Abstract
Despite recent technological advancements allowing the characterization of cancers at a molecular level along with biomarkers for cancer diagnosis, the management of ovarian cancers (OC) remains challenging. Proteins assume functions encoded by the genome and the complete set of proteins, termed the proteome, reflects the health state. Comprehending the circulatory proteomic profiles for OC subtypes, therefore, has the potential to reveal biomarkers with clinical utility concerning early diagnosis or to predict response to specific therapies. Furthermore, characterization of the proteomic landscape of tumor-derived tissue, cell lines, and PDX models has led to the molecular stratification of patient groups, with implications for personalized therapy and management of drug resistance. Here, we review single and multiple marker panels that have been identified through proteomic investigations of patient sera, effusions, and other biospecimens. We discuss their clinical utility and implementation into clinical practice.
1. Introduction
Ovarian cancer (OC) is often used as an umbrella term referring to malignancies caused by ovarian epithelial inclusion cysts that are trapped beneath the surface of the epithelium of the ovary as well as malignancies in the peritoneum and fallopian tube [1]. Advanced OC is one of the deadliest malignancies in women with a 5-year survival rate below 30% and high incidences of occurrence in the Eastern and Central European population (11.4 per 100,000 and 6.0 per 100,000, respectively) [2]. Although the incidence varies across populations, the average lifetime risk of developing OC is 1.3% [3].
Most OC are epithelial (90%), and it is a heterogeneous disease comprising of a range of subtypes [4]. The most frequent subtype is high-grade serous carcinoma (HGSC) corresponding to around 60 % of cases, whereas low-grade serous carcinoma, mucinous, clear cell, and endometrioid OC are all less abundant [4]. The spread of OC is frequently systematically categorized using a scoring scheme outlined by the International Federation of Gynecology and Obstetrics (FIGO). FIGO scoring is based on the tumor-node-metastasis (T-N-M) approach which systematically describes the extent of the tumor (T) as well as its spread to lymph nodes (N) and potential metastasis (M) and categorizes OC into 4 stages (denoted I, II, III, and IV). Stage I is characterized OC only in the ovary(s) or fallopian tube(s) and Stage II by its spread to a close organ such as the uterus, bladder, or rectum. Stage III is defined by the spread to the abdomen and/or lymph nodes and stage IV by distant metastasis. i.e., pleura. While Stage I tumors are associated with a good prognosis most OC cases are not diagnosed at this stage. Stage II and III OCs are removed by debulking surgery followed by treatment with a combination of platinum and taxane chemotherapy which leads to considerable improvement in survival [5]. Stage III tumors are categorized by the spread to the adjacent peritoneum through metastasis. Stage IV is defined through distant metastasis and frequently treated by a combination of debulking surgery to remove the primary tumor and chemotherapy to target metastases. Due to the lack of efficient tools for early diagnosis, around 10–20% of the OCs are detected at this stage and treatment options remain limited along with poor survival rates [6]. OC tumors are typically also categorized as low or high grade, which reflects the differentiation state of tumor cells. The less differentiated low-grade tumors are typically associated with a better prognosis.Several genetic studies have linked dysregulated gene expression and mutations to OC. However, not all OCs display a similar pattern, emphasizing that the disease is heterogeneous also at the molecular level. For example, and by analogy with malignant breast cancer, mutations in BRCA1 and BRCA2 are linked to OC [7]. Moreover, the high-grade serous OCs display a high frequency of TP53 mutations and other OC histologic subgroups have frequent mutations in ARID1A, PIK3CA, PTEN, CTNNB1, KRAS, and RPL22 [7,8,9,10].
One of the major challenges associated with the diagnosis of OC is the asymptomatic nature of the disease. Early-stage (I and II) OC are therefore challenging to detect. Late-stage (III and IV) OC is associated with more severe symptoms, and invasive surgery is the most viable option for disease management [11]. Although primary complete debulking surgery (PDS) strikingly increases survival for advanced-stage OC, it is not a perfect approach and many patients suffer from the recurring disease. In certain cases, the tumor burden needs to be reduced before PDS. This is frequently achieved through neo-adjuvant chemotherapy and referred to as interval debulking surgery [12].
While the 5-year survival rates for early-stage (I and II) OC can be up to 90% with clinical interventions like cytoreductive surgery and combination chemotherapy, the late-stage (III and IV) OC 5-year survival rate is below 30% [13]. Therefore, diagnostic biomarkers that distinguish benign from malignant tumors at an early stage would be of tremendous value (Figure 1a). Moreover, as OC is a complex heterogeneous disease, biomarkers predicting the responsiveness of tumors to drugs, which would thereby guide personalized treatment, would be of great clinical utility (Figure 1b).
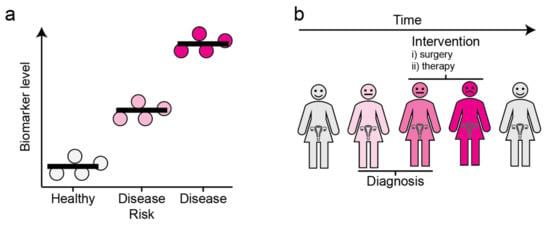
Figure 1.
The concept and utility of biomarkers. (a) Disease biomarkers. Molecules of which the level is associated with a disease state are referred to as biomarkers. (b) Clinical utility of biomarkers. The levels of biomarkers can be monitored over time, allowing for early diagnosis and informed decisions regarding clinical interventions. Grey: Healthy, Light Pink: Individuals with disease risk, Dark Pink: Individuals harboring the disease.
