Abstract
Human skin is the most exposed part of the human body that needs constant protection and care from heat, light, dust, and direct exposure to other harmful radiation, such as UV rays. Skin cancer is one of the dangerous diseases found in humans. Melanoma is a form of skin cancer that begins in the cells (melanocytes) that control the pigment in human skin. Early detection and diagnosis of skin cancer, such as melanoma, is necessary to reduce the death rate due to skin cancer. In this paper, the classification of acral lentiginous melanoma, a type of melanoma with benign nevi, is being carried out. The proposed stacked ensemble method for melanoma classification uses different pre-trained models, such as Xception, Inceptionv3, InceptionResNet-V2, DenseNet121, and DenseNet201, by employing the concept of transfer learning and fine-tuning. The selection of pre-trained CNN architectures for transfer learning is based on models having the highest top-1 and top-5 accuracies on ImageNet. A novel stacked ensemble-based framework is presented to improve the generalizability and increase robustness by fusing fine-tuned pre-trained CNN models for acral lentiginous melanoma classification. The performance of the proposed method is evaluated by experimenting on a Figshare benchmark dataset. The impact of applying different augmentation techniques has also been analyzed through extensive experimentations. The results confirm that the proposed method outperforms state-of-the-art techniques and achieves an accuracy of 97.93%.
1. Introduction
Skin is the most curious and outer layer of the human body, protecting the body from heat, light, dust, and other harmful radiations, such as ultra-violet. Human skin is made up of two layers called the dermis and epidermis. The outermost layer of the skin is called the epidermis, composed of three types of scaly and flat cells on the surface called squamous cells. The cells that protect the skin from damage and provide skin color are basal cells and melanocytes cells. Many diseases can harm the skin, and cancer is one of the most aggressive and deadly diseases in human skin. In skin cancer, melanoma and non-melanoma are the two most known types [1]. Melanoma is the deadliest and most severe skin cancer that is the cause of almost all types of skin cancers whose growth starts with the cells of melanocytes present on the outermost layer of the skin. Melanoma is also called malignant melanoma, which can grow and affect nearby healthy cells. This process is commonly known as metastasis. Malignant melanoma has four major subtypes: superficial spreading melanoma, nodular melanoma, lentigo malignant melanoma, and acral lentiginous melanoma [2]. The acral lentiginous melanoma is most commonly found in people with darker skin, such as Hispanic, African, and Asian ancestries. As compared to men, this type of melanoma frequently occurs more in women [3]. One reason for the increase in melanoma cases is due to UV radiation from sunshine or burning of the skin in sun rays. Acral lentiginous melanoma appears as a tiny (about 6 mm) flat spot of discolored skin, often black or dark brown. It usually grows on the soles, palms, or under nails sometimes and mainly occurs on the back of men and fingers and legs in women [4]. It has poor diagnosis because it is hard to differentiate between an acral melanoma and an acral nevus. Usually, it is identified at the later stages of melanoma development that reduces the survival rates of patients [5]. Melanoma is a curable disease if it is diagnosed at an earlier stage [6]. Early diagnosis techniques for melanoma include biopsy, pathology report, and medical imaging analysis, such as dermoscopy. Dermoscopy is a non-invasive imaging technique usually used to diagnose melanoma early to improve survival chances. In the dermoscopy, a magnified image of the cancerous region is taken at high resolution to locate the region on the skin, which is then analyzed by the dermatologists for melanoma detection [7]. The analysis of dermoscopy images by dermatologists is expensive and requires a high level of expertise to precisely determine the disease [8]. This issue has raised the need for developing accurate computer-aided diagnosis techniques that could assist in the early detection of melanoma from dermoscopy images. However, it is a challenging task due to several reasons. First of all, melanoma may include a high degree of visual similarity between cancerous and non-cancerous cells, making it hard to discriminate between melanoma and non-melanoma skin cancer. Secondly, it is difficult to segment the skin lesion from normal skin regions because of the low contrast. Thirdly, melanoma and non-melanoma are visually similar, and the skin conditions in different peoples have visually different melanoma. Third, the high intra-class variation of melanoma size, color, shape, and location in dermoscopic images makes it hard to detect melanoma. In addition, other artifacts, such as color calibration charts, hair, ruler marks, and veins, also cause blurriness and occlusions, making this problem more complicated [1,9,10]. Numerous automated techniques have been proposed to assist dermatologists in melanoma diagnosis in recent years. These techniques include traditional machine learning and deep learning-based methods [5,11]. Recently, deep learning-based methods have produced excellent results in medical image analysis, such as segmentation, detection, and classification. Hence, more attention is being paid to deep learning-based methods for melanoma detection. This research proposes a transfer learning-based approach for acral lentiginous melanoma identification from dermoscopy images. The main contribution of this paper is as follows:
- A novel stacked ensemble framework based on transfer learning is presented to address the task of acral lentiginous melanoma classification;
- Extensive experiments have been performed on the benchmark dataset with and without data augmentation to show the impact of data augmentation in improving the accuracy of the proposed model;
- The proposed method outperforms state-of-the-art methods for acral lentiginous melanoma classification.
The rest of the paper is organized as follows: In Section 2, an extensive literature review of the existing studies based on deep learning, transfer learning, and deep ensemble learning. In Section 3, the proposed stacked ensemble approach for the classification of acral lentiginous melanoma is elaborated. Section 4 details experiments performed on the dermoscopy imaging dataset. Finally, the paper is concluded in Section 5.
