Classification of Alzheimer’s Disease Based on Core-Large Scale Brain Network Using Multilayer Extreme Learning Machine
Abstract
:1. Introduction
2. Materials
2.1. fMRI Dataset
2.2. Subjects
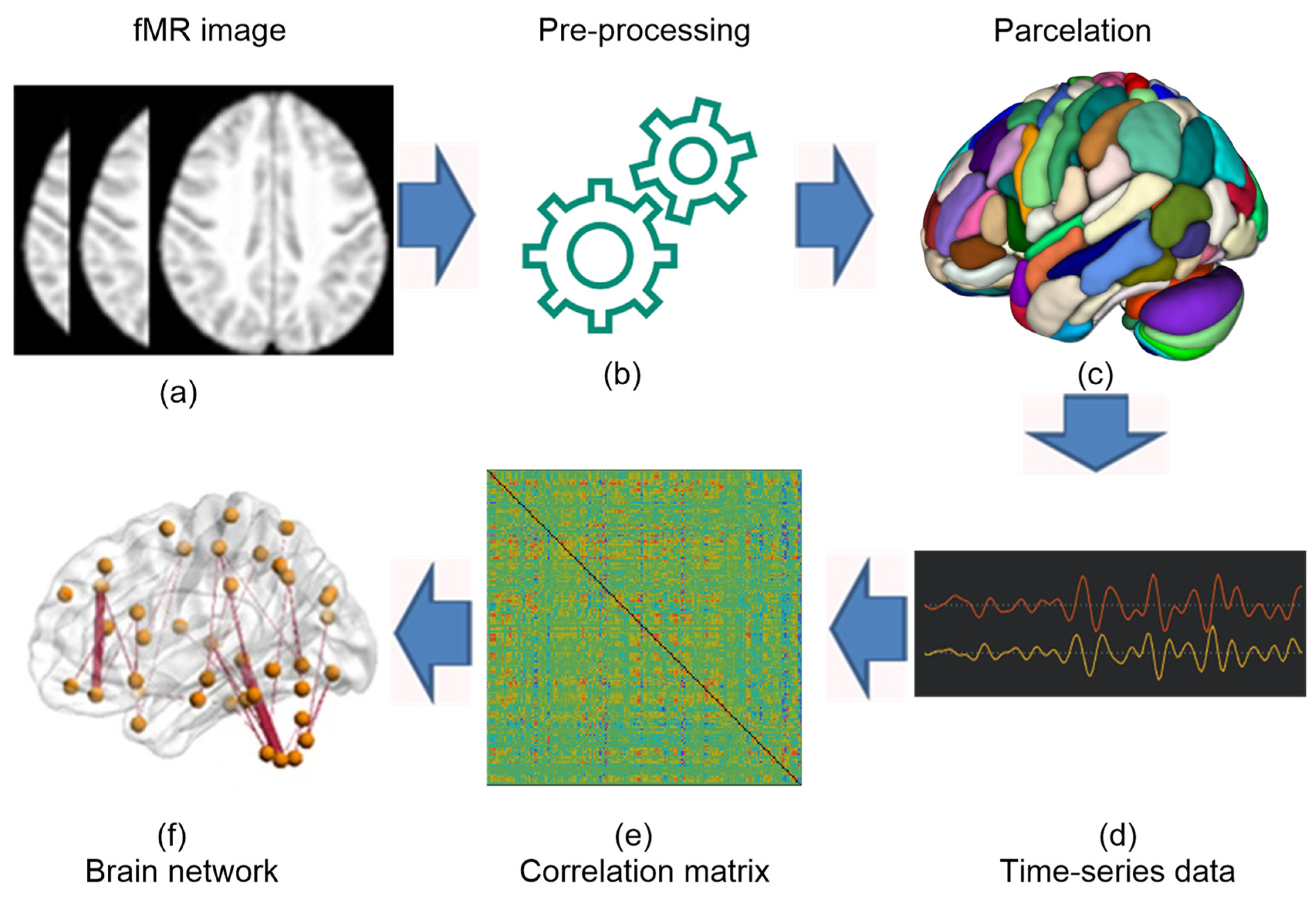
2.3. Data Preprocessing
2.4. Functional Connectivity Measures
2.5. ROI-to-ROI Connectivity (RRC) Matrices
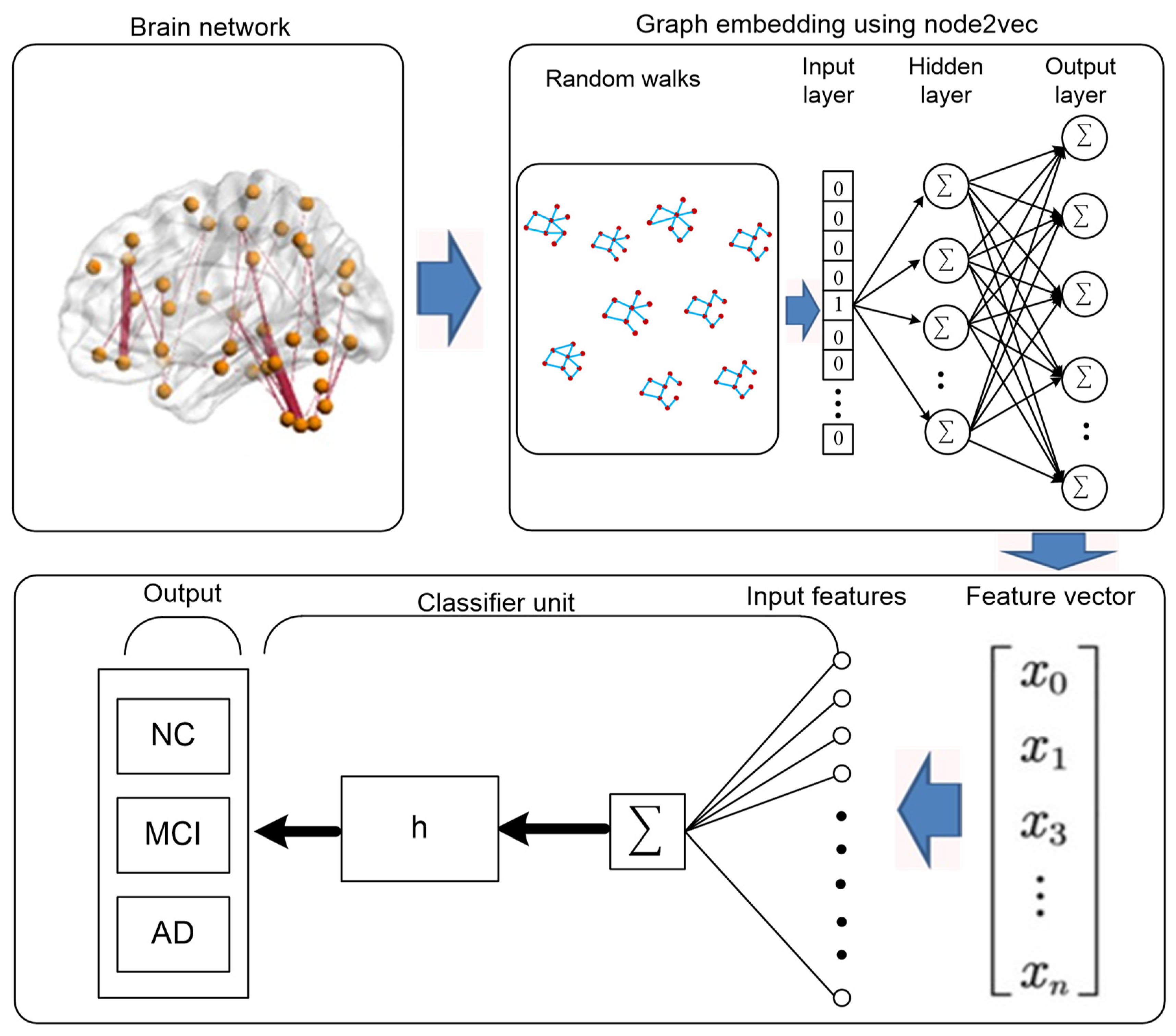
2.6. Proposed Framework
- Construction of brain networks including large scale brain network, whole brain network and combined brain network.
- Convert graph data to feature vector using graph embedding.
- Perform the feature selection on embedded data.
- Perform the classification using single layered regularized extreme learning machine (SL-RELM) and multilayered regularized extreme learning machine (ML-RELM).