2. Protein Biomarkers Associated with OC
The identification of biomarkers for improved OC diagnosis and informed clinical decision-making would represent great value for both patients and the healthcare system. Protein markers are most frequently analyzed in the tumor, tumor effusions, or circulating fluids such as blood plasma (Figure 2). Early studies have reported the use of single markers in blood serum such as CA125 (Uniprot ID Q8WXI7, also known as Mucin-16) [14] and HE4 (Uniprot ID Q14508) [14]. With high-throughput semi-automated systems for sample handling and analysis, as well as the implementation of machine learning-based AI approaches, the use of biomarker panels comprising multiple markers has emerged as a superior approach. For example, panels of analytes such as the one proposed by Mor G et al., consisting of Leptin, Prolactin, Osteopontin, and insulin-like growth factor-II (IGF-II) have been proven to be useful for discriminating cancer and non-cancer patients as well as the assessment of stage I/II disease [15]. In a recent study, the assessment of multiple biomarkers, along with CA125, considerably improved the performance of the predictive model for early diagnosis. This panel comprised of CA125, HE4, CHI3L1, PEBP4, and/or AGR2, provided 85.7% sensitivity at 95.4% specificity up to one-year before diagnosis [16]. Moreover, a study by Enroth et al., recently revealed a candidate 11-protein biomarker panel for early OC diagnostics [17]. Currently, studies aimed at uncovering OC biomarkers are increasingly implementing similar multiple-marker models for predictive analysis (Table 1).
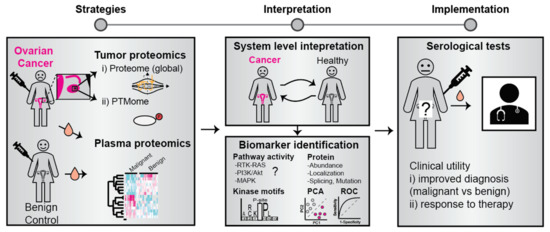
Figure 2.
Identification and clinical use of OC protein biomarkers. Biomarkers can be identified by a comparative analysis of proteins and their modification state in tumor material and blood plasma from patients and controls. The bioinformatic analysis may involve cellular pathway activity mapping, principal component analysis (PCA), and receiver operator characteristics (ROC). Identified biomarkers have the potential to improve disease diagnosis and predict response to therapy.

Table 1.
Examples of key protein markers associated with Ovarian Cancer.
Proteomic characterization of tumor tissue specimens has also revealed molecular aberrations that contribute to the onset and progression of OC [28,29]. Immunohistochemistry-based examination of tumor specimens using members of the cytokeratin family (CK7 and CK20) helps in distinguishing serous OC from other gastrointestinal malignancies [30]. In-depth analysis of genetic and histopathological signatures has also led to categorizing OCs into two types, Type I (Low grade) and Type II (High grade). While Type I tumors have a high frequency of KRAS and BRAF mutation, Type II tumors have a high frequency of TP53 mutations [31,32,33,34]. Other biospecimens such as effusions, pap smear fluids, and cervical swabs, are also valuable for understanding OC pathobiology and represent sources of markers that can predict clinical outcomes [35,36]. For example, a 9-biomarker panel in ovarian cyst fluids has been shown to discriminate between type 1 and type 2 tumors [37]. Moreover, a pilot study has depicted the utility of vaginal lysophosphatidic acid (LPA) levels as a non-invasive diagnostic marker for OC in post-menopausal women [38]. Another study investigating OC effusions revealed prominent involvement of cell-cell adhesion molecules like FAK, Erk, and P-Cadherin [39]. The study also suggested that cell adhesion molecules can comprise a prognostic signature that can be utilized to predict tumor aggressiveness as well as patient segregation. Cell adhesion protein expression, when correlated with clinicopathological parameters, has also been used to identify patient cohorts for clinical trials with small molecule inhibitors of FAK and other upstream effectors [40,41,42]. While these markers have yielded insights into OC development and the molecular pathways associated with it, they are still in the early stages of investigation and are yet to be implemented for disease management.
The emergence of ‘liquid biopsies’ has indeed ushered in a new era in diagnostics [43]. There is now tremendous potential for identifying biomarkers for improved OC diagnosis by mining such liquid biopsies with state-of-the-art (prote)-omics technologies [44,45]. Mostly the liquid biopsies are probed for circulating tumor DNA, tumor cells, exosomes, or tumor microRNA. In OCs, circulating tumor cells (CTC) are often present and useful as surrogate markers of minimal residual disease. In a study by Zhang et al., wherein nearly 100 patients were screened and subjected to CA125 measurements; CTCs were detected in nearly 90% of the newly diagnosed patients. The number of CTC also correlated with the stage of the OC. However, the ratio of CTC in comparison to other components in plasma is low and the choice of detection technique influences the number of CTCs identified. Although major strides have been made through the implementation of liquid biopsies for several cancers, more research is required to assess the full utility of CTC determination for OC, which primarily metastasizes directly through the abdominal cavity [46,47,48,49,50].
With recent advances in high-throughput omics technology and automated handling of large sample cohorts, the scope of establishing multi-marker panels has increased considerably. An ideal scenario for the effective clinical management of OCs would implement an integrated approach where blood-based markers and imaging analysis are collectively used for diagnosis and guiding clinical decisions on surgery and choice of therapy.
3. Candidate Markers for OC Diagnosis in Circulatory Fluids
CA125 (Uniprot ID Q8WXI7) is a large glycoprotein with a molecular weight of over 1.5 M Da, and is one of the most extensively used tumor markers for OC. Its link to OC was first described in 1981 by Bast RC et al., and its level is elevated in 90% of high-grade OC patients [13,51]. CA125 is considered a ‘gold standard’ marker for OC and is widely used for clinical assessment regarding disease progression and therapeutic efficacy in OC patients [51]. However, its utility in early diagnosis is limited as its expression is only found to be elevated in the later stages of OC and is often found to be elevated even in cases of benign endometriosis [52,53]. Therefore, the quest for identifying markers that can aid in early detection remains an area of active research.