2. Background
In this section, a brief introduction of transfer learning followed by an overview of each pre-trained CNN architectures that are used in the methodology is being discussed. Deep learning-based models can achieve promising results when large datasets are available for training the model. However, it is not always possible to increase training samples for some domains, such as medical imaging, due to the scarcity of data. In these domains, transfer learning can be useful. In transfer learning, a model trained on a large dataset, such as ImageNet, can be used for applications similar to domains with comparatively smaller datasets, as is shown in Figure 1.
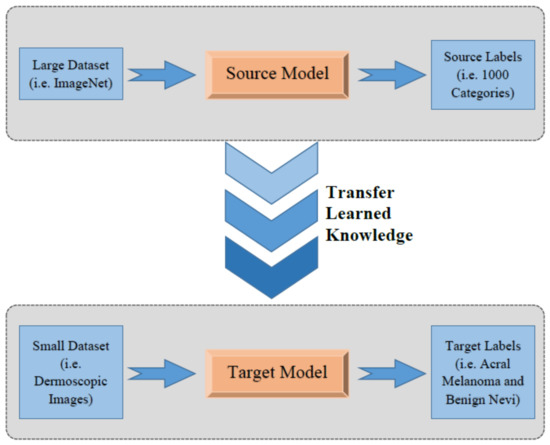
Figure 1.
Concept of transfer learning.
In transfer learning, the knowledge gained by training on an extremely large dataset with thousands of classes is transferred to similar problems through weight sharing. In this way, the weights that are already being trained and adjusted can be utilized and shared with another problem, such as the classification of acral lentiginous melanoma in this case. Transfer learning has been successfully and widely used for different applications, such as video analytics, automation, manufacturing, medical imaging, and baggage screening [12].
In this paper, different pre-trained models including, VGG16 [13], Xception [14], InceptionResnetV2 [15], DenseNet121 [16], DenseNet169 [16], and DenseNet210 [16] are fine-tuned for melanoma classification. Instead of designing a CNN architecture from scratch, the proposed methodology is based on fine-tuning a few top layers in which weights in early layers are frozen. Early layers of any CNN-based models are responsible for extracting low-level features, such as edges, lines, blobs, etc. The efficient extraction of these low-level features is extremely important for any image classification problem. Since pre-trained deep CNN architecture weights are already highly optimized on a large dataset, the proposed methodology is based on the fine-tuning of top layers only to optimize high-level features while keeping initial layers frozen. Then, an ensemble of the models mentioned above is created to achieve excellent results. The background information on each model is presented in the subsequent sections.
2.1. Pre-Trained Xception Model
The first model chosen for the methodology is a pre-trained Xception network [14], also known as an extreme version of Inception. Xception is a deep CNN architecture developed by Google researchers having a total depth of 71 layers. It is a modified version of Inception-V3 architecture that has surpassed VGG16, ResNet, and Inception-V3 in many classification tasks. It consists of a modified version of depthwise separable convolution and max-pooling layers, all linked together as a residual network. These modified depthwise separable convolutions in Xception consist of pointwise convolutions (1 × 1 convolution) followed by depthwise convolutions (n × n convolution). The illustration of the idea of modified depthwise separable convolutions is shown in Figure 2. The architecture diagram of Xception comprises three important sections; Entry flow, Middle flow, and Exit flow, as shown in Figure 3. The input image is passed into the entry flow, followed by a middle flow that is repeated eight times, and finally, it is passed into the exit flow for classification at the end.
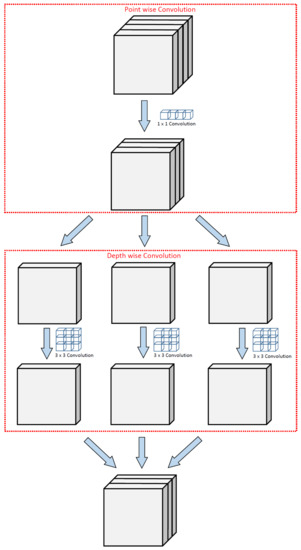
Figure 2.
Illustration of modified depthwise separable convolution in Xception network.
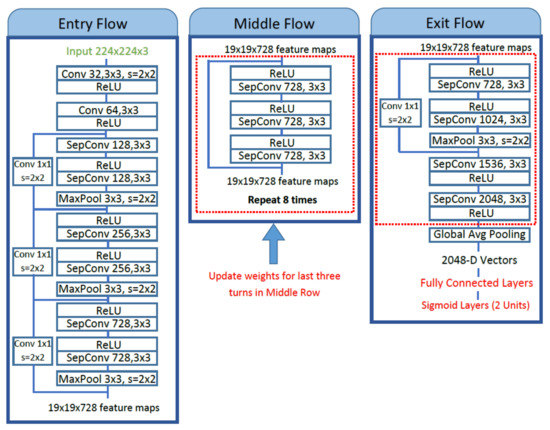
Figure 3.
The fine-tuning of proposed method of pre-trained Xception architecture.
Fine-tuning of the pre-trained Xception model is carried out by unfreezing the top five Xception blocks while keeping other bottom layers frozen to extract the most relevant and detailed features from dermoscopic images. Fine-tuning is performed on the middle flow and an exit flow of Xception network architecture, as shown in Figure 3. The separable convolution layers in the middle flow are retrained followed by exit flow, and weights are updated to extract relevant features. After global average pooling, these extracted features are passed to the top model containing four fully connected layers with 1024, 512, 256, and 128 units, respectively, each with ReLU activation and an output layer with sigmoid activation for binary classification.