2.7. Construction of Brain Networks
2.8. Graph-Embedding
3. Feature Selection
3.1. Least Absolute Shrinkage and Selection Operator (LASSO)
3.2. Features Selection with Adaptive Structure Learning (FSASL)
3.3. Local Learning and Clustering Based Feature Selection (LLCFS)
3.4. Pairwise Correlation-Based Feature Selection (CFS)
4. Classification
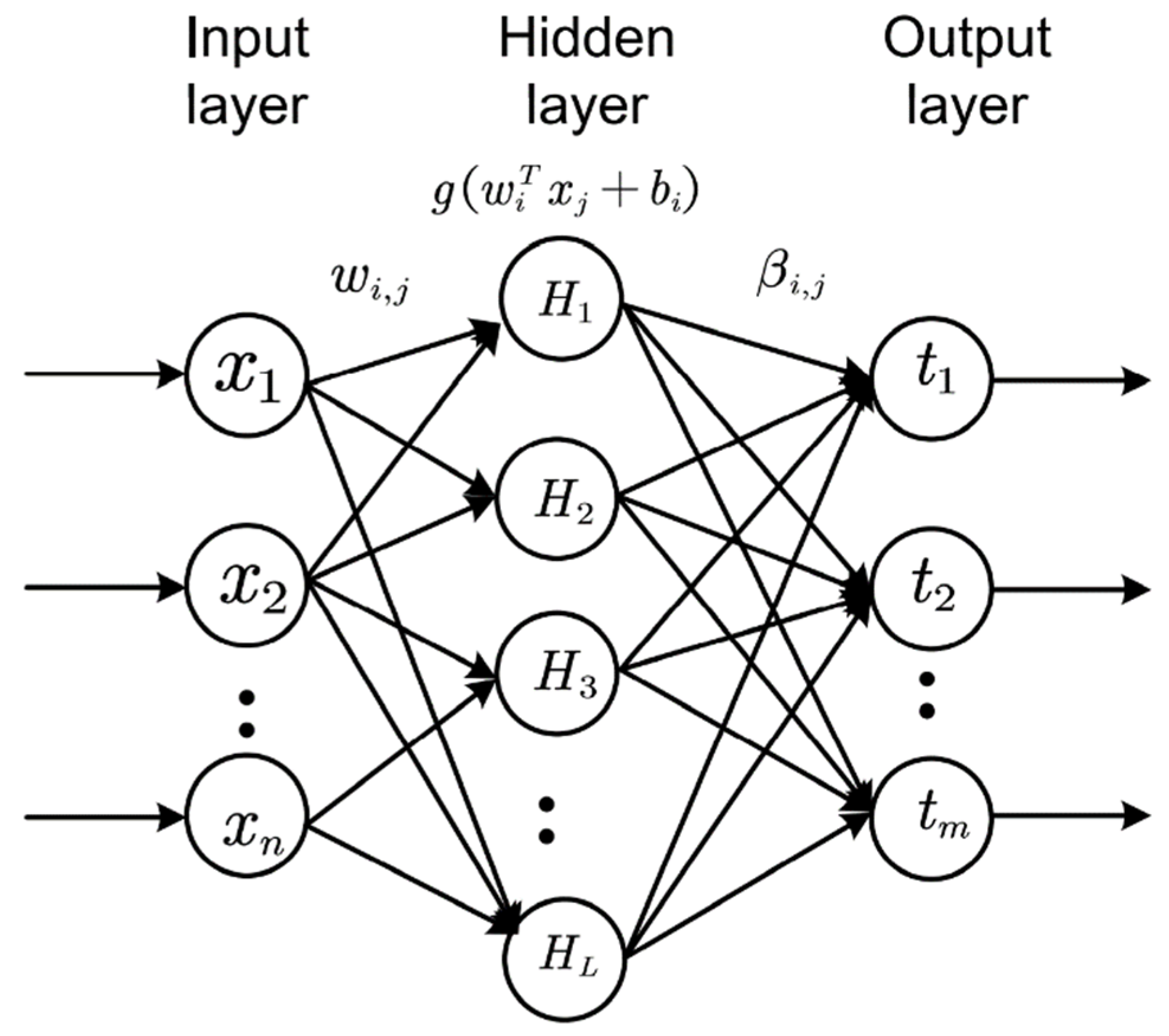
4.1. Extreme Learning Machine (ELM)
| Algorithm 1. Pseudocode for multiple hidden layer ELM. |
| Input: feature matrix , output matrix , regularization for all layers, input weights , biases and activation and the number of layers Output: hidden layer feature representation and output weight |
| Step1: Let , calculate Step2: For Step3: calculate Step4: Step5: Let , calculate and |
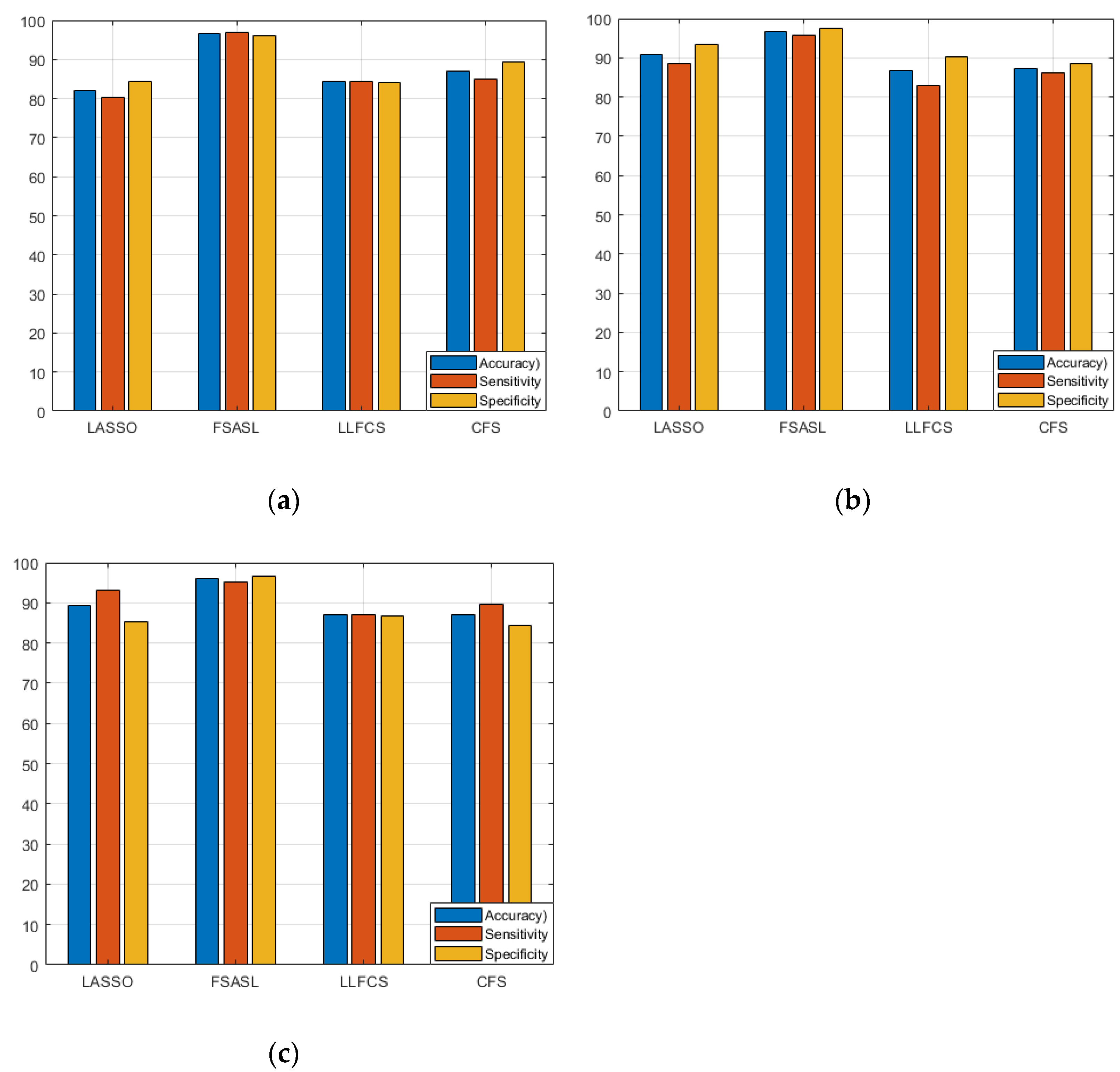
4.2. Experiment and Performance Evaluation
5. Discussion
6. Limitations
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- American Psychiatric Association. Task Force on DSM-IV. In Diagnostic and Statistical Manual of Mental Disorders, 4th ed.; DSM-IV; American Psychiatric Association: Washington, DC, USA, 1994; Volume 25. [Google Scholar]
- Schmitter, D.; Roche, A.; Maréchal, B.; Ribes, D.; Abdulkadir, A.; Bach-Cuadra, M.; Daducci, A.; Granziera, C.; Klöppel, S.; Maeder, P.; et al. An evaluation of volume-based morphometry for prediction of mild cognitive impairment and Alzheimer’s disease. NeuroImage Clin. 2015, 7, 7–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alzheimer’s Association. 2016 Alzheimer’s disease facts and figures. Alzheimer’s Dementia 2016, 12, 459–509. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhou, L.; Shen, C.; Yin, J. Multiple kernel learning in the primal for multimodal Alzheimer’s disease classification. IEEE J. Biomed. Health Inform. 2014, 18, 984–990. [Google Scholar] [CrossRef] [PubMed]
- Wong, W. Economic burden of Alzheimer disease and managed care considerations. Am. J. Manag. Care 2020, 26, S177–S183. [Google Scholar]
- Lama, R.K.; Gwak, J.S.; Park, J.S.; Lee, S.W. Diagnosis of Alzheimer’s disease based on structural MRI images using a regularized extreme learning machine and PCA features. J. Healthc. Eng. 2017, 2017, 5485080. [Google Scholar] [CrossRef]