A series of other markers have subsequently been identified from OC patient sera, including the Human epididymis protein 4 (HE4) (Uniprot ID Q14508), which is a secreted glycoprotein that is overexpressed in both serous and endometrioid OCs and thus might be useful in specific clinical scenarios [45,54,55]. In the same vein, line protein markers like EGFR, ErbB2, and osteopontin, in combination with the above-mentioned, have been reported to be of relevance in OC. These markers exhibit greater sensitivity and specificity when compared to single biomarker assays for the early detection of OCs [56,57]. However, the utility of these markers in isolation and combination remains a challenge when it comes to early diagnosis of the advent of ovarian malignancy [1].
The development of multivariate index assays (MIA) that comprise panels of biomarkers to assess the extent of malignancy has greatly facilitated the identification markers that can also aid in early diagnosis. For example, the FDA-approved MIA test Ova1® is based on a 5-protein biomarker panel (CA125, TF, B2M, Transthyretin, APOA1) that serves to aid clinical decision-making regarding surgery in the case of ovarian adnexal mass [8,58]. Similarly, the ROMA® test comprises two proteins (CA1254 and HE4) that predict the risk of finding a malignancy during surgery in the case of an increased ovarian adnexal mass, but the ROMA test has established predictive power for both pre-and postmenopausal women [1,52,53,59]. The above-mentioned panels highlight the utility of plasma biomarkers for clinical assessment of OCs, and add confidence to the prospects of taking markers identified in proteomics experiments from bench side to bedside. Notably, none of the approved tests have yet shown utility for screening purposes [60,61].
Blood is the preferred source of biomarkers as it is routinely extracted and handled at primary care units and hospitals. Therefore, blood-based biomarkers can be readily assessed which is a considerable advantage over biospecimens extracted with invasive methods, such as tumor biopsies or effusions. Consequently, a large number of clinical studies have been undertaken using proteomics platforms to assess both diagnostic and prognostic markers from patient sera and plasma [62,63]. Investigation of serum proteome using mass spectrometry platforms has led to the identification of many differential markers including the three- biomarker panel consisting of APOA1, transthyretin (downregulated), and inter-α-trypsin inhibitor heavy chain H4 (cleavage fragment) (upregulated) as well as CTAPIII and PF4 [26,64,65,66] (Table 1). Many of these markers are now being screened using highly sensitive targeted mass spectrometry-based methods such as selected reaction monitoring (SRM) [14]. A recent study using an SRM assay was able to identify a 5 protein panel consisting of IGHG2, LGALS3BP, DSG2, L1CAM, and THBS1 which yielded almost 94% specificity, along with CA125, for distinguishing OC patients from healthy controls [67]. These developments highlight the potential of integrating multi-protein panels identified by mass spectrometry studies into mainstream clinical diagnostics modules.
4. Proteomic Profiling of Solid Tumors and Clinically Relevant Protein Markers
Recent advances in mass spectrometry-based workflows have contributed to the identification of several proteins linked to OC in both tumor biospecimens as well as in plasma and serum [68,69,70]. These studies have employed a plethora of strategies that include label-free data-dependent acquisition (DDA), label-free SWATH, and isobaric protein labeling approaches denoted TMT [37] or iTRAQ [70,71]. This has resulted in the identification of several proteins that previously had not been well studied in the context of OCs. This includes transtherytin (TTR), apolipoprotein A1 (APOA1), and casein kinase II alpha 1 subunit isoform-a (CSNK2A1), as well as markers indicative of drug resistance such as Destrin (DSTN), Tumor rejection antigen (gp96) (HSP90B1), and EGF-containing fibulin-like extracellular matrix protein 1 (EFEMP1) [62]. These markers were shown to correlate with the clinical outcomes of the tumor. For instance, elevated levels of glycoprotein tumor rejection antigen (gp96) were observed in a human OC cell line that was resistant to paclitaxel vs nonresistant OC cell lines [72]. Similarly, HGSCs, which account for around 70–80% of morbidities, often exhibit resistance to conventional chemotherapy. System-wide analyses of both genomic and proteomic evidence have revealed that inhibition of PI3K/AKT/mTOR pathway components can be an alternative approach for patient management [73]. Quantitative proteomics studies on cancer cell lines as well as more sophisticated approaches using PDX models are pivotal in uncovering cancer cell vulnerabilities that can then be targeted by specific inhibitors for efficient management of aggressive tumors [73,74,75]. The proteomic profiling of tumor tissue has also revealed widespread aberration in key cellular pathways such as the p38-MAPK stress-activated signaling pathway and cytoskeletal components [76]. A dysregulated pathway can represent a tumor vulnerability and is key in designing therapeutic intervention strategies. In a recognized initiative by the Clinical Proteomic Tumor Analysis Consortium (CPTAC), a comprehensive study of nearly 100 patients with HGSC was performed along with 25 controls consisting of normal tissues. Using both label-free and isobaric labeling quantitative proteomics approaches to this dataset has enabled a deeper understanding of the protein level alterations in HGSC. For example, the proteomic characterization linked post-translational modifications (PTM), including phosphorylation and glycosylation, to OC progression. The CPTAC data have also enhanced our understanding of OC heterogeneity and enabled the classification of HGSC subgroups based on gene changes using the TCGA database and proteome composition from the CPTAC data. Further investigation revealed a group of kinases that had elevated activity in tumors, including several mitotic kinases, AURKA and cyclin dependent kinases, CDK1, CDK4, and CDK7. These represent potential intervention points for HGSC therapy and can pave the way for better treatment regimens [76,77,78]. Ultimately, the deep proteomic profiling of the tumor tissues has enabled the subtyping of tumors and the prediction of overall survival of HGSCs. For instance, the global proteomic profile of the HGSCs corresponded to TCGA transcriptomic subtypes, namely the mesenchymal, proliferative, immunoreactive, and differentiative [69]. Tong et al. has further extrapolated the data from the CPTAC to identify subtypes based on the patient phosphoproteome, as well as key druggable kinases. The activity of eight kinases including CDK9, CDC7, BRAF, and AXL, which were part of the Ph4 and Ph5 subtypes, also correlated with patient survival [79]. Collectively, these studies have pin-pointed clinically actionable targets.