2.2. Pre-Trained InceptionResNet-v2 Model
The second model is pre-trained InceptionResNet-V2, based on inception networks and has 164 layers. It integrates residual connections as in ResNet [17] architectures to increase the performance with low computational costs. After the summation of residual connections, batch normalization is added with each block. To stabilize the training process, residual connections are scaled-down before feeding into the activations of the previous layers. For this work, the top two blocks of this model are fine-tuned, and weights are updated. Global average pooling layer is applied, and the last four fully connected layers with 1024, 512, 256, and 128 units, respectively, and ReLU activation is used. For the last layer, the sigmoid activation function is used for binary classification as shown in Figure 4.
Figure 4.
The fine-tuning of proposed method of pre-trained Inception-ResNet-V2 architecture.
2.3. Pre-Trained DenseNet121 Model
The third pre-trained model is DenseNet121 [16]. DenseNets simplifies the connectivity pattern by ensuring information flow between layers compared to other state-of-the-art Deep CNN architectures. This exploits network potential through feature reuse instead of drawing feature representational capability from extremely deep or wide architecture reuse. It requires fewer parameters than an equivalent traditional CNN, as there is no need to learn redundant feature maps. The feature maps in DenseNets are concatenated after each Dense block that acts as an input for the next dense block. This model contains four dense blocks followed by a transition block. The top layer containing dense blocks is fine-tuned, and weights are updated. Global average pooling layer followed by four fully connected layers with 1024, 512, and 256 units, respectively, with ReLU activation are added on top of the pre-trained model. Lastly, a sigmoid layer with two units is used as the output layer. The proposed methodology for fine-tuning of pre-trained models is shown in Figure 5.
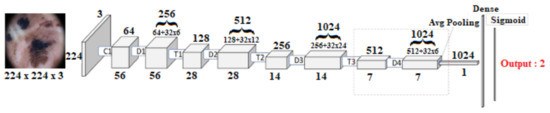
Figure 5.
The fine-tuning of proposed method of pre-trained DenseNet121 and DenseNet201 architecture.
2.4. Pre-Trained DenseNet201 Model
The last pre-trained model is DenseNet201. It has 201 convolutional layers. The fine-tuning of this model is also carried out by un-freezing Dense block 4. Global average pooling layer followed by four fully connected layers with 1024, 512, and 256 units, respectively, with ReLU activation are added on top of the model. Lastly, a sigmoid layer with two units is used as the output layer. The proposed methodology for fine-tuning this model is shown in Figure 5.
3. Related Work
Skin cancer is a life-threatening disease that must be classified and diagnosed in its early stage. Before the deep learning era, classical machine learning approaches were used, dependent on hand-crafted feature engineering. In recent years, the emergence of deep learning in medical imaging enables the model to learn complex features automatically. This section presents a comprehensive literature review of the existing methods based on deep learning, transfer learning, and ensemble learning.
3.1. Deep Learning-Based Techniques
In [18], authors survey about 19 studies conducted on skin lesions classification which use CNN based classifier and then compared their performance with clinicians. These experiments were conducted on single images of suspicious lesions. In [19], the author surveyed automatic skin cancer detection and the application of image processing and machine learning in cancer detection. In [20], authors surveyed about integrating patient data into skin lesion classification using CNN. Another study [21] presented a survey on the latest research efforts in detecting skin lesion and their classification through CNN, transfer learning, and ensemble approaches. Several deep learning-based techniques have been proposed for skin cancer detection using dermoscopic images, such as [5], a CNN-based approach was proposed. In which, researchers created their dataset and used data augmentation to enhance the quality of the dataset and achieved 80.23% accuracy. For better results, another study used multiple CNN models for melanoma classification [9]. They trained VGG-16, VGG-19 pre-trained models on their dataset and achieved an accuracy of 76%, but their accuracy was not good enough. To cope with this issue, ref. [22] used deep learning architecture. The main focus of their research was lesion attribute detection, lesion boundary segmentation, and lesion diagnosis. They used multiple pre-trained models such as AlexNet [23], Xception, ResNet [17], and VGGNet, and obtained the best accuracy of 92.74% on ResNet. Another study [24] used deep learning models for three main tasks: segmentation, feature extraction, and classification; these tasks were performed on the ISIC-2017 dataset. Experimental results illustrate auspicious accuracy, 75% for segmentation and 91% for classification. Several studies [25,26,27,28] used deep learning with different architectures and algorithms on a well-known ISIC dataset for lesion classification. Training deep learning-based models from scratch is time-consuming and needs more computational resources. To overcome this issue, ref. [29] used pre-trained models such as VGG-16, AlexNet, and ResNet for classification and achieved 83.83% accuracy on ISIC 2017 dataset. Another study [30] used VGG-16, VGG-19, and DCNN for training with different types of data augmentation on the HAM 10000 dataset [31]. Some other studies, such as [32], used CNN with GAN to improve the performance on the ISIC dataset [33]. GAN was used to generate synthetic medical images to overcome the deficiency of data and achieved 71% accuracy. In another study [34], melanoma skin cancer was detected by machine learning and imaging biomarker cues on datasets provided by IBC’s and achieved 77% accuracy. Furthermore, ref. [35] used pixel-based fusion and multilevel feature reduction to perform two experiments on ISBI-2016, and ISIC-2017 datasets, for segmentation and classification and achieved an accuracy of 95% melanoma classification. In [36], additional features of skin lesions images were extracted for classification of melanoma type and to decrease the false-positive rate. They applied SVM, neural network, and Random-Forest classifiers on the heraldic13 dataset, and the highest accuracy of 90% was achieved with the random forest classifier.