- Lama, R.K.; Kwon, G.R. Diagnosis of Alzheimer’s Disease Using Brain Network. Front. Neurosci. 2021, 15, 605115. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, Y.; Zhou, L.; Yuan, H.; Shen, D. Multimodal classification of Alzheimer’s disease and mild cognitive impairment. NeuroImage 2011, 55, 856–867. [Google Scholar] [CrossRef] [Green Version]
- Phillips, J.S.; Da Re, F.; Dratch, L.; Xie, S.X.; Irwin, D.J.; McMillan, C.T.; Vaishnavi, S.N.; Ferrarese, C.; Lee, E.B.; Shaw, L.M.; et al. Neocortical origin and progression of gray matter atrophy in nonamnestic Alzheimer’s disease. Neurobiol. Aging 2018, 63, 75–87. [Google Scholar] [CrossRef]
- Du, A.T.; Schuff, N.; Kramer, J.H.; Rosen, H.J.; Gorno-Tempini, M.L.; Rankin, K.; Miller, B.L.; Weiner, M.W. Different regional patterns of cortical thinning in Alzheimer’s disease and frontotemporal dementia. Brain 2007, 130, 1159–1166. [Google Scholar] [CrossRef]
- Good, C.D.; Scahill, R.I.; Fox, N.C.; Ashburner, J.; Friston, K.J.; Chan, D.; Crum, W.R.; Rossor, M.N.; Frackowiak, R.S. Automatic differentiation of anatomical patterns in the human brain: Validation with studies of degenerative dementias. NeuroImage 2002, 17, 29–46. [Google Scholar] [CrossRef]
- Shi, F.; Liu, B.; Zhou, Y.; Yu, C.; Jiang, T. Hippocampal volume and asymmetry in mild cognitive impairment and Alzheimer’s disease: Meta-analyses of MRI studies. Hippocampus 2009, 19, 1055–1064. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, J.; Friston, K.J. Voxel-based morphometry-The methods. NeuroImage 2000, 11, 805–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trivedi, M.A.; Wichmann, A.K.; Torgerson, B.M.; Ward, M.A.; Schmitz, T.W.; Ries, M.L.; Koscik, R.L.; Asthana, S.; Johnson, S.C. Structural MRI discriminates individuals with Mild Cognitive Impairment from age-matched controls: A combined neuropsychological and voxel based morphometry study. Alzheimer’s Dementia 2006, 2, 296–302. [Google Scholar] [CrossRef] [Green Version]