5. Identification of PTMs in Ovarian Cancer and Their Clinical Implications
Proteins are frequently modified by so-called PTM, examples of which include glycosylation, phosphorylation, and methylation, as a means to regulate and tune their function [80,81]. All PTMs involve an increase in the mass of the modified protein and are almost invariably analyzed in scale using mass spectrometry-based workflows [82]. The identification of several high molecular glycoproteins associated with OC like CA-125 and HE4 suggests a link between glycosylation and the disease pathobiology. Indeed, glycoproteomic profiling of high-grade serous ovarian carcinomas (HGSC) has revealed differential glycosylation patterns between OC subtypes. This can aid in patient stratification and promote the understanding of the role of glycosylation in tumor etiology and development [72]. Therefore, the implementation of comprehensive glycoproteomic profiling has great potential in future OC biomarker studies.
A recent study that employed high-density HuProt™ arrays and used a multiplexed PTM analysis approach, revealed several dysregulated signaling proteins, including members of the Src kinase family and focal adhesion kinases, in HGSC. An in-depth analysis revealed prominent alterations of key PTMs, including tyrosine phosphorylation, and identified key kinases such as SYK and PTK2B that represent potential drug targets [83].
Another prominent PTM linked to OC is the site-specific methylation of 70 kDa heat shock protein (Hsp70) at Lys561 [84]. The site is specifically and exclusively modified by the enzyme METTL21A [85,86] as a means to modulate the chaperone activity of Hsp70 [87], and loss of methylation at the site has been linked to poor prognosis and reduced overall survival [84]. Notably, Hsp70-K561 methylation constitutes a part of the so-called ‘chaperone code’ [88,89], where multiple PTMs, individually or collectively, act to regulate chaperone function. Taken together, this suggests that precise regulation and PTM-mediated tuning of Hsp70 activity is important in OC biology.
A deep proteomic investigation on patient-derived primary cell lines from epithelial OC revealed that there are differentially expressed proteins as well as phosphorylation sites that can discriminate between cancer and healthy cells. In detail, the phosphorylating kinase enzyme CDK7 was identified as a druggable target, representing a novel therapeutic entry point and strategy [90].
Collectively, these recent insights from proteomic characterization of PTMs in patient biospecimens and cancer cell lines highlight the value of analyzing protein modifications, in addition to protein levels, in biomarker studies (Table 2). As PTM proteomics technology develops, we anticipate an increased focus on such studies. Protein PTM status will likely represent a key feature in next-generation OC diagnostics.

Table 2.
Example biomarkers linked to PTM and drug resistance in OC.
6. Ovarian Cancer Drug Resistance and Proteomics
The standard method of care for OC involves surgery and chemotherapy with carboplatin and paclitaxel [93]. However, drug resistance is a major problem for many patients. Proteomic investigations have attempted to decipher the underlying mechanism of drug resistance in the patient cohorts and have indicated aberrations in the ATP synthesis and RAN GTPase binding components [94]. Moreover, analysis of serum samples of OC patients has suggested the involvement of FN1, SERPINA1, and ORM1 [95]. These markers may potentially be used as a panel to identify resistant patients prior to chemotherapy (See also Table 2). Other studies link aberration of key signaling cascades such as PI3K/Akt/mTOR pathway to OC drug resistance [96,97,98,99]. Overexpression of EGFR and subsequent EGFR-mediated angiogenesis and further metastasis is also very common among high-grade patients [100,101].
It is clear that OC can obtain resistance through a range of pathways, many of which are linked to oncogenic kinase signaling [102]. Recent advances in single-cell technologies such as spatial profiling [103] and single-cell proteomics [104] have great potential for uncovering tumor heterogeneity and shedding light on mechanisms for inherent and acquired drug resistance. Mechanisms underlying drug resistance can be delineated using preclinical OC cell line models [105,106]. We envision that this will also be a focal point for OC research in the coming decade.
7. High-Density Protein Microarrays and OC Biomarkers
The development and utility of high-density protein microarrays have led to significant progress in the field of cancer proteomics [14,107]. For example, human proteome arrays comprising nearly 17000 full-length proteins in duplicate have enabled studies uncovering specific and differential autoantibody responses between OC patients and controls [108,109]. Using this full-length protein array technology, studies have found that markers such as Lamin A/C, SSRP1, and RALBP1 are elevated in OC patients [110]. Antibody microarrays can also be useful for screening disease-associated markers based on the specificity of the antigen-antibody interactions. Using high-density arrays with immobilized antibodies provides a rapid approach to screen for cancer-specific markers from complex proteomes, such as human serum [107,111]. Antibody-microarrays have developed to the point where very high test accuracy can be achieved, as is illustrated by the detection of early-stage pancreatic cancer [112]. This technology also provides a way to validate markers for clinical utility identified by orthogonal techniques, such as mass spectrometry.