3.2. Transfer Learning-Based Techniques
Transfer learning-based techniques have achieved high accuracy and significantly reduced the need for large datasets for different classification tasks. For instance, ref. [37] utilized a transfer learning-based method for Skin Lesion Classification and achieved 85.8% accuracy. Another study [38] proposed two-stage frameworks; in the first stage, the inter-class difference of data distribution was carried out, while in the second stage, training of deep CNN on the ISIC-2016 dataset was performed and achieved a 94% of F-score. In several other studies, such as [10,39,40], transfer learning has been applied using AlexNet for classification on the HAM1000 dataset and achieved an accuracy of 96.87%. Some other studies [37], and [41], used the transfer learning technique using VGG-16 for feature extraction and SVM, decision tree, linear discriminant analysis, and K-nearest neighbor algorithms for classification on HAM10000 and ISIC dataset.
3.3. Ensemble Learning-Based Techniques
Recent studies focued on making an ensemble of different models to achieve high accuracy using dermoscopic images. The Ensemble technique has proven to be successful in increasing the overall accuracy of different applications. Ensemble of a Deep Neural Networks models, such as AlexNet, VGGNet, GoogLeNet was used in [42] for skin cancer classification and achieved 84.8% accuracy on ISIC 2017 dataset. Other studies [43,44,45], used an ensemble of different techniques for classification on ISIC 2017 datasets and achieved an accuracy of 76%. The recent studies [5,11,22] suggest that little attention has been paid to diagnose acral melanoma because of its infrequent occurrences. The prior researches [7,24,46] mainly focused on the classification of skin lesions images into some cancer types and did not provide further information about the subtype of cancer. For example, the study [47] classified skin lesions into melanoma and non-melanoma. Another research [1] was focused on the classification of skin lesions into different categories. The classification of melanomas in subtypes is very important for better diagnosis, and it can increase the patient’s survival rates [5,11]. This work is focused on acral lentiginous melanoma detection from dermoscopy images.
4. Methodology
This section elaborates on the methodology adopted to classify acral melanoma and benign nevi from dermoscopy images. Figure 6 shows a block diagram of the proposed method. First, data augmentation is applied to increase the number of training samples for each category. Second, pre-trained models, VGG16, Inception-V3, Xception and InceptionResNetV2, DenseNet121, DenseNet169, and DenseNet201 are fine-tuned to make them inline for classification. Finally, ensemble learning is applied to detect acral lentiginous melanoma by creating a stacked ensemble of fine-tuned models. These steps have been discussed in the subsequent sections.
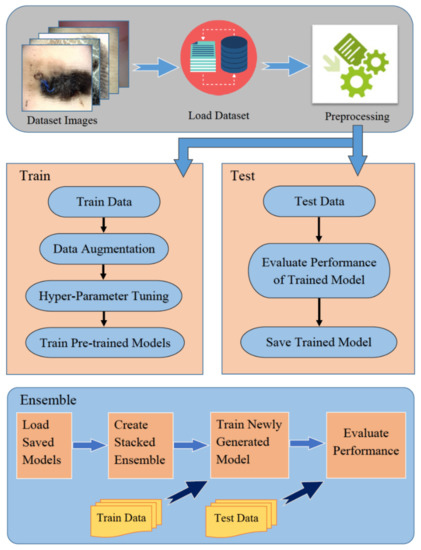
Figure 6.
Block diagram of the proposed methodology.
4.1. Preprocessing
Before passing images into the CNN for training, preprocessing is applied to the dermoscopy images. First of all, due to the difference in image dimension, all images are resized automatically to a fixed 224 × 224 dimension using the open-cv library to make it compatible to fit these images in the pre-defined shape of input tensors of selected pre-trained CNN architectures. It has been observed that resizing images has little impact on the prediction capability of the model. However, if we use the default image size, the total number of parameters will increase exponentially, making our model computationally expensive. Color channels of images are then transformed from BGR to RGB format. Finally, all images are normalized to scale the pixel intensity values from 0–255 to 0–1. The class labels are encoded to 0 and 1 for acral melanoma and benign nevi, respectively. For medical imaging, especially for the classification of acral lentiginous melanoma, dataset samples are not large enough to train the deep learning-based models. Literature suggests data augmentation techniques, such as image translation, rotation, sheering, mirroring, width shift, height shift, horizontal, and vertical flipping can be applied on dermoscopy images to increase dataset samples [48,49]. Five augmentation techniques are applied in this work, including rotation, width shift, height shift, vertical flipping, and horizontal flipping.