- Karas, G.B.; Scheltens, P.; Rombouts, S.A.; Visser, P.J.; van Schijndel, R.A.; Fox, N.C.; Barkhof, F. Global and local gray matter loss in mild cognitive impairment and Alzheimer’s disease. NeuroImage 2004, 23, 708–716. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Ward, B.D.; Xie, C.; Li, W.; Wu, Z.; Jones, J.L.; Franczak, M.; Antuono, P.; Li, S.J. Classification of Alzheimer disease, mild cognitive impairment, and normal cognitive status with large-scale network analysis based on resting-state functional MR imaging. Radiology 2011, 259, 213–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Jiang, T.; Liang, M.; Wang, L.; Tian, L.; Zhang, X.; Li, K.; Liu, Z. Discriminative analysis of early Alzheimer’s disease based on two intrinsically anti-correlated networks with resting-state fMRI. Int. Conf. Med. Image Comput. Comput. Assist. Interv. 2006, 4191, 340–347. [Google Scholar] [CrossRef] [Green Version]
- Challis, E.; Hurley, P.; Serra, L.; Bozzali, M.; Oliver, S.; Cercignani, M. Gaussian process classification of Alzheimer’s disease and mild cognitive impairment from resting-state fMRI. NeuroImage 2015, 112, 232–243. [Google Scholar] [CrossRef] [Green Version]
- Jie, B.; Zhang, D.; Gao, W.; Wang, Q.; Wee, C.Y.; Shen, D. Integration of network topological and connectivity properties for neuroimaging classification. IEEE Trans. Biomed. Eng. 2014, 61, 576–589. [Google Scholar] [CrossRef]
- Khazaee, A.; Ebrahimzadeh, A.; Babajani-Feremi, A. Identifying patients with Alzheimer’s disease using resting-state fMRI and graph theory. J. Int. Fed. Clin. Neurophysiol. 2015, 126, 2132–2141. [Google Scholar] [CrossRef]
- Greicius, M.D.; Srivastava, G.; Reiss, A.L.; Menon, V. Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: Evidence from functional MRI. Proc. Natl. Acad. Sci. USA 2004, 101, 4637–4642. [Google Scholar] [CrossRef] [Green Version]
- Menon, V. Large-scale brain networks and psychopathology: A unifying triple network model. Trends Cogn. Sci. 2011, 15, 483–506. [Google Scholar] [CrossRef] [PubMed]
- Joo, S.H.; Lim, H.K.; Lee, C.U. Three large-scale functional brain networks from resting-state functional MRI in subjects with different levels of cognitive impairment. Psychiatry Investig. 2016, 13, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Vecchio, F.; Miraglia, F.; Rossini, P.M. Connectome: Graph theory application in functional brain network architecture. Clin. Neurophysiol. Pract. 2017, 2, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://adni.loni.usc.edu/ (accessed on 27 October 2021).
