Data Sets
4.2.1. Detection Performance of Label Noise
4.2.2. Classification Performance after Editing
4.2.3. Time Efficiency Comparison
4.3. Experimental Results on Multi-Class UCI Data Sets
4.3.1. Performance of Detection of Label Noise in Multi-Class Data Sets
4.3.2. Classification Performance on Multi-Class Data Sets after Editing
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Parameters of the Classification Algorithms
- LR (https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegression.html, accessed on 1 April 2022): norm used in the penalization penalty = “l2”, algorithm used in the optimization process solver = “lbfgs”, maximum number of iterations max_iter = 100, and others were set defaults;
- DT (https://scikit-learn.org/stable/modules/generated/sklearn.tree.DecisionTreeClassifier.html, accessed on 1 April 2022): function to measure the quality of a split criterion = “gini”, strategy used to choose the split at each node splitter = “best”, minimum number of samples required to split an internal node min_samples_split = 2, minimum number of samples required to be at a leaf node min_samples_leaf = 1, and others were set to defaults;
- Adaboost (https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.AdaBoostClassifier.html, accessed on 1 April 2022): base estimator from which the boosted ensemble is built base_estimator = “DecisionTreeClassifier”, maximum number of estimators at which boosting is terminated n_estimators = 50, weight applied to each classifier learning_rate = 1.0, use the SAMME.R real boosting algorithm and set algorithm = “SAMME.R”, and others were set defaults;
- kNN (https://scikit-learn.org/stable/modules/generated/sklearn.neighbors.NearestNeighbors.html, accessed on 1 April 2022): number of neighbors to use n_neighbors = 5, and others were set defaults;
- GBDT (https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.GradientBoostingClassifier.html, accessed on 1 April 2022): boosting learning rate learning_rate = 0.1, number of boosting stages to perform n_estimators = 100, function to measure the quality of a split criterion = “friedman_mse”, minimum number of samples required to split an internal node min_samples_split = 2, and others were set defaults.
References
- Zhu, X.; Wu, X. Class noise vs. attribute noise: A quantitative study. Artif. Intell. Rev. 2004, 22, 177–210. [Google Scholar] [CrossRef]
- Frénay, B.; Verleysen, M. Classification in the presence of label noise: A survey. IEEE Trans. Neural Netw. Learn. Syst. 2013, 25, 845–869. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Jeske, D.R. The efficiency of logistic regression compared to normal discriminant analysis under class-conditional classification noise. J. Multivar. Anal. 2010, 101, 1622–1637. [Google Scholar] [CrossRef] [Green Version]
- Laird, P.D. Learning from Good and Bad Data; Springer Science & Business Media: New York, NY, USA, 2012; Volume 47. [Google Scholar]
- Abellán, J.; Masegosa, A.R. Bagging decision trees on data sets with classification noise. In Proceedings of the International Symposium on Foundations of Information and Knowledge Systems, Sofia, Bulgaria, 15–19 February 2010; Springer: Berlin/Heidelberg, Germany, 2010; pp. 248–265. [Google Scholar]
- Bross, I. Misclassification in 2 × 2 tables. Biometrics 1954, 10, 478–486. [Google Scholar] [CrossRef]
- Wilson, D.R.; Martinez, T.R. Reduction techniques for instance-based learning algorithms. Mach. Learn. 2000, 38, 257–286. [Google Scholar] [CrossRef]