4.2. Stacked Ensemble of Fine-Tuned Pre-Trained CNN Architectures
Pre-trained Deep CNN architecture has a different depth and network structure. Thus, the performance varies on different problems. Each pre-trained model has its strength and limitation while applied to medical images. Multiple models are trained on the same dataset, predictions are made on each model, and results are combined using the staked ensemble learning method to achieve the best performance. The ensemble learning can reduce variance and significantly improve the performance [50]. The simplest way to combine the predictions of multiple trained models is to take an average of predictions made by each model on a similar set of training and testing data. Averaging ensemble equally combines predictions from multiple trained models and generates combined predictions [50]. On the other hand, the weighted average ensemble technique, also known as model blending, assigns weights to the predictions of an individual model that is optimized using validation data [51]. Stacked generalization or stacking is the modified version of an averaging ensemble that involves post-training the newly generated model generated by combining multiple sub-models. The proposed methodology consists of ensemble learning performed by stacking of four fine-tuned, pre-trained models; Xception, Inception-ResNet-V2, DenseNet121, and DenseNet201, as shown in Figure 7.
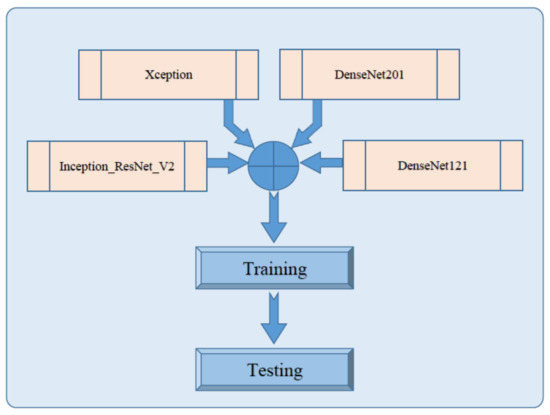
Figure 7.
Block diagram of proposed stacked ensemble model.
In this regard, each pre-trained model is fine-tuned, retrained, evaluated, and saved independently, as shown in Figure 3, Figure 4 and Figure 5 for Xception, Inception-ResNet-V2, DenseNet121, and DenseNet20, respectively. These saved models are then loaded independently and combined to form a new architecture by stacking mechanism. This design is efficient in terms of complexity because late fusion is used instead of early fusion for staking different models. Top layers of a newly formed stacked ensemble model consist of global average pooling followed by a fully connected layer with 10 neurons and a sigmoid activation function for classification.
5. Experimentations and Results
This section presents the experimental setup adopted to classify acral lentiginous melanoma with benign nevi and compares the proposed method with state-of-the-art methods.
5.1. Experimental Setup
Different hyper-parameters are tuned to train the model for achieving high accuracy, as shown in Table 1, and binary cross-entropy is used as a loss function. Adam [52] is used as an optimizer for all pre-trained models, and the learning rate is set to 0.00001 (1 × 10) with a batch size of 32. The number of epochs varies for the top 4 fine-tuned pre-trained CNN architectures. The ensemble of these pre-trained models is trained for ten epochs. All these hyper-parameters are learned empirically.

Table 1.
Hyper-parameters settings.
The dataset used for the experimentation has been taken from [53]. This dataset consists of 724 dermoscopy images; out of these, 350 belong to the acral lentiginous class, and 347 belong to benign nevi class. The sample images from this dataset are shown in Figure 8. This dataset was collected from January 2013 to March 2014 at Severance Medical Clinic, Yonsei University, Seoul, Korea, and from March 2015 to April 2016 at Dongsan Clinic in KeiMyung University Daegu, Korea. For experimentation, the dataset is divided into a 70:10:20 ratio for training, validating, and testing, respectively, where 506 images are randomly selected for the training set and 72 images for the validation set while the test set included 146 images. The class-wise distribution of the dataset is shown in Table 2.
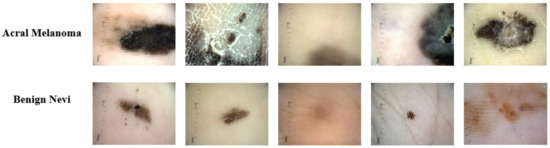
Figure 8.
Acral melanoma and benign nevi sample images.

Table 2.
Class-wise distribution of dataset.
Two experiments are carried out to compare the impact of data augmentation on the performance of pre-trained CNN architectures. In this regard, Table 3 shows the performance of fine-tuned pre-trained models without data augmentation. In contrast, Table 4 shows the performance of fine-tuned pre-trained models with data augmentation. Data augmentation increases the overall performance of fine-tuned pre-trained models, especially in the case of the Xception model, accuracy is increased from 90% to 95%, which is a significant gain in performance. Data augmentation is also helpful in avoiding overfitting and improving the model’s overall performance as models are trained on small datasets leads to overfitting.

Table 3.
Pre-trained model results without data augmentation.

Table 4.
Pre-trained model results with data augmentation.
To validate the performance of the proposed model, accuracy, precision, recall, F1 score, sensitivity, and specificity are used as performance metrics, as shown in Table 5.

Table 5.
Class-wise performance of the proposed method.
The accuracy metric is the standard metrics in terms of classification problems, and is defined by:
The results confirm that the proposed stacked ensemble model successfully classified acral melanoma and benign nevi. In comparison, benign nevi is the most correctly classified with an accuracy of 98.79% and acral melanoma with an accuracy of 96.77%. The sample classification results of acral melanoma and benign nevi through the proposed model are shown in Figure 9.
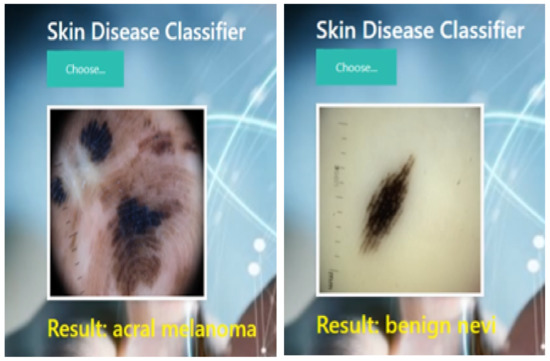
Figure 9.