- Nieto-Castanon, A. Handbook of Functional Connectivity Magnetic Resonance Imaging Methods in CONN; Hilbert Press: Boston, MA, USA, 2020. [Google Scholar]
- Penny, W.D.; Friston, K.J.; Ashburner, J.T.; Kiebel, S.J.; Nichols, T.E. Statistical Parametric Mapping: The Analysis of Functional Brain Images; Elsevier: Amsterdam, The Netherlands, 2007. [Google Scholar]
- Andersson, J.L.; Hutton, C.; Ashburner, J.; Turner, R.; Friston, K. Modeling geometric deformations in EPI time series. NeuroImage 2001, 13, 903–919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henson, R.N.A.; Buechel, C.; Josephs, O.; Friston, K.J. The slice-timing problem in event-related fMRI. NeuroImage 1999, 9, 125. [Google Scholar] [CrossRef] [Green Version]
- Ashburner, J.; Friston, K.J. Unified segmentation. NeuroImage 2005, 26, 839–851. [Google Scholar] [CrossRef]
- Behzadi, Y.; Restom, K.; Liau, J.; Liu, T.T. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. NeuroImage 2007, 37, 90–101. [Google Scholar] [CrossRef] [Green Version]
- Grover, A.; Leskovec, J. Node2vec: Scalable feature learning for networks. In Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Las Vegas, NV, USA, 13–17 August 2016; pp. 855–864. [Google Scholar] [CrossRef] [Green Version]
- Tibshirani, R. Regression Shrinkage and Selection via the Lasso. J. R. Stat. Soc. Ser. B 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Du, L.; Shen, Y.D. Unsupervised Feature Selection with Adaptive Structure Learning. 2015. Available online: http://arxiv.org/abs/1504.00736 (accessed on 14 November 2021).
- Zeng, H.; Cheung, Y.M. Feature selection and kernel learning for local learning-based clustering. IEEE Trans. Pattern Anal. Mach. Intell. 2011, 33, 1532–1547. [Google Scholar] [CrossRef] [Green Version]
- Hall, M.A. Correlation-based Feature Selection for Machine Learning. Ph.D. Thesis, The University of Waikato, Hamilton, New Zealand, 1999. [Google Scholar]
- Cao, J.; Zhang, K.; Luo, M.; Yin, C.; Lai, X. Extreme learning machine and adaptive sparse representation for image classification. Neural Netw. 2016, 81, 91–102. [Google Scholar] [CrossRef]
- Zhang, W.; Shen, H.; Ji, Z.; Meng, G.; Wang, B. Identification of mild cognitive impairment using extreme learning machines model. In Proceedings of the International Conference on Intelligent Computing, Fuzhou, China, 20–23 August 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 589–600. [Google Scholar]
- Peng, X.; Lin, P.; Zhang, T.; Wang, J. Extreme learning machine-based classification of ADHD using brain structural MRI data. PLoS ONE 2013, 8, e79476. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, M.N.I.; Min, B.; Jo, H.J.; Lee, B. Multiclass classification for the differential diagnosis on the ADHD subtypes using recursive feature elimination and hierarchical extreme learning machine: Structural MRI study. PLoS ONE 2016, 11, e0160697. [Google Scholar] [CrossRef] [Green Version]
- Cambria, E.; Huang, G.B.; Kasun, L.L.C.; Zhou, H.; Vong, C.M.; Lin, J.; Yin, J.; Cai, Z.; Liu, Q.; Li, K.; et al. Extreme learning machines [trends & controversies. IEEE Intell. Syst. 2013, 28, 30–59. [Google Scholar] [CrossRef]
- De Vos, F.; Koini, M.; Schouten, T.M.; Seiler, S.; van der Grond, J.; Lechner, A.; Schmidt, R.; de Rooij, M.; Rombouts, S.A. A comprehensive analysis of resting state fMRI measures to classify individual patients with Alzheimer’s disease. Neuroimage 2018, 167, 62–72. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Greicius, M.D.; Gennatas, E.D.; Growdon, M.E.; Jang, J.Y.; Rabinovici, G.D.; Kramer, J.H.; Weiner, M.; Miller, B.L.; Seeley, W.W. Divergent network connectivity changes in behavioural variant frontotemporal dementia and Alzheimer’s disease. Brain 2010, 133, 1352–1367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Zeng, W.; Deng, J.; Nie, W.; Zhang, Y. The identification of Alzheimer’s disease using functional connectivity between activity voxels in resting-state fMRI data. IEEE J. Transl. Eng. Health Med. 2020, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Eavani, H.; Satterthwaite, T.D.; Gur, R.E.; Gur, R.C.; Davatzikos, C. Unsupervised learning of functional network dynamics in resting state fMRI. Int. Conf. Inf. Processing Med. Imaging 2013, 7917, 426–437. [Google Scholar] [CrossRef] [Green Version]