8. Perspective on Existing Biomarker Panels: Their Utility, Limitations, and Future Scope
The journey of identifying markers for early diagnosis of OCs, from CA125 & HE4 to current multi-marker models, has indeed come a long way [107]. Notable progress has been made in uncovering molecular features and mechanisms of the heterogeneous malignancy. Several studies have shown that multivariate biomarker panels outperform single marker analysis. For example, the 6 marker panels comprising leptin, prolactin, osteopontin, insulin-like growth factor 2, macrophage inhibitory factor, and CA125 outperformed CA125 alone, with a specificity of 99.4% [14,56]. Many biomarkers display good performance in single cohorts, but most fail in independent validation studies [113]. To cope with this, a recent effort by the Mann Lab systematically identified plasma proteins related to sample handling bias to generate a resource of “low quality” markers [114].
Recently, Enroth et al., described an 11-protein biomarker signature that separated ovarian cancer stages I–IV from benign controls with a specificity of 93% in an independent validation cohort [17]. However, no report has so far demonstrated high sensitivity for early-stage OC. Current challenges include increasing the specificity of the multi-marker panels, as well as the establishment of robust technological platforms that can easily quantify these markers from patients in a clinical setting.
The development and clinical implementation of complex multivariate algorithms that combine age, menopausal status, imaging, and serum-based biomarkers into a single index for estimation of the risk of malignancy have benefitted patients. For example, strategies such as Risk of Malignancy Index (RMI) [115], Risk of Malignancy Algorithm [20] (ROMA), and OVA1 [20] have been useful to aid decisions regarding treatment strategy, and specifically whether to refer the case to specialized cancer surgery in the case of increased pelvic mass [116,117]. Such multi-variate marker panels will now also be developed for early diagnosis, and there is currently a window of opportunity for high throughput (prote)-omics technologies to uncover such markers.
The biomarker diagnostic test format is a key question for clinical implementation. Today, single or multiplexed antibody-based tests, such as ELISA, represent the most frequently used method. Antibody-based tests have two clear benefits compared to MS-based platforms. First, they can readily target low abundant proteins that a mass spectrometer is typically blind to due to abundance detection bias and the vast dynamic concentration range of proteins in plasma [63]. Second, most biomedical hospital labs have the instrumentation and knowledge to perform such tests, but they rely on commercial tests based on highly specific antibodies. In that respect, MS is a more versatile method that allows the targeting of any protein, although it is still a niche field and few clinics are equipped with the instrumentation and expertise required for its conduction. For MS to be widely applied, the establishment of tailored reference laboratories that are dedicated to such analysis would be a solution. Hospitals would then send samples to be analyzed in such expert biomarker laboratories, a logistic procedure most hospitals have already implemented.
9. Conclusions and Outlook
Circulating protein biomarkers display great potential to discriminate between patients with benign and malignant ovarian cysts, while also guiding treatment decisions [8,118]. In recent years, proteomics characterization of plasma, effusions, and solid tumors has uncovered molecular mechanisms and a plethora of candidate biomarkers for OC, although these still need to be validated to show clinical utility. We foresee a wealth of studies in the coming years validating these candidate markers, while also identifying additional markers. We also anticipate that basic OC research will focus on the single-cell resolved analysis of tumor protein and PTMs. The integrated analysis of tumor specimens with matched blood samples is particularly interesting and has the potential to reveal accessible surrogate blood-based biomarkers that reflect tumor biology and can be used in personalized treatment.
Author Contributions
S.M. and M.E.J. co-wrote the review with input from K.S. and C.A.K.B. All authors have read and agreed to the published version of the manuscript.
Funding
Research in the Jakobsson Lab is supported by Stiftelsen Cancera, Mats Paulssons Stiftelse for Research, Innovation and Community Development, the Crafoord Foundation [ref 20200526], EU-MSCA-COFUND (847583), Fru Berta Kamprads Stiftelse (FBKS-2021-31-(330) and CREATE Health Cancer Center. Research in the Sundfeldt group is supported by the Swedish Cancer Foundation, Sjöbergsstiftelsen, and ALF-VGR. The APC was funded by the Lund University APC fund.
Conflicts of Interest
The authors declare no conflict of interest in the current study.