- Wilson, D.L. Asymptotic properties of nearest neighbor rules using edited data. IEEE Trans. Syst. Man Cybern. 1972, SMC-2, 408–421. [Google Scholar] [CrossRef] [Green Version]
- Tomek, I. An Experiment with the Edited Nearest-Neighbor Rule. IEEE Trans. Syst. Man Cybern. 1976, SMC-6, 448–452. [Google Scholar] [CrossRef]
- Sánchez, J.S.; Barandela, R.; Marqués, A.I.; Alejo, R.; Badenas, J. Analysis of new techniques to obtain quality training sets. Pattern Recognit. Lett. 2003, 24, 1015–1022. [Google Scholar] [CrossRef]
- Koplowitz, J.; Brown, T.A. On the relation of performance to editing in nearest neighbor rules. Pattern Recognit. 1981, 13, 251–255. [Google Scholar] [CrossRef]
- Pan, Z.; Wang, Y.; Ku, W. A new general nearest neighbor classification based on the mutual neighborhood information. Knowl.-Based Syst. 2017, 121, 142–152. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, S. Noisy data elimination using mutual k-nearest neighbor for classification mining. J. Syst. Softw. 2012, 85, 1067–1074. [Google Scholar] [CrossRef]
- Gowda, K.C.; Krishna, G. The condensed nearest neighbor rule using the concept of mutual nearest neighborhood. IEEE Trans. Inf. Theory 1979, 25, 488–490. [Google Scholar] [CrossRef] [Green Version]
- Gowda, K.C.; Krishna, G. Agglomerative clustering using the concept of mutual nearest neighbourhood. Pattern Recognit. 1978, 10, 105–112. [Google Scholar] [CrossRef]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, J.S.; Pla, F.; Ferri, F.J. Prototype selection for the nearest neighbour rule through proximity graphs. Pattern Recognit. Lett. 1997, 18, 507–513. [Google Scholar] [CrossRef]
- Chaudhuri, B. A new definition of neighborhood of a point in multi-dimensional space. Pattern Recognit. Lett. 1996, 17, 11–17. [Google Scholar] [CrossRef]
- Devijver, P.A. On the editing rate of the multiedit algorithm. Pattern Recognit. Lett. 1986, 4, 9–12. [Google Scholar] [CrossRef]
- Hattori, K.; Takahashi, M. A new edited k-nearest neighbor rule in the pattern classification problem. Pattern Recognit. 2000, 33, 521–528. [Google Scholar] [CrossRef]
- Hart, P. The condensed nearest neighbor rule. IEEE Trans. Inf. Theory 1968, 14, 515–516. [Google Scholar] [CrossRef]
- Kubat, M.; Matwin, S. Addressing the curse of imbalanced training sets: One-sided selection. In Proceedings of the ICML, Citeseer, Nashville, TN, USA, 8–12 July 1997; Volume 97, p. 179. [Google Scholar]
- Tomek, I. Two Modifications of CNN. IEEE Trans. Syst. Man Cybern. 1976, SMC-6, 769–772. [Google Scholar]
- Laurikkala, J. Improving identification of difficult small classes by balancing class distribution. In Proceedings of the Conference on Artificial Intelligence in Medicine in Europe, Cascais, Portugal, 1–4 July 2001; Springer: Berlin/Heidelberg, Germany; pp. 63–66. [Google Scholar]
- Xia, S.; Xiong, Z.; Luo, Y.; Dong, L.; Xing, C. Relative density based support vector machine. Neurocomputing 2015, 149, 1424–1432. [Google Scholar] [CrossRef]
- Xia, S.; Chen, B.; Wang, G.; Zheng, Y.; Gao, X.; Giem, E.; Chen, Z. mCRF and mRD: Two classification methods based on a novel multiclass label noise filtering learning framework. IEEE Trans. Neural Netw. Learn. Syst. 2021, 33, 2916–2930. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Xia, S.; Chen, Z.; Wang, B.; Wang, G. RSMOTE: A self-adaptive robust SMOTE for imbalanced problems with label noise. Inf. Sci. 2021, 553, 397–428. [Google Scholar] [CrossRef]