- Wee, C.Y.; Yap, P.T.; Zhang, D.; Wang, L.; Shen, D. Group-constrained sparse fMRI connectivity modeling for mild cognitive impairment identification. Brain Struct. Funct. 2014, 219, 641–656. [Google Scholar] [CrossRef] [Green Version]
- Ju, R.; Hu, C.; Li, Q. Early diagnosis of Alzheimer’s disease based on resting-state brain networks and deep learning. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017, 16, 244–257. [Google Scholar] [CrossRef]
- Suk, H.I.; Wee, C.Y.; Lee, S.W.; Shen, D. State-space model with deep learning for functional dynamics estimation in resting-state fMRI. NeuroImage 2016, 129, 292–307. [Google Scholar] [CrossRef] [Green Version]






| HC (31) | MCI (31) | AD (33) | |
|---|---|---|---|
| Mean (Standard Deviation) | Mean (Standard Deviation) | Mean (Standard Deviation) | |
| Age | 73.4 ± 4.5 | 74.1 ± 4.9 | 73.2 ± 5.6 |
| Global CDR | 0.05 ± 0.21 | 0.52 ± 0.2 | 0.97 ± 0.29 |
| MMSE | 27.5 ± 1.9 | 26.5 ± 2.12 | 20.6 ± 2.5 |
| Feature Selection Method | Performance Metrics | Accuracy | Sensitivity | Specificity | F-Measure |
|---|---|---|---|---|---|
| LASSO | Mean (%) | 82.06 | 78.58 | 85.58 | 0.86 |
| Standard deviation | 2.67 | 2.751 | 4.25 | ||
| FSASL | Mean (%) | 86.51 | 85.25 | 88.00 | 0.92 |
| Standard deviation | 3.670 | 6.18 | 4.75 | ||
| LLCFS | Mean (%) | 85.24 | 78.66 | 91.91 | 0.85 |
| Standard deviation | 4.06 | 7.59 | 5.65 | ||
| CFS | Mean (%) | 86.28 | 82.33 | 90.08 | 0.86 |
| Standard deviation | 3.27 | 6.51 | 4.88 |
| Feature Selection Method | Performance Metrics | Accuracy | Sensitivity | Specificity | F-Measure |
|---|---|---|---|---|---|
| LASSO | Mean (%) | 90.64 | 83.33 | 98.08 | 0.995 |
| Standard deviation | 2.05 | 4.27 | 3.19 | ||
| FSASL | Mean (%) | 96.14 | 95.16 | 97.08 | 0.97 |
| Standard deviation | 1.71 | 2.74 | 1.89 | ||
| LLCFS | Mean (%) | 85.40 | 81.0 | 89.83 | 0.95 |
| Standard deviation | 4.03 | 4.33 | 6.67 | ||
| CFS | Mean (%) | 89.09 | 86.33 | 92.00 | 0.89 |
| Standard deviation | 4.10 | 6.54 | 4.12 |
| Feature Selection Method | Performance Metrics | Accuracy | Sensitivity | Specificity | F-Measure |
|---|---|---|---|---|---|
| LASSO | Mean (%) | 90.05 | 93.33 | 86.67 | 0.96 |
| Standard deviation | 2.50 | 3.19 | 3.98 | ||
| FSASL | Mean (%) | 95.19 | 94.16 | 96.16 | 1 |
| Standard deviation | 2.63 | 3.62 | 2.81 | ||
| LLCFS | Mean (%) | 86.86 | 87.16 | 86.5 | 0.79 |
| Standard deviation | 5.51 | 6.67 | 6.66 | ||
| CFS | Mean (%) | 87.91 | 88.41 | 87.58 | 0.93 |
| Standard deviation | 2.87 | 6.81 | 6.16 |
| Feature Selection Method | Performance Metrics | Accuracy | Sensitivity | Specificity | F-Measure |
|---|---|---|---|---|---|
| LASSO | Mean (%) | 84.06 | 81.58 | 86.75 | 0.81 |
| Standard deviation | 3.48 | 4.48 | 5.32 | ||
| FSASL | Mean (%) | 95.42 | 94.5 | 96.41 | 0.97 |
| Standard deviation | 2.14 | 2.58 | 2.48 | ||
| LLCFS | Mean (%) | 85.01 | 81.66 | 88.41 | 0.93 |
| Standard deviation | 3.86 | 5.37 | 5.29 | ||
| CFS | Mean (%) | 88.38 | 84.25 | 92.41 | 0.91 |
| Standard deviation | 2.36 | 4.25 | 2.55 |
| Feature Selection Method | Performance Metrics | Accuracy | Sensitivity | Specificity | F-Measure |
|---|---|---|---|---|---|
| LASSO | Mean (%) | 90.12 | 83.0 | 97.16 | 0.97 |
| Standard deviation | 1.89 | 3.89 | 2.69 | ||
| FSASL | Mean (%) | 96.47 | 95.33 | 97.66 | 0.97 |
| Standard deviation | 1.46 | 2.122 | 1.61 | ||
| LLCFS | Mean (%) | 87.02 | 82.25 | 91.75 | 0.82 |
| Standard deviation | 4.37 | 4.02 | 6.55 | ||
| CFS | Mean (%) | 88.38 | 84.25 | 92.42 | 0.91 |
| Standard deviation | 2.36 | 4.25 | 2.56 |
| Feature Selection Method | Performance Metrics | Accuracy | Sensitivity | Specificity | F-Measure |