References
- Ueland, F. A Perspective on Ovarian Cancer Biomarkers: Past, Present and Yet-To-Come. Diagnostics 2017, 7, 14. [Google Scholar] [CrossRef]
- Reid, B.M.; Permuth, J.B.; Sellers, T.A. Epidemiology of ovarian cancer: A review. Cancer Biol. Med. 2017, 14, 9–32. [Google Scholar] [PubMed]
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Miller, K.D.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian cancer statistics, 2018. CA. Cancer J. Clin. 2018, 68, 284–296. [Google Scholar] [CrossRef]
- Koshiyama, M.; Matsumura, N.; Konishi, I. Subtypes of Ovarian Cancer and Ovarian Cancer Screening. Diagnostics 2017, 7, 12. [Google Scholar] [CrossRef]
- Cristea, M.; Han, E.; Salmon, L.; Morgan, R.J. Review: Practical considerations in ovarian cancer chemotherapy. Ther. Adv. Med. Oncol. 2010, 2, 175–187. [Google Scholar] [CrossRef]
- Javadi, S.; Ganeshan, D.M.; Qayyum, A.; Iyer, R.B.; Bhosale, P. Ovarian Cancer, the Revised FIGO Staging System, and the Role of Imaging Women’s Imaging Review. AJR 2016, 206. [Google Scholar] [CrossRef] [PubMed]
- King, M.-C.; Marks, J.H.; Mandell, J.B.; New, T.; Breast, Y. Breast and Ovarian Cancer Risks Due to Inherited Mutations in BRCA1 and BRCA2. Science 2003, 302, 643–646. [Google Scholar] [CrossRef]
- Zhang, Z.; Chan, D.W. The road from discovery to clinical diagnostics: Lessons learned from the first FDA-cleared in vitro diagnostic multivariate index assay of proteomic biomarkers. Cancer Epidemiol. Biomark. Prev. 2010, 19, 2995–2999. [Google Scholar] [CrossRef] [PubMed]
- Testa, U.; Petrucci, E.; Pasquini, L.; Castelli, G.; Pelosi, E. Ovarian Cancers: Genetic Abnormalities, Tumor Heterogeneity and Progression, Clonal Evolution and Cancer Stem Cells. Medicines 2018, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Parmigiani, G. Meta-analysis of BRCA1 and BRCA2 penetrance. J. Clin. Oncol. 2007, 25, 1329–1333. [Google Scholar] [CrossRef] [PubMed]
- Lawrenson, K.; Gayther, S.A. Ovarian cancer: A clinical challenge that needs some basic answers. PLoS Med. 2009, 6, 126–129. [Google Scholar] [CrossRef] [PubMed]
- Tangjitgamol, S.; Manusirivithaya, S.; Laopaiboon, M.; Lumbiganon, P.; Bryant, A. Interval debulking surgery for advanced epithelial ovarian cancer. Cochrane Database Syst. Rev. 2013, 4, 1–37. [Google Scholar] [CrossRef]
- Van der Burg, M.E.L.; Lammes, F.B.; Verweij, J. CA 125 in ovarian cancer. Neth. J. Med. 1992, 40, 36–51. [Google Scholar] [PubMed]
- Picotti, P.; Aebersold, R. Selected reaction monitoring-based proteomics: Workflows, potential, pitfalls and future directions. Nat. Methods 2012, 9, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Mor, G.; Visintin, I.; Lai, Y.; Zhao, H.; Schwartz, P.; Rutherford, T.; Yue, L.; Bray-Ward, P.; Ward, D.C. Serum protein markers for early detection of ovarian cancer. Proc. Natl. Acad. Sci. USA 2005, 102, 7677–7682. [Google Scholar] [CrossRef]
- Whitwell, H.J.; Worthington, J.; Blyuss, O.; Gentry-Maharaj, A.; Ryan, A.; Gunu, R.; Kalsi, J.; Menon, U.; Jacobs, I.; Zaikin, A.; et al. Improved early detection of ovarian cancer using longitudinal multimarker models. Br. J. Cancer 2020, 122. [Google Scholar] [CrossRef]
- Enroth, S.; Berggrund, M.; Lycke, M.; Broberg, J.; Lundberg, M.; Assarsson, E.; Olovsson, M.; Stålberg, K.; Sundfeldt, K.; Gyllensten, U. High throughput proteomics identifies a high-accuracy 11 plasma protein biomarker signature for ovarian cancer. Commun. Biol. 2019, 2, 1–12. [Google Scholar] [CrossRef]
- Bast, R.C. Early detection of ovarian cancer: New technologies in pursuit of a disease that is neither common nor rare. Trans. Am. Clin. Climatol. Assoc. 2004, 115, 233. [Google Scholar] [PubMed]
- Zhang, L.; Chen, Y.; Wang, K. Comparison of CA125, HE4, and ROMA index for ovarian cancer diagnosis. Curr. Probl. Cancer 2019, 43, 135–144. [Google Scholar] [CrossRef]
- Moore, R.G.; McMeekin, D.S.; Brown, A.K.; DiSilvestro, P.; Miller, M.C.; Allard, W.J.; Gajewski, W.; Kurman, R.; Bast, R.C.; Skates, S.J. A novel multiple marker bioassay utilizing HE4 and CA125 for the prediction of ovarian cancer in patients with a pelvic mass. Gynecol. Oncol. 2009, 112, 40–46. [Google Scholar] [CrossRef]
- Sölétormos, G.; Duffy, M.J.; Othman Abu Hassan, S.; Verheijen, R.H.M.; Tholander, B.; Bast, R.C.; Gaarenstroom, K.N.; Sturgeon, C.M.; Bonfrer, J.M.; Petersen, P.H.; et al. Clinical Use of Cancer Biomarkers in Epithelial Ovarian Cancer: Updated Guidelines from the European Group on Tumor Markers. Lippincott Williams and Wilkins. Int. J. Gynecol. Cancer 2016, 26, 43–51. [Google Scholar] [CrossRef]
- Feng, Y.; Xiao, M.; Zhang, Z.; Cui, R.; Jiang, X.; Wang, S.; Bai, H.; Liu, C.; Zhang, Z. Potential interaction between lysophosphatidic acid and tumor-associated macrophages in ovarian carcinoma. J. Inflamm. 2020, 17, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Gadducci, A.; Ferdeghini, M.; Ceccarini, T.; Prontera, C.; Facchini, V.; Bianchi, R.; Fioretti, P. A comparative evaluation of the ability of serum CA 125, CA 19-9, CA 15-3, CA 50, CA 72-4 and TATI assays in reflecting the course of disease in patients with ovarian carcinoma. Eur. J. Gynaecol. Oncol. 1990, 11, 127–133. [Google Scholar]