- Liu, Y.; Xia, S.; Yu, H.; Luo, Y.; Chen, B.; Liu, K.; Wang, G. Prediction of Aluminum Electrolysis Superheat Based on Improved Relative Density Noise Filter SMO. In Proceedings of the 2018 IEEE International Conference on Big Knowledge (ICBK), Singapore, 17–18 November 2018; pp. 376–381. [Google Scholar]
- Huang, L.; Shao, Y.; Peng, J. An Adaptive Voting Mechanism Based on Relative Density for Filtering Label Noises. In Proceedings of the 2022 IEEE 5th International Conference on Electronics Technology (ICET), Chengdu, China, 13–16 May 2022. [Google Scholar]
- Brodley, C.E.; Friedl, M.A. Identifying mislabeled training data. J. Artif. Intell. Res. 1999, 11, 131–167. [Google Scholar] [CrossRef]
- Garcia, L.P.F.; Lorena, A.C.; Carvalho, A.C. A study on class noise detection and elimination. In Proceedings of the 2012 Brazilian Symposium on Neural Networks, Curitiba, Brazil, 20–25 October 2012; pp. 13–18. [Google Scholar]
- Sluban, B.; Gamberger, D.; Lavraě, N. Advances in class noise detection. In Proceedings of the ECAI 2010, Lisbon, Portugal, 16–20 August 2010; IOS Press: Amsterdam, The Netherlands, 2010; pp. 1105–1106. [Google Scholar]
- Zhu, X.; Wu, X.; Chen, Q. Eliminating class noise in large datasets. In Proceedings of the Proceedings of the 20th International Conference on Machine Learning (ICML-03), Washington, DC, USA, 21–24 August 2003; pp. 920–927. [Google Scholar]
- Khoshgoftaar, T.M.; Rebours, P. Improving software quality prediction by noise filtering techniques. J. Comput. Sci. Technol. 2007, 22, 387–396. [Google Scholar] [CrossRef]
- Biggio, B.; Nelson, B.; Laskov, P. Support vector machines under adversarial label noise. In Proceedings of the Asian Conference on Machine Learning, PMLR, Taoyuan, Taiwan, 13–15 November 2011; pp. 97–112. [Google Scholar]
- Jin, R.; Liu, Y.; Si, L.; Carbonell, J.G.; Hauptmann, A. A new boosting algorithm using input-dependent regularizer. In Proceedings of the Twentieth International Conference on Machine Learning, Washington, DC, USA, 21–24 August 2003; Carnegie Mellon University: Pittsburgh, PA, USA, 2003. [Google Scholar]
- Khardon, R.; Wachman, G. Noise Tolerant Variants of the Perceptron Algorithm. J. Mach. Learn. Res. 2007, 8, 227–248. [Google Scholar]
- Wu, X.; Kumar, V.; Ross Quinlan, J.; Ghosh, J.; Yang, Q.; Motoda, H.; McLachlan, G.J.; Ng, A.; Liu, B.; Yu, P.S.; et al. Top 10 algorithms in data mining. Knowl. Inf. Syst. 2008, 14, 1–37. [Google Scholar] [CrossRef] [Green Version]
- Han, H.; Wang, W.Y.; Mao, B.H. Borderline-SMOTE: A new over-sampling method in imbalanced data sets learning. In Proceedings of the International Conference on Intelligent Computing, Hefei, China, 23–26 August 2005; Springer: Berlin/Heidelberg, Germany, 2005; pp. 878–887. [Google Scholar]
- Morales, P.; Luengo, J.; Garcia, L.P.F.; Lorena, A.C.; de Carvalho, A.C.; Herrera, F. The NoiseFiltersR Package: Label Noise Preprocessing in R. R J. 2017, 9, 219. [Google Scholar] [CrossRef] [Green Version]
- Xia, S.; Huang, L.; Wang, G.; Gao, X.; Shao, Y.; Chen, Z. An adaptive and general model for label noise detection using relative probabilistic density. Knowl.-Based Syst. 2022, 239, 107907. [Google Scholar] [CrossRef]
- Fawcett, T. ROC graphs: Notes and practical considerations for researchers. Mach. Learn. 2004, 31, 1–38. [Google Scholar]
- Demšar, J. Statistical Comparisons of Classifiers over Multiple Data Sets. J. Mach. Learn. Res. 2006, 7, 1–30. [Google Scholar]