Sample classification results of pre-trained model.
5.2. Comparison with State-of-the-Art Methods
The proposed method of fine-tuning pre-trained Xception and Inception-ResNet-V2, DenseNet121, and DenseNet201 achieved overall test accuracy of 95.17%, 95.17%, 94.48%, and 95.86% respectively. The stacked ensemble of these models is generated to increase the model’s overall performance. As shown in Table 6, the proposed ensemble technique obtained a test accuracy of 97.93% which shows significant improvement in terms of accuracy. The confusion matrix of the proposed model is shown in Figure 10.

Table 6.
Performance comparison of each pre-trained model and stacked ensemble model.
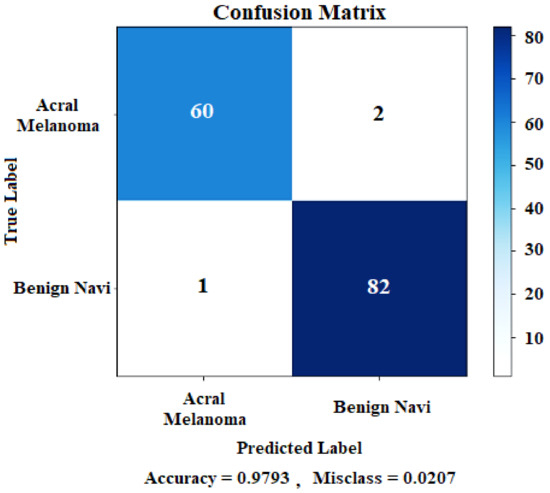
Figure 10.
Stacked ensemble model confusion matrix.
The confusion matrix is also known as the error matrix. It is a specific type of tabular layout that gives information about the ground truth class and a predicted class, showing the model’s performance for each class. The ground truths are shown along the y-axis and predicted class labels are along the x-axis of the confusion matrix.
The comparison of the performance of the proposed model is shown in Table 7, which confirms that the proposed method outperforms state-of-the-art methods.

Table 7.
Comparison of stacked ensemble model with existing models.
6. Conclusions
This research proposed a stacked ensemble-based method for acral lentiginous melanoma classification, the most common type of melanoma in Asians. Four pre-trained models, i.e., InceptionV3, Xception, InceptionresnetV2, and DenseNet121, are trained and ensembled to achieve excellent results. The ensemble-based approach has significantly outperformed all four individual models in terms of accuracy on the acral melanoma dataset. As the size of the dataset is not big enough, data augmentation and transfer learning is applied to train all these models. The proposed model achieved 97.83% sensitivity, 97.50% specificity, and 97.93% accuracy for the classification of acral melanoma and benign nevi dermoscopy images. It has been concluded that the proposed method can be helpful for dermatologists in identifying skin lesions effectively. This technique can be extended for other skin cancer diseases as future work. In addition to this, segmentation of skin lesions can also be considered to assist dermatologists in identifying an affected skin region.
Author Contributions
R.R., F.Z. and A.B.S. conceived and designed the experiment; R.R. and F.Z., performed the experiments with data augmentation; S.T. and G.B.A., performed the experiments without data augmentation. A.B.S. and Z.H. analyzed the data; A.B.S., Z.H., F.Z. and R.R. contributed reagents/materials/analysis tools. All the authors equally contributed in manuscript writeup and review. All authors have read and agreed to the published version of the manuscript.
Funding
This research is supported by the PDE-GIR project which has received funding from the European Unions Horizon 2020 research and innovation programme under the Marie Skodowska-Curie grant agreement No. 778035.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Codella, N.C.; Nguyen, Q.B.; Pankanti, S.; Gutman, D.A.; Helba, B.; Halpern, A.C.; Smith, J.R. Deep learning ensembles for melanoma recognition in dermoscopy images. IBM J. Res. Dev. 2017, 61, 5:1–5:15. [Google Scholar] [CrossRef]
- Alasadi, A.H.H.; Alsafy, B.M. Diagnosis of Malignant Melanoma of Skin Cancer Types. Int. J. Interact. Multimed. Artif. Intell. 2017, 4, 44–49. [Google Scholar] [CrossRef][Green Version]
- Darmawan, C.C.; Jo, G.; Montenegro, S.E.; Kwak, Y.; Cheol, L.; Cho, K.H.; Mun, J.H. Early detection of acral melanoma: A review of clinical, dermoscopic, histopathologic, and molecular characteristics. J. Am. Acad. Dermatol. 2019, 81, 805–812. [Google Scholar] [CrossRef]
- Vocaturo, E.; Perna, D.; Zumpano, E. Machine learning techniques for automated melanoma detection. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019; pp. 2310–2317. [Google Scholar]
- Yu, C.; Yang, S.; Kim, W.; Jung, J.; Chung, K.Y.; Lee, S.W.; Oh, B. Acral melanoma detection using a convolutional neural network for dermoscopy images. PLoS ONE 2018, 13, e0193321. [Google Scholar]
- Nezhadian, F.K.; Rashidi, S. Melanoma skin cancer detection using color and new texture features. In Proceedings of the 2017 Artificial Intelligence and Signal Processing Conference (AISP), Shiraz, Iran, 25–27 October 2017; pp. 1–5. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 1–48. [Google Scholar] [CrossRef]
- Ganster, H.; Pinz, P.; Rohrer, R.; Wildling, E.; Binder, M.; Kittler, H. Automated melanoma recognition. IEEE Trans. Med. Imaging 2001, 20, 233–239. [Google Scholar] [CrossRef]