|---|---|---|---|---|---|
| LASSO | Mean (%) | 84.95 | 86.75 | 83.08 | 0.84 |
| Standard deviation | 4.81 | 5.188 | 5.18 | ||
| FSASL | Mean (%) | 98.38 | 97.16 | 99.66 | 1 |
| Standard deviation | 1.51 | 2.69 | 1.05 | ||
| LLCFS | Mean (%) | 88.83 | 90.91 | 87.0 | 0.91 |
| Standard deviation | 4.60 | 3.89 | 8.30 | ||
| CFS | Mean (%) | 88.07 | 87.66 | 88.5 | 0.97 |
| Standard deviation | 4.18 | 7.70 | 6.22 |
| Feature Selection Method | Performance Metrics | Accuracy | Sensitivity | Specificity | F-Measure |
|---|---|---|---|---|---|
| LASSO | Mean (%) | 84.88 | 81.83 | 88.0 | 0.93 |
| Standard deviation | 1.76 | 3.68 | 4.12 | ||
| FSASL | Mean (%) | 85.82 | 85.0 | 86.91 | 0.86 |
| Standard deviation | 2.88 | 5.29 | 4.332 | ||
| LLCFS | Mean (%) | 82.58 | 82.41 | 82.91 | 0.88 |
| Standard deviation | 2.83 | 3.75 | 5.43 | ||
| CFS | Mean (%) | 70.15 | 70.66 | 69.33 | 0.73 |
| Standard deviation | 7.37 | 6.28 | 11.26 |
| Feature Selection Method | Performance Metrics | Accuracy | Sensitivity | Specificity | F-Measure |
|---|---|---|---|---|---|
| LASSO | Mean (%) | 96.75 | 97.75 | 95.83 | 0.94 |
| Standard deviation | 1.52 | 2.22 | 3.04 | ||
| FSASL | Mean (%) | 90.12 | 91.16 | 89.25 | 0.94 |
| Standard deviation | 3.64 | 5.58 | 4.39 | ||
| LLCFS | Mean (%) | 78.57 | 81.0 | 76.0 | 0.78 |
| Standard deviation | 3.06 | 5.93 | 3.98 | ||
| CFS | Mean (%) | 74.03 | 73.58 | 74.5 | 0.73 |
| Standard deviation | 5.13 | 9.77 | 9.74 |
| Feature Selection Method | Performance Metrics | Accuracy | Sensitivity | Specificity | F-Measure |
|---|---|---|---|---|---|
| LASSO | Mean (%) | 86.35 | 85.08 | 87.5 | 0.86 |
| Standard deviation | 3.00 | 5.037 | 4.79 | ||
| FSASL | Mean (%) | 88.19 | 91.58 | 84.91 | 0.93 |
| Standard deviation | 3.10 | 4.77 | 3.35 | ||
| LLCFS | Mean (%) | 82.5 | 81.66 | 83.16 | 0.86 |
| Standard deviation | 4.02 | 6.56 | 5.14 | ||
| CFS | Mean (%) | 70.55 | 65.83 | 75.25 | 0.96 |
| Standard deviation | 6.01 | 5.77 | 7.61 |
| Number of Subjects | Classification Method | fMRI Features | Classification Accuracy (%) | |
|---|---|---|---|---|
| AD | HC | |||
| 34 | 45 | Naïve Bayes | Directed graph features [20] | 93.3 |
| 77 | 173 | Area under curve | Combination of functional connectivity matrices, functional connectivity dynamics, Amplitude of low-frequency fluctuation [42] | 85 |
| 12 | 12 | Linear Discriminant Analysis | Default mode network and salience network map difference [43] | 92 |
| 67 | 76 | Support Vector Machine | ROI-to ROI correlation with significant difference [44] | 92.9 |
| Number of Subjects | Classification Method | fMRI Features | Classification Accuracy (%) | |
|---|---|---|---|---|
| MCI | HC | |||
| 31 | 31 | Support Vector Machine | Covariance matrix of whole brain network [45] | 62.90 |
| 31 | 31 | Support Vector Machine | fMRI time series of ROI [46] | 66.13 |
| 91 | 79 | Deep Auto Encoder | ROI-to ROI Correlation [47] | 86.5 |
| 31 | 31 | Support Vector Machine | Mean time series of ROI [48] | 72.58 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lama, R.K.; Kim, J.-I.; Kwon, G.-R. Classification of Alzheimer’s Disease Based on Core-Large Scale Brain Network Using Multilayer Extreme Learning Machine. Mathematics 2022, 10, 1967. https://doi.org/10.3390/math10121967
Lama RK, Kim J-I, Kwon G-R. Classification of Alzheimer’s Disease Based on Core-Large Scale Brain Network Using Multilayer Extreme Learning Machine. Mathematics. 2022; 10(12):1967. https://doi.org/10.3390/math10121967
Chicago/Turabian StyleLama, Ramesh Kumar, Ji-In Kim, and Goo-Rak Kwon. 2022. "Classification of Alzheimer’s Disease Based on Core-Large Scale Brain Network Using Multilayer Extreme Learning Machine" Mathematics 10, no. 12: 1967. https://doi.org/10.3390/math10121967
APA StyleLama, R. K., Kim, J.-I., & Kwon, G.-R. (2022). Classification of Alzheimer’s Disease Based on Core-Large Scale Brain Network Using Multilayer Extreme Learning Machine. Mathematics, 10(12), 1967. https://doi.org/10.3390/math10121967