- Galar, M.; Fernández, A.; Barrenechea, E.; Bustince, H.; Herrera, F. An overview of ensemble methods for binary classifiers in multi-class problems: Experimental study on one-vs-one and one-vs-all schemes. Pattern Recognit. 2011, 44, 1761–1776. [Google Scholar] [CrossRef]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]





| Data | Classes | Features | Samples | Train | Pos | Neg | Test |
|---|---|---|---|---|---|---|---|
| bupa | 2 | 6 | 345 | 276 | 160 | 116 | 69 |
| diabetes | 2 | 8 | 768 | 614 | 214 | 400 | 154 |
| ecoli | 2 | 7 | 336 | 268 | 28 | 240 | 68 |
| fourclass | 2 | 2 | 862 | 689 | 245 | 444 | 173 |
| haberman | 2 | 3 | 306 | 244 | 65 | 179 | 62 |
| heart | 2 | 13 | 270 | 216 | 96 | 120 | 54 |
| image | 2 | 18 | 2086 | 1668 | 950 | 718 | 418 |
| newthyroid | 2 | 5 | 215 | 172 | 28 | 144 | 43 |
| pima | 2 | 8 | 768 | 614 | 214 | 400 | 154 |
| votes | 2 | 16 | 435 | 348 | 134 | 214 | 87 |
| Recall | False | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ENN | RENN | AllkNN | NCR | RD | IPF | Dynamic CF | EIRS- GNN | ENN | RENN | AllkNN | NCR | RD | IPF | Dynamic CF | EIRS- GNN | |
| 96.86 | 96.86 | 88.20 | 86.36 | 25.12 | 80.45 | 83.95 | 86.61 | 81.39 | 81.50 | 74.27 | 77.39 | 90.55 | 57.56 | 52.34 | 55.50 | |
| bupa | 85.19 | 85.19 | 37.04 | 62.96 | 33.33 | 44.44 | 51.85 | 62.96 | 88.56 | 88.56 | 88.89 | 87.50 | 91.09 | 88.79 | 83.91 | 82.60 |
| ecoli | 100.00 | 100.00 | 96.15 | 88.46 | 11.54 | 100.00 | 100.00 | 94.61 | 81.56 | 82.07 | 72.83 | 79.82 | 94.12 | 46.94 | 50.94 | 46.03 |
| fourclass | 100.00 | 100.00 | 100.00 | 97.06 | 17.65 | 77.94 | 97.06 | 100.00 | 72.24 | 72.47 | 52.78 | 61.85 | 84.62 | 68.07 | 10.81 | 3.41 |
| haberman | 95.65 | 95.65 | 95.65 | 95.65 | 56.52 | 73.91 | 73.91 | 77.39 | 87.85 | 87.85 | 83.33 | 86.98 | 86.17 | 76.39 | 79.52 | 74.55 |
| heart | 100.00 | 100.00 | 80.95 | 80.95 | 38.10 | 80.95 | 71.43 | 90.48 | 84.33 | 84.33 | 76.06 | 83.96 | 86.44 | 75.71 | 72.22 | 65.55 |
| image | 97.59 | 97.59 | 95.18 | 94.58 | 15.06 | 92.17 | 96.99 | 94.46 | 73.83 | 73.83 | 59.80 | 54.23 | 90.08 | 26.44 | 24.41 | 38.59 |
| newthyroid | 100.00 | 100.00 | 100.00 | 100.00 | 6.25 | 93.75 | 100.00 | 100.00 | 75.00 | 75.38 | 65.96 | 72.88 | 96.15 | 6.25 | 15.79 | 36.00 |
| pima | 95.08 | 95.08 | 86.89 | 81.97 | 40.98 | 70.49 | 75.41 | 72.79 | 86.51 | 86.51 | 83.84 | 86.23 | 89.08 | 76.88 | 74.01 | 76.50 |
| votes | 100.00 | 100.00 | 100.00 | 88.24 | 8.82 | 97.06 | 94.12 | 97.06 | 77.33 | 77.33 | 75.36 | 74.14 | 94.83 | 35.29 | 38.46 | 54.89 |
| diabetes | 95.08 | 95.08 | 90.16 | 73.77 | 22.95 | 73.77 | 78.69 | 76.39 | 86.70 | 86.70 | 83.82 | 86.28 | 92.93 | 74.86 | 73.33 | 76.85 |
| 95.26 | 95.45 | 77.35 | 82.95 | 34.89 | 82.46 | 82.17 | 82.43 | 71.89 | 72.22 | 64.15 | 69.07 | 79.84 | 43.20 | 42.78 | 40.89 | |
| bupa | 89.09 | 89.09 | 58.18 | 63.64 | 47.27 | 63.64 | 69.09 | 68.36 | 77.73 | 77.73 | 73.98 | 77.27 | 78.69 | 72.44 | 66.37 | 69.19 |
| ecoli | 98.11 | 100.00 | 90.57 | 92.45 | 28.30 | 86.79 | 86.79 | 88.68 | 65.56 | 65.81 | 57.89 | 70.48 | 82.35 | 20.69 | 31.34 | 29.21 |
| fourclass | 99.27 | 99.27 | 98.54 | 90.51 | 28.47 | 78.10 | 96.35 | 99.27 | 67.70 | 67.70 | 51.61 | 59.74 | 76.79 | 54.08 | 20.00 | 0.73 |
| haberman | 95.83 | 95.83 | 85.42 | 85.42 | 47.92 | 72.92 | 66.67 | 63.75 | 77.00 | 77.00 | 73.72 | 77.72 | 79.09 | 59.77 | 64.04 | 63.58 |
| heart | 88.37 | 88.37 | 79.07 | 93.02 | 39.53 | 83.72 | 79.07 | 83.72 | 75.16 | 75.16 | 63.04 | 69.70 | 78.75 | 52.63 | 51.43 | 41.16 |
| image | 98.20 | 98.20 | 76.28 | 87.39 | 26.73 | 91.89 | 92.79 | 93.99 | 68.31 | 68.31 | 53.90 | 50.00 | 79.73 | 19.69 | 19.53 | 21.86 |