- Rezaoana, N.; Hossain, M.S.; Andersson, K. Detection and Classification of Skin Cancer by Using a Parallel CNN Model. In Proceedings of the 2020 IEEE International Women in Engineering (WIE) Conference on Electrical and Computer Engineering (WIECON-ECE), Bhubaneswar, India, 26–27 December 2020; pp. 380–386. [Google Scholar]
- Hosny, K.M.; Kassem, M.A.; Foaud, M.M. Classification of skin lesions using transfer learning and augmentation with Alex-net. PLoS ONE 2019, 14, e0217293. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Chu, Y.; Yoo, S.; Choi, S.; Choe, S.; Koh, S.; Chung, K.; Xing, L.; Oh, B.; Yang, S. Augmented decision-making for acral lentiginous melanoma detection using deep convolutional neural networks. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 1842–1850. [Google Scholar] [CrossRef]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Chollet, F. Deep learning with separable convolutions. arXiv 2016, arXiv:1610.02357. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A.A. Inception-v4, inception-resnet and the impact of residual connections on learning. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Haggenmüller, S.; Maron, R.C.; Hekler, A.; Utikal, J.S.; Barata, C.; Barnhill, R.L.; Beltraminelli, H.; Berking, C.; Betz-Stablein, B.; Blum, A.; et al. Skin cancer classification via convolutional neural networks: Systematic review of studies involving human experts. Eur. J. Cancer 2021, 156, 202–216. [Google Scholar] [CrossRef]
- Razmjooy, N.; Ashourian, M.; Karimifard, M.; Estrela, V.V.; Loschi, H.J.; Do Nascimento, D.; França, R.P.; Vishnevski, M. Computer-aided diagnosis of skin cancer: A review. Curr. Med. Imaging 2020, 16, 781–793. [Google Scholar] [CrossRef]
- Höhn, J.; Hekler, A.; Krieghoff-Henning, E.; Kather, J.N.; Utikal, J.S.; Meier, F.; Gellrich, F.F.; Hauschild, A.; French, L.; Schlager, J.G.; et al. Integrating patient data into skin cancer classification using convolutional neural networks: Systematic review. J. Med. Internet Res. 2021, 23, e20708. [Google Scholar] [CrossRef] [PubMed]
- Saeed, J.; Zeebaree, S. Skin lesion classification based on deep convolutional neural networks architectures. J. Appl. Sci. Technol. Trends 2021, 2, 41–51. [Google Scholar] [CrossRef]
- Kassani, S.H.; Kassani, P.H. A comparative study of deep learning architectures on melanoma detection. Tissue Cell 2019, 58, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Li, Y.; Shen, L. Skin lesion analysis towards melanoma detection using deep learning network. Sensors 2018, 18, 556. [Google Scholar] [CrossRef]
- Barata, C.; Marques, J.S. Deep learning for skin cancer diagnosis with hierarchical architectures. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 841–845. [Google Scholar]
- Adegun, A.A.; Viriri, S. Deep learning-based system for automatic melanoma detection. IEEE Access 2019, 8, 7160–7172. [Google Scholar] [CrossRef]
- Demir, A.; Yilmaz, F.; Kose, O. Early detection of skin cancer using deep learning architectures: Resnet-101 and inception-v3. In Proceedings of the 2019 Medical Technologies Congress (TIPTEKNO), Izmir, Turkey, 3–5 October 2019; pp. 1–4. [Google Scholar]
- Daghrir, J.; Tlig, L.; Bouchouicha, M.; Sayadi, M. Melanoma skin cancer detection using deep learning and classical machine learning techniques: A hybrid approach. In Proceedings of the 2020 5th International Conference on Advanced Technologies for Signal and Image Processing (ATSIP), Sousse, Tunisia, 2–5 September 2020; pp. 1–5. [Google Scholar]
- Mahbod, A.; Schaefer, G.; Wang, C.; Ecker, R.; Ellinger, I. Skin Lesion Classification Using Hybrid Deep Neural Networks. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 1229–1233. [Google Scholar] [CrossRef]
- Aburaed, N.; Panthakkan, A.; Al-Saad, M.; Amin, S.A.; Mansoor, W. Deep Convolutional Neural Network (DCNN) for Skin Cancer Classification. In Proceedings of the 2020 27th IEEE International Conference on Electronics, Circuits and Systems (ICECS), Glasgow, UK, 23–25 November 2020; pp. 1–4. [Google Scholar]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 2018, 180161. [Google Scholar] [CrossRef]
- Sedigh, P.; Sadeghian, R.; Masouleh, M.T. Generating synthetic medical images by using GAN to improve CNN performance in skin cancer classification. In Proceedings of the 2019 7th International Conference on Robotics and Mechatronics (ICRoM), Tehran, Iran, 20–21 November 2019; pp. 497–502. [Google Scholar]
- Combalia, M.; Codella, N.C.; Rotemberg, V.; Helba, B.; Vilaplana, V.; Reiter, O.; Carrera, C.; Barreiro, A.; Halpern, A.C.; Puig, S.; et al. BCN20000: Dermoscopic lesions in the wild. arXiv 2019, arXiv:1908.02288. [Google Scholar]