- Zhang, Z.; Yu, Y.; Xu, F.; Berchuck, A.; van Haaften-Day, C.; Havrilesky, L.J.; de Bruijn, H.W.A.; van der Zee, A.G.J.; Woolas, R.P.; Jacobs, I.J.; et al. Combining multiple serum tumor markers improves detection of stage I epithelial ovarian cancer. Gynecol. Oncol. 2007, 107, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Woolas, R.P.; Conaway, M.R.; Xu, F.; Jacobs, I.J.; Yu, Y.; Daly, L.; Davies, A.P.; O’Briant, K.; Berchuck, A.; Soper, J.T.; et al. Combinations of multiple serum markers are superior to individual assays for discriminating malignant from benign pelvic masses. Gynecol. Oncol. 1995, 59, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Bast, R.C.; Yu, Y.; Li, J.; Sokoll, L.J.; Rai, A.J.; Rosenzweig, J.M.; Cameron, B.; Wang, Y.Y.; Meng, X.Y.; et al. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res. 2004, 64, 5882–5890. [Google Scholar] [CrossRef] [PubMed]
- Coscia, F.; Lengyel, E.; Duraiswamy, J.; Ashcroft, B.; Bassani-Sternberg, M.; Wierer, M.; Johnson, A.; Wroblewski, K.; Montag, A.; Yamada, S.D.; et al. Multi-level Proteomics Identifies CT45 as a Chemosensitivity Mediator and Immunotherapy Target in Ovarian Cancer. Cell 2018, 175, 159–170.e16. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson, S.; Krogh, M.; Szigyarto, C.A.K.; Uhlen, M.; Schedvins, K.; Silfverswärd, C.; Linder, S.; Auer, G.; Alaiya, A.; James, P. Large-scale proteomics analysis of human ovarian cancer for biomarkers. J. Proteome Res. 2007, 6, 1440–1450. [Google Scholar] [CrossRef]
- Dieters-Castator, D.Z.; Rambau, P.F.; Kelemen, L.E.; Siegers, G.M.; Lajoie, G.A.; Postovit, L.M.; Kobel, M. Proteomics-derived biomarker panel improves diagnostic precision to classify endometrioid and high-grade serous ovarian carcinoma. Clin. Cancer Res. 2019, 25, 4309–4319. [Google Scholar] [CrossRef]
- Kriplani, D.; Patel, M.M. Immunohistochemistry: A diagnostic aid in differentiating primary epithelial ovarian tumors and tumors metastatic to the ovary. South Asian J. Cancer 2013, 2, 254–258. [Google Scholar] [CrossRef]
- Shih, I.M.; Kurman, R.J. Ovarian Tumorigenesis: A Proposed Model Based on Morphological and Molecular Genetic Analysis. Am. J. Pathol. 2004, 164, 1511–1518. [Google Scholar] [CrossRef]
- Dehari, R.; Kurman, R.J.; Logani, S.; Shih, I.M. The development of high-grade serous carcinoma from atypical proliferative (borderline) serous tumors and low-grade micropapillary serous carcinoma: A morphologic and molecular genetic analysis. Am. J. Surg. Pathol. 2007, 31, 1007–1012. [Google Scholar] [CrossRef]
- Wong, K.-K.; Gershenson, D.; Tsai, C.-C. BRAF mutational analysis in ovarian tumors: Recent perspectives. Pathol. Lab. Med. Int. 2015, 7, 75. [Google Scholar] [CrossRef]
- Kurman, R.J.; Shih, I.M. The origin and pathogenesis of epithelial ovarian cancer: A proposed unifying theory. Am. J. Surg. Pathol. 2010, 34, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Davidson, B.; Smith, Y.; Nesland, J.M.; Kærn, J.; Reich, R.; Tropè, C.G. Defining a prognostic marker panel for patients with ovarian serous carcinoma effusion. Hum. Pathol. 2013, 44, 2449–2460. [Google Scholar] [CrossRef] [PubMed]
- Boylan, K.L.M.; Afiuni-Zadeh, S.; Geller, M.A.; Argenta, P.A.; Griffin, T.J.; Skubitz, A.P.N. Evaluation of the potential of Pap test fluid and cervical swabs to serve as clinical diagnostic biospecimens for the detection of ovarian cancer by mass spectrometry-based proteomics. Clin. Proteom. 2021, 18, 4. [Google Scholar] [CrossRef] [PubMed]
- Marcišauskas, S.; Ulfenborg, B.; Kristjansdottir, B.; Waldemarson, S.; Sundfeldt, K. Univariate and classification analysis reveals potential diagnostic biomarkers for early stage ovarian cancer Type 1 and Type 2. J. Proteom. 2019, 196, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Minis, E.; Holcomb, K.; Sisti, G.; Nasioudis, D.; Kanninen, T.T.; Athanasiou, A.; Frey, M.K.; Chapman-Davis, E.; Caputo, T.A.; Witkin, S.S. Evaluation of lysophosphatidic acid in vaginal fluid as a biomarker for ovarian cancer: A pilot study. Eur. J. Obstet. Gynecol. Reprod. Biol. X 2019, 2, 100012. [Google Scholar] [CrossRef]
- Kim, G.; Davidson, B.; Henning, R.; Wang, J.; Yu, M.; Annunziata, C.; Hetland, T.; Kohn, E.C. Adhesion molecule protein signature in ovarian cancer effusions is prognostic of patient outcome. Cancer 2012, 118, 1543–1553. [Google Scholar] [CrossRef] [PubMed]
- Tancioni, I.; Uryu, S.; Sulzmaier, F.J.; Shah, N.R.; Lawson, C.; Miller, N.L.G.; Jean, C.; Chen, X.L.; Ward, K.K.; Schlaepfer, D.D. FAK inhibition disrupts a β5 integrin signaling axis controlling anchorage-independent ovarian carcinoma growth. Mol. Cancer Ther. 2014, 13, 2050–2061. [Google Scholar] [CrossRef]