| newthyroid | 100.00 | 100.00 | 90.91 | 81.82 | 24.24 | 93.94 | 90.91 | 90.30 | 62.92 | 65.98 | 62.96 | 68.97 | 80.49 | 16.22 | 28.57 | 25.13 |
| pima | 93.44 | 93.44 | 60.66 | 72.13 | 36.89 | 78.69 | 72.95 | 69.83 | 77.02 | 77.02 | 71.21 | 76.72 | 81.40 | 57.71 | 59.17 | 62.27 |
| votes | 98.53 | 98.53 | 69.12 | 85.29 | 29.41 | 97.06 | 94.12 | 89.71 | 70.61 | 70.61 | 62.40 | 64.85 | 80.58 | 19.51 | 28.89 | 35.51 |
| diabetes | 91.80 | 91.80 | 64.75 | 77.87 | 40.16 | 77.87 | 72.95 | 76.72 | 76.86 | 76.86 | 70.74 | 75.26 | 80.48 | 59.23 | 58.41 | 60.30 |
| 91.83 | 91.83 | 69.00 | 75.16 | 40.20 | 77.47 | 78.93 | 79.54 | 65.18 | 65.18 | 57.56 | 63.01 | 70.97 | 37.80 | 38.26 | 31.67 | |
| bupa | 92.68 | 92.68 | 63.41 | 68.29 | 43.90 | 64.63 | 71.95 | 66.83 | 68.72 | 68.72 | 64.63 | 65.22 | 72.73 | 58.27 | 52.03 | 53.37 |
| ecoli | 91.25 | 91.25 | 96.25 | 73.75 | 31.25 | 90.00 | 83.75 | 85.25 | 61.78 | 61.78 | 50.32 | 65.09 | 75.73 | 21.74 | 34.31 | 19.35 |
| fourclass | 97.09 | 97.09 | 71.84 | 84.47 | 36.89 | 78.64 | 89.32 | 99.51 | 61.61 | 61.61 | 50.99 | 57.14 | 69.35 | 36.96 | 23.65 | 0.68 |
| haberman | 86.11 | 86.11 | 76.39 | 69.44 | 40.28 | 73.61 | 65.28 | 63.89 | 68.69 | 68.69 | 63.58 | 70.93 | 72.90 | 46.46 | 57.27 | 52.08 |
| heart | 89.06 | 89.06 | 53.12 | 73.44 | 39.06 | 70.31 | 67.19 | 76.87 | 65.66 | 65.66 | 60.92 | 64.93 | 75.25 | 43.75 | 53.26 | 34.04 |
| image | 96.00 | 96.00 | 72.60 | 82.20 | 42.80 | 78.00 | 92.40 | 89.60 | 63.96 | 63.96 | 52.80 | 50.12 | 69.52 | 41.79 | 23.89 | 19.86 |
| newthyroid | 94.12 | 94.12 | 82.35 | 78.43 | 41.18 | 94.12 | 96.08 | 93.73 | 62.79 | 62.79 | 52.81 | 63.30 | 68.18 | 11.11 | 12.50 | 19.17 |
| pima | 88.04 | 88.04 | 54.89 | 71.74 | 43.48 | 72.83 | 71.74 | 63.59 | 67.92 | 67.92 | 63.93 | 67.49 | 71.43 | 47.86 | 48.44 | 51.49 |
| votes | 94.23 | 94.23 | 57.69 | 75.96 | 37.50 | 88.46 | 83.65 | 86.73 | 63.97 | 63.97 | 57.45 | 59.49 | 66.38 | 22.03 | 28.69 | 19.04 |
| diabetes | 89.67 | 89.67 | 61.41 | 73.91 | 45.65 | 64.13 | 67.93 | 69.46 | 66.73 | 66.73 | 58.15 | 66.42 | 68.18 | 48.02 | 48.56 | 47.60 |
| Average | 94.65 | 94.71 | 78.18 | 81.49 | 33.40 | 80.13 | 81.68 | 82.86 | 72.82 | 72.97 | 65.32 | 69.82 | 80.45 | 46.19 | 44.46 | 42.69 |
| NSY | ENN | RENN | AllkNN | NCR | RD | IPF | Dynamic CF | EIRS- kGNN | |
|---|---|---|---|---|---|---|---|---|---|
| 0.1 | |||||||||
| bupa | 67.78 | 68.48 | 68.55 | 69.15 | 68.23 | 65.78 | 52.15 | 62.17 | 64.78 |
| ecoli | 81.29 | 81.92 | 0.00 | 83.40 | 83.30 | 80.44 | 75.36 | 81.71 | 87.52 |
| fourclass | 92.92 | 94.14 | 93.98 | 94.05 | 94.44 | 94.97 | 81.92 | 94.60 | 94.83 |
| haberman | 57.65 | 65.92 | 65.92 | 58.74 | 65.64 | 66.33 | 68.57 | 65.46 | 70.10 |
| heart | 78.44 | 83.69 | 83.69 | 82.51 | 87.38 | 86.86 | 91.49 | 87.19 | 91.38 |
| image | 92.31 | 94.07 | 94.10 | 94.58 | 94.73 | 94.87 | 94.17 | 95.01 | 92.59 |
| newthyroid | 91.43 | 91.35 | 89.76 | 89.29 | 91.27 | 90.40 | 91.39 | 90.08 | 89.48 |
| pima | 74.36 | 74.65 | 75.03 | 75.08 | 77.86 | 78.65 | 72.00 | 77.91 | 76.52 |
| votes | 93.50 | 95.66 | 95.95 | 93.63 | 95.66 | 96.65 | 95.77 | 96.23 | 95.50 |
| diabetes | 77.15 | 79.16 | 79.35 | 78.37 | 79.13 | 79.29 | 77.36 | 80.25 | 80.79 |
| Average | 80.68 | 82.90 | 74.63 | 81.88 | 83.76 | 83.42 | 80.02 | 83.06 | 84.35 |
| 0.2 | |||||||||
| bupa | 71.52 | 65.66 | 65.66 | 70.86 | 68.31 | 69.85 | 57.18 | 72.34 | 70.47 |
| ecoli | 82.76 | 77.07 | 82.04 | 81.08 | 82.99 | 80.94 | 82.20 | 73.79 | 79.86 |