- Gareau, D.S.; Browning, J.; Da Rosa, J.C.; Suarez-Farinas, M.; Lish, S.; Zong, A.M.; Firester, B.; Vrattos, C.; Renert-Yuval, Y.; Gamboa, M.; et al. Deep learning-level melanoma detection by interpretable machine learning and imaging biomarker cues. J. Biomed. Opt. 2020, 25, 112906. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.; Khan, M.A.; Mehmood, Z.; Saba, T.; Sardaraz, M.; Rashid, M. Microscopic melanoma detection and classification: A framework of pixel-based fusion and multilevel features reduction. Microsc. Res. Tech. 2020, 83, 410–423. [Google Scholar] [CrossRef]
- Parmar, B.; Talati, B. Automated Melanoma Types and Stages Classification for dermoscopy images. In Proceedings of the 2019 Innovations in Power and Advanced Computing Technologies (i-PACT), Vellore, India, 22–23 March 2019; Volume 1, pp. 1–7. [Google Scholar]
- Rahman, Z.; Ami, A.M. A Transfer Learning Based Approach for Skin Lesion Classification from Imbalanced Data. In Proceedings of the 2020 11th International Conference on Electrical and Computer Engineering (ICECE), Dhaka, Bangladesh, 17–19 December 2020; pp. 65–68. [Google Scholar]
- Zunair, H.; Hamza, A.B. Melanoma detection using adversarial training and deep transfer learning. Phys. Med. Biol. 2020, 65, 135005. [Google Scholar] [CrossRef] [PubMed]
- Al Nazi, Z.; Abir, T.A. Automatic skin lesion segmentation and melanoma detection: Transfer learning approach with u-net and dcnn-svm. In Proceedings of the International Joint Conference on Computational Intelligence; Springer: Singapore, 2020; pp. 371–381. [Google Scholar]
- Younis, H.; Bhatti, M.H.; Azeem, M. Classification of skin cancer dermoscopy images using transfer learning. In Proceedings of the 2019 15th International Conference on Emerging Technologies (ICET), Peshawar, Pakistan, 2–3 December 2019; pp. 1–4. [Google Scholar]
- Islam, M.K.; Ali, M.S.; Ali, M.M.; Haque, M.F.; Das, A.A.; Hossain, M.M.; Duranta, D.; Rahman, M.A. Melanoma Skin Lesions Classification using Deep Convolutional Neural Network with Transfer Learning. In Proceedings of the 2021 1st International Conference on Artificial Intelligence and Data Analytics (CAIDA), Riyadh, Saudi Arabia, 6–7 April 2021; pp. 48–53. [Google Scholar]
- Harangi, B.; Baran, A.; Hajdu, A. Classification of skin lesions using an ensemble of deep neural networks. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 2575–2578. [Google Scholar]
- Osowski, S.; Les, T. Deep Learning Ensemble for Melanoma Recognition. In Proceedings of the 2020 International Joint Conference on Neural Networks (IJCNN), Glasgow, UK, 19–24 July 2020; pp. 1–7. [Google Scholar]
- Milton, M.A.A. Automated skin lesion classification using ensemble of deep neural networks in ISIC 2018: Skin lesion analysis towards melanoma detection challenge. arXiv 2019, arXiv:1901.10802. [Google Scholar]
- Ashraf, R.; Kiran, I.; Mahmood, T.; Butt, A.U.R.; Razzaq, N.; Farooq, Z. An efficient technique for skin cancer classification using deep learning. In Proceedings of the 2020 IEEE 23rd International Multitopic Conference (INMIC), Bahawalpur, Pakistan, 5–7 November 2020; pp. 1–5. [Google Scholar]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Han, S.S.; Moon, I.J.; Lim, W.; Suh, I.S.; Lee, S.Y.; Na, J.I.; Kim, S.H.; Chang, S.E. Keratinocytic skin cancer detection on the face using region-based convolutional neural network. JAMA Dermatol. 2020, 156, 29–37. [Google Scholar] [CrossRef]
- Zhuang, J.; Yang, J.; Gu, L.; Dvornek, N. Shelfnet for fast semantic segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision Workshops, Seoul, Korea, 27–28 October 2019. [Google Scholar]
- Mikolajczyk, A.; Grochowski, M. Data augmentation for improving deep learning in image classification problem. In 2018 International Interdisciplinary PhD Workshop (IIPhDW); IEEE: Piscatvey, NJ, USA, 2018; pp. 117–122. [Google Scholar]
- Lu, Y.; Zhang, L.; Wang, B.; Yang, J. Feature ensemble learning based on sparse autoencoders for image classification. In Proceedings of the 2014 International Joint Conference on Neural Networks (IJCNN), Beijing, China, 6–11 July 2014; pp. 1739–1745. [Google Scholar]
- Izmailov, P.; Podoprikhin, D.; Garipov, T.; Vetrov, D.; Wilson, A.G. Averaging weights leads to wider optima and better generalization. arXiv 2018, arXiv:1803.05407. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Acral Melanoma and Benign Data Set. Available online: https://figshare.com/s/a8c22c09f999f60a81bd (accessed on 15 March 2021).
- Ann, A.J.; Ruiz, C. Using deep learning for melanoma detection in dermoscopy images. Int. J. Mach. Learn. Comput. 2018, 8, 61–68. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).