| fourclass | 90.84 | 93.71 | 93.71 | 95.57 | 93.90 | 95.81 | 74.66 | 95.01 | 96.35 |
| haberman | 68.74 | 73.19 | 73.19 | 70.60 | 73.15 | 71.66 | 0.00 | 72.23 | 73.30 |
| heart | 73.82 | 78.11 | 77.28 | 79.00 | 78.67 | 80.04 | 83.69 | 77.76 | 84.74 |
| image | 90.28 | 93.52 | 93.52 | 93.25 | 94.64 | 94.84 | 95.33 | 95.63 | 94.35 |
| newthyroid | 89.28 | 96.98 | 75.71 | 75.71 | 97.62 | 94.29 | 86.31 | 87.06 | 93.06 |
| pima | 72.22 | 71.35 | 71.48 | 76.20 | 75.14 | 76.06 | 74.02 | 76.74 | 74.87 |
| votes | 92.87 | 97.10 | 97.10 | 92.74 | 97.36 | 96.36 | 97.86 | 98.50 | 97.51 |
| diabetes | 71.37 | 70.31 | 71.05 | 69.69 | 74.15 | 69.95 | 72.77 | 75.34 | 73.08 |
| Average | 80.37 | 81.70 | 80.07 | 80.47 | 83.59 | 82.98 | 72.40 | 82.44 | 83.76 |
| 0.3 | |||||||||
| bupa | 54.68 | 61.95 | 61.07 | 56.90 | 57.27 | 59.07 | 51.65 | 63.66 | 65.25 |
| ecoli | 80.87 | 96.02 | 95.93 | 88.27 | 89.32 | 88.83 | 0.00 | 88.15 | 95.63 |
| fourclass | 85.96 | 86.18 | 86.20 | 86.89 | 90.32 | 90.70 | 79.52 | 92.34 | 95.65 |
| haberman | 52.76 | 50.69 | 50.39 | 58.06 | 56.35 | 56.45 | 0.00 | 62.04 | 61.10 |
| heart | 81.86 | 81.17 | 80.75 | 84.26 | 86.57 | 90.10 | 82.24 | 87.44 | 95.26 |
| image | 84.04 | 88.52 | 88.52 | 88.73 | 91.46 | 89.87 | 78.24 | 93.56 | 93.12 |
| newthyroid | 74.25 | 87.50 | 86.63 | 94.40 | 91.83 | 92.66 | 98.49 | 96.27 | 90.87 |
| pima | 70.61 | 71.94 | 71.90 | 72.31 | 73.44 | 68.82 | 72.16 | 78.46 | 74.52 |
| votes | 86.71 | 91.22 | 91.19 | 87.25 | 92.45 | 91.27 | 95.90 | 93.31 | 96.70 |
| diabetes | 65.74 | 67.66 | 67.44 | 65.94 | 67.31 | 70.68 | 69.25 | 70.80 | 68.65 |
| Average | 73.75 | 78.29 | 78.00 | 78.30 | 79.63 | 79.84 | 62.74 | 82.60 | 83.23 |
| ENN | RENN | AllkNN | NCR | RD | IPF | DynamicCF | EIRS-kGNN | |
|---|---|---|---|---|---|---|---|---|
| bupa | 0.0040 | 0.0040 | 0.0040 | 0.0171 | 0.5731 | 2.8240 | 4.6631 | 15.5996 |
| ecoli | 0.0046 | 0.0082 | 0.0145 | 0.0138 | 0.4846 | 1.5313 | 3.6875 | 11.9451 |
| fourclass | 0.0050 | 0.0084 | 0.0116 | 0.0364 | 3.5548 | 1.9304 | 8.4703 | 46.4174 |
| haberman | 0.0033 | 0.0039 | 0.0089 | 0.0138 | 0.4798 | 1.8660 | 3.6170 | 11.7543 |
| heart | 0.0049 | 0.0076 | 0.0053 | 0.0176 | 0.3460 | 5.5721 | 10.2261 | 9.1362 |
| image | 0.0754 | 0.0819 | 0.0774 | 0.2014 | 20.0588 | 2.1662 | 1.2292 | 252.4761 |
| newthyroid | 0.0040 | 0.0096 | 0.0110 | 0.0103 | 0.2554 | 1.6695 | 2.3169 | 6.1508 |
| pima | 0.0108 | 0.0083 | 0.0083 | 0.0449 | 2.5337 | 2.6234 | 12.0903 | 49.4904 |
| votes | 0.0049 | 0.0050 | 0.0088 | 0.0196 | 0.8574 | 2.5050 | 9.0699 | 19.4140 |
| diabetes | 0.0128 | 0.0119 | 0.0144 | 0.0401 | 2.7852 | 2.4052 | 12.5797 | 52.3045 |
| Average | 0.0130 | 0.0149 | 0.0164 | 0.0415 | 3.1929 | 2.5093 | 6.7950 | 47.4688 |
| Data | Samples | Classes | Features | Each Class |
|---|---|---|---|---|
| glass | 214 | 6 | 9 | [7, 23, 56, 60, 13, 10] |
| iris | 150 | 3 | 4 | [40, 40, 40] |
| newthyroid | 215 | 3 | 5 | [120, 28, 24] |
| seeds | 210 | 3 | 7 | [56, 56, 56] |
| segmentation | 2310 | 7 | 16 | [264, 264, 264, 264, 264, 264, 264] |
| vertebralColumn | 310 | 3 | 6 | [48, 80, 120] |
| wine | 178 | 3 | 13 | [47, 56, 38] |
| yeast | 1484 | 10 | 8 | [370, 343, 195, 35, 130, 24, 16, 40, 28, 4] |
| Recall | False | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Data | ENN | RENN | AllNN | NCR | RD | IPF | Dynamic CF | EIRS- GNN | ENN | RENN | AllNN | NCR | RD | IPF | Dynamic CF | EIRS- GNN |
| 0.10 | 100.00 | 100.00 | 93.67 | 97.34 | 27.82 | 97.30 | 96.18 | 93.90 | 80.68 | 80.68 | 67.64 | 54.18 | 90.12 | 47.78 | 51.00 | 52.24 |
| glass | 100.00 | 100.00 | 93.33 | 93.33 | 40.00 | 100.00 | 100.00 | 88.00 | 86.73 | 86.73 | 79.10 | 77.05 | 91.04 | 75.41 | 77.94 | 80.45 |
| iris | 100.00 | 100.00 | 91.67 | 100.00 | 25.00 | 83.33 | 100.00 | 100.00 | 73.91 | 73.91 | 57.69 | 40.00 | 83.33 | 44.44 | 25.00 | 28.53 |
| newthyroid | 100.00 | 100.00 | 87.50 | 100.00 | 18.75 | 100.00 | 93.75 | 91.25 | 76.12 | 76.12 | 61.11 | 40.74 | 88.46 | 40.74 | 40.00 | 50.89 |
| seeds | 100.00 | 100.00 | 100.00 | 100.00 | 6.67 | 100.00 | 93.33 | 97.33 | 79.45 | 79.45 | 57.14 | 46.43 | 96.30 | 37.50 | 46.15 | 47.86 |
| segmentation | 100.00 | 100.00 | 89.56 | 98.90 | 15.38 | 99.45 | 98.35 | 97.80 | 74.76 | 74.76 | 55.71 | 35.71 | 90.11 | 23.63 | 31.68 | 36.51 |
| vertebralColumn | 100.00 | 100.00 | 91.67 | 91.67 | 45.83 | 100.00 | 95.83 | 89.17 | 86.29 | 86.29 | 77.55 | 77.55 | 87.91 | 60.66 | 63.49 | 73.86 |
| wine | 100.00 | 100.00 | 100.00 | 100.00 | 8.33 | 100.00 | 91.67 | 100.00 | 79.66 | 79.66 | 69.23 | 33.33 | 94.12 | 20.00 | 42.11 | 18.46 |
| yeast | 100.00 | 100.00 | 95.65 | 94.78 | 62.61 | 95.65 | 96.52 | 87.66 | 88.53 | 88.53 | 83.58 | 82.64 | 89.68 | 79.85 | 81.62 | 81.39 |
| 0.20 | 99.08 | 99.08 | 89.73 | 91.05 | 38.87 | 95.30 | 94.14 | 91.19 | 72.23 | 72.23 | 57.16 | 46.53 | 80.39 | 37.68 | 39.54 | 37.51 |
| glass | 100.00 | 100.00 | 100.00 | 96.88 | 43.75 | 100.00 | 96.88 | 89.37 | 75.38 | 75.38 | 62.79 | 62.65 | 84.27 | 59.49 | 64.77 | 63.62 |
| iris | 100.00 | 100.00 | 91.67 | 83.33 | 20.83 | 95.83 | 95.83 | 100.00 | 70.37 | 70.37 | 51.11 | 42.86 | 86.11 | 28.12 | 14.81 | 15.42 |
| newthyroid | 100.00 | 100.00 | 96.97 | 93.94 | 39.39 | 96.97 | 90.91 | 88.49 | 68.27 | 68.27 | 53.62 | 32.61 | 75.00 | 21.95 | 34.78 | 35.11 |
| seeds | 96.97 | 96.97 | 78.79 | 81.82 | 27.27 | 84.85 | 90.91 | 90.30 | 71.68 | 71.68 | 51.85 | 38.64 | 79.55 | 30.00 | 28.57 | 32.27 |
| segmentation | 99.73 | 99.73 | 89.29 | 98.08 | 30.49 | 99.73 | 98.08 | 97.58 | 67.56 | 67.56 | 46.81 | 29.59 | 79.93 | 13.16 | 18.86 | 18.15 |
| vertebralColumn | 95.92 | 95.92 | 75.51 | 81.63 | 53.06 | 91.84 | 95.92 | 84.08 | 75.52 | 75.52 | 67.26 | 61.54 | 76.36 | 43.75 | 50.00 | 57.44 |
| wine | 100.00 | 100.00 | 96.30 | 100.00 | 33.33 | 100.00 | 88.89 | 100.00 | 70.97 | 70.97 | 51.85 | 35.71 | 80.43 | 41.30 | 35.14 | 10.00 |
| yeast | 100.00 | 100.00 | 89.32 | 92.74 | 62.82 | 93.16 | 95.73 | 79.66 | 78.07 | 78.07 | 71.95 | 68.64 | 81.49 | 63.67 | 69.40 | 68.07 |
| 0.30 | 98.57 | 98.57 | 84.23 | 89.06 | 54.93 | 92.58 | 93.89 | 90.82 | 64.83 | 64.83 | 51.61 | 42.61 | 69.09 | 34.66 | 30.56 | 27.12 |
| glass | 97.92 | 97.92 | 97.92 | 93.75 | 75.00 | 97.92 | 91.67 | 89.17 | 68.87 | 68.87 | 53.47 | 51.61 | 66.67 | 51.55 | 57.69 | 49.99 |
| iris | 97.22 | 97.22 | 80.56 | 88.89 | 50.00 | 86.11 | 97.22 | 96.11 | 64.29 | 64.29 | 45.28 | 36.00 | 66.67 | 24.39 | 20.45 | 5.47 |
| newthyroid | 98.04 | 98.04 | 84.31 | 96.08 | 35.29 | 96.08 | 90.20 | 85.88 | 63.77 | 63.77 | 50.57 | 31.94 | 75.68 | 22.22 | 31.34 | 21.79 |
| seeds | 100.00 | 100.00 | 79.17 | 85.42 | 47.92 | 79.17 | 89.58 | 92.09 | 63.36 | 63.36 | 48.65 | 37.88 | 67.14 | 35.59 | 15.69 | 23.25 |
| segmentation | 100.00 | 100.00 | 88.61 | 95.48 | 45.93 | 99.64 | 98.55 | 97.65 | 61.52 | 61.52 | 44.25 | 30.80 | 71.23 | 14.71 | 16.15 | 12.31 |
| vertebralColumn | 95.95 | 95.95 | 74.32 | 77.03 | 56.76 | 90.54 | 94.59 | 82.16 | 66.67 | 66.67 | 58.65 | 57.46 | 70.83 | 41.23 | 35.19 | 45.90 |
| wine | 100.00 | 100.00 | 78.05 | 82.93 | 56.10 | 95.12 | 92.68 | 100.00 | 61.68 | 61.68 | 50.77 | 38.18 | 63.49 | 36.07 | 11.63 | 4.20 |
| yeast | 99.43 | 99.43 | 90.91 | 92.90 | 72.44 | 96.02 | 96.59 | 83.52 | 68.50 | 68.50 | 61.21 | 57.03 | 70.99 | 51.51 | 56.35 | 54.02 |
| Data | NSY | ENN | RENN | AllkNN | NCR | RD | IPF | Dynamic CF | EIRS- kGNN |
|---|---|---|---|---|---|---|---|---|---|
| glass | 69.29 | 62.07 | 62.07 | 70.48 | 69.27 | 66.54 | 63.20 | 65.87 | 65.34 |
| iris | 91.50 | 89.83 | 89.83 | 90.67 | 91.00 | 91.00 | 91.00 | 88.50 | 94.83 |
| newthyroid | 77.43 | 82.40 | 82.49 | 79.22 | 73.82 | 73.85 | 75.83 | 78.34 | 76.14 |
| seeds | 87.38 | 87.50 | 86.90 | 91.67 | 90.83 | 90.83 | 88.45 | 91.07 | 90.24 |
| segmentation | 92.77 | 94.32 | 94.25 | 94.60 | 95.19 | 95.30 | 95.39 | 95.39 | 94.73 |
| vertebralColumn | 78.15 | 76.50 | 76.69 | 80.70 | 82.61 | 81.33 | 79.41 | 81.56 | 83.35 |
| wine | 91.93 | 89.96 | 88.88 | 87.88 | 90.35 | 90.13 | 90.97 | 90.59 | 92.62 |
| yeast | 66.15 | 68.10 | 68.03 | 70.03 | 69.29 | 69.89 | 63.87 | 70.03 | 67.01 |
| Average | 81.83 | 81.33 | 81.14 | 83.16 | 82.79 | 82.36 | 81.02 | 82.67 | 83.03 |
| glass | 60.58 | 57.03 | 57.12 | 64.37 | 61.65 | 62.73 | 61.14 | 63.64 | 64.19 |
| iris | 90.17 | 91.50 | 91.50 | 90.33 | 90.33 | 91.00 | 94.17 | 91.17 | 94.83 |
| newthyroid | 74.92 | 72.77 | 71.75 | 76.80 | 75.83 | 76.43 | 74.46 | 75.83 | 74.20 |
| seeds | 85.83 | 87.50 | 88.10 | 89.88 | 89.40 | 89.40 | 90.00 | 87.86 | 88.57 |
| segmentation | 89.07 | 92.46 | 92.35 | 93.70 | 94.53 | 94.43 | 94.31 | 94.97 | 94.61 |
| vertebralColumn | 74.56 | 75.67 | 75.60 | 81.07 | 79.80 | 77.64 | 79.47 | 77.97 | 80.43 |
| wine | 84.15 | 86.87 | 85.56 | 89.89 | 90.76 | 90.05 | 90.45 | 92.84 | 92.26 |
| yeast | 63.28 | 64.98 | 65.03 | 69.14 | 69.09 | 69.51 | 64.07 | 67.99 | 67.94 |
| Average | 77.82 | 78.60 | 78.38 | 81.90 | 81.42 | 81.40 | 81.01 | 81.53 | 82.13 |
| glass | 60.45 | 57.50 | 58.87 | 67.56 | 64.97 | 66.69 | 59.94 | 68.17 | 64.15 |
| iris | 82.83 | 85.50 | 86.33 | 87.33 | 90.67 | 90.33 | 93.33 | 92.00 | 94.83 |
| newthyroid | 68.41 | 71.31 | 70.67 | 73.39 | 71.93 | 73.21 | 72.18 | 77.55 | 74.68 |
| seeds | 80.71 | 85.60 | 86.07 | 84.29 | 88.10 | 86.19 | 89.29 | 84.64 | 88.10 |
| segmentation | 83.81 | 91.05 | 91.21 | 92.25 | 93.83 | 94.26 | 93.99 | 94.79 | 94.37 |
| vertebralColumn | 70.20 | 72.05 | 72.61 | 77.74 | 71.98 | 78.79 | 78.25 | 76.95 | 79.31 |
| wine | 83.98 | 86.89 | 86.23 | 83.66 | 85.74 | 88.96 | 91.12 | 90.55 | 93.48 |
| yeast | 57.85 | 58.65 | 59.02 | 64.32 | 68.52 | 67.28 | 61.38 | 64.41 | 66.71 |
| Average | 73.53 | 76.07 | 76.38 | 78.82 | 79.47 | 80.71 | 79.94 | 81.13 | 81.95 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, B.; Huang, L.; Chen, Z.; Wang, G. An Ensemble and Iterative Recovery Strategy Based kGNN Method to Edit Data with Label Noise. Mathematics 2022, 10, 2743. https://doi.org/10.3390/math10152743
Chen B, Huang L, Chen Z, Wang G. An Ensemble and Iterative Recovery Strategy Based kGNN Method to Edit Data with Label Noise. Mathematics. 2022; 10(15):2743. https://doi.org/10.3390/math10152743
Chicago/Turabian StyleChen, Baiyun, Longhai Huang, Zizhong Chen, and Guoyin Wang. 2022. "An Ensemble and Iterative Recovery Strategy Based kGNN Method to Edit Data with Label Noise" Mathematics 10, no. 15: 2743. https://doi.org/10.3390/math10152743
APA StyleChen, B., Huang, L., Chen, Z., & Wang, G. (2022). An Ensemble and Iterative Recovery Strategy Based kGNN Method to Edit Data with Label Noise. Mathematics, 10(15), 2743. https://doi.org/10.3390/math10152743