- Xu, B.; Lefringhouse, J.; Liu, Z.; West, D.; Baldwin, L.A.; Ou, C.; Chen, L.; Napier, D.; Chaiswing, L.; Brewer, L.D.; et al. Inhibition of the integrin/FAK signaling axis and c-Myc synergistically disrupts ovarian cancer malignancy. Oncogenesis 2017, 6, 295. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Lei, S.; Zhang, Q.; Ma, L.; Li, N.; Wang, J. Deregulation of cell adhesion molecules is associated with progression and poor outcomes in endometrial cancer: Analysis of the Cancer Genome Atlas data. Oncol. Lett. 2020, 19, 1906–1914. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sundfeldt, K.; Mateoiu, C.; Shih, I.M.; Kurman, R.J.; Schaefer, J.; Silliman, N.; Kinde, I.; Springer, S.; Foote, M.; et al. Diagnostic potential of tumor DNA from ovarian cyst fluid. Elife 2016, 5, e15175. [Google Scholar] [CrossRef]
- Wang, Y.; Li, L.; Douville, C.; Cohen, J.D.; Yen, T.T.; Kinde, I.; Sundfelt, K.; Kjær, S.K.; Hruban, R.H.; Shih, I.M.; et al. Evaluation of liquid from the Papanicolaou test and other liquid biopsies for the detection of endometrial and ovarian cancers. Sci. Transl. Med. 2018, 10. [Google Scholar] [CrossRef]
- Aktas, B.; Kasimir-Bauer, S.; Heubner, M.; Kimmig, R.; Wimberger, P. Molecular profiling and prognostic relevance of circulating tumor cells in the blood of ovarian cancer patients at primary diagnosis and after platinum-based chemotherapy. Int. J. Gynecol. Cancer 2011, 21, 822–830. [Google Scholar] [CrossRef]
- Mari, R.; Mamessier, E.; Lambaudie, E.; Provansal, M.; Birnbaum, D.; Bertucci, F.; Sabatier, R. Liquid biopsies for ovarian carcinoma: How blood tests may improve the clinical management of a deadly disease. Cancers 2019, 11, 774. [Google Scholar] [CrossRef]
- Zhang, X.; Li, H.; Yu, X.; Li, S.; Lei, Z.; Li, C.; Zhang, Q.; Han, Q.; Li, Y.; Zhang, K.; et al. Cellular Physiology and Biochemistry Cellular Physiology and Biochemistry Analysis of Circulating Tumor Cells in Ovarian Cancer and Their Clinical Value as a Biomarker. Cell Physiol. Biochem. 2018, 48, 1983–1994. [Google Scholar] [CrossRef]
- Van Berckelaer, C.; Brouwers, A.J.; Peeters, D.J.E.; Tjalma, W.; Trinh, X.B.; van Dam, P.A. Current and future role of circulating tumor cells in patients with epithelial ovarian cancer. Eur. J. Surg. Oncol. 2016, 42, 1772–1779. [Google Scholar] [CrossRef]
- Asante, D.B.; Calapre, L.; Ziman, M.; Meniawy, T.M.; Gray, E.S. Liquid biopsy in ovarian cancer using circulating tumor DNA and cells: Ready for prime time? Cancer Lett. 2020, 468, 59–71. [Google Scholar] [CrossRef]
- Bast, R.C.; Feeney, M.; Lazarus, H.; Nadler, L.M.; Colvin, R.B.; Knapp, R.C. Reactivity of a monoclonal antibody with human ovarian carcinoma. J. Clin. Invest 1981, 68, 1331–1337. [Google Scholar] [CrossRef] [PubMed]
- Rustin, G.J.S.; Nelstrop, A.E.; Tuxen, M.K.; Lambert, H.E. Defining progression of ovarian carcinoma during follow-up according to CA 125: A North Thames Ovary Group study. Ann. Oncol. 1996, 7, 361–364. [Google Scholar] [CrossRef] [PubMed]
- Kulasingam, V.; Pavlou, M.P.; Diamandis, E.P. Integrating high-throughput technologies in the quest for effective biomarkers for ovarian cancer. Nat. Rev. Cancer 2010, 10, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Drapkin, R.; Von Horsten, H.H.; Lin, Y.; Mok, S.C.; Crum, C.P.; Welch, W.R.; Hecht, J.L. Human epididymis protein 4 (HE4) is a secreted glycoprotein that is overexpressed by serous and endometrioid ovarian carcinomas. Cancer Res. 2005, 65, 2162–2169. [Google Scholar] [CrossRef]
- Lycke, M.; Ulfenborg, B.; Kristjansdottir, B.; Sundfeldt, K. Increased Diagnostic Accuracy of Adnexal Tumors with A Combination of Established Algorithms and Biomarkers. J. Clin. Med. 2020, 9, 299. [Google Scholar] [CrossRef] [PubMed]
- Muinao, T.; Deka Boruah, H.P.; Pal, M. Multi-biomarker panel signature as the key to diagnosis of ovarian cancer. Heliyon 2019, 5, e02826. [Google Scholar] [CrossRef]
- Yurkovetsky, Z.; Skates, S.; Lomakin, A.; Nolen, B.; Pulsipher, T.; Modugno, F.; Marks, J.; Godwin, A.; Gorelik, E.; Jacobs, I.; et al. Development of a multimarker assay for early detection of ovarian cancer. J. Clin. Oncol. 2010, 28, 2159–2166. [Google Scholar] [CrossRef]
- Zhang, Z. An In Vitro Diagnostic Multivariate Index Assay (IVDMIA) for Ovarian Cancer: Harvesting the Power of Multiple Biomarkers. Rev. Obstet. Gynecol. 2012, 5, 35–41. [Google Scholar]
- Wei, S.; Li, H.; Zhang, B. The diagnostic value of serum HE4 and CA-125 and ROMA index in ovarian cancer. Biomed. Rep. 2016, 5, 41–44. [Google Scholar] [CrossRef]
- Mai, P.L.; Wentzensen, N.; Greene, M.H. Challenges related to developing serum-based biomarkers for early ovarian cancer detection. Cancer Prev. Res. 2011, 4, 303–306. [Google Scholar] [CrossRef]
- Franier, B.D.L.; Thompson, M. Early stage detection and screening of ovarian cancer: A research opportunity and significant challenge for biosensor technology. Biosens. Bioelectron. 2019, 135, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Swiatly, A.; Plewa, S.; Matysiak, J.; Kokot, Z.J. Mass spectrometry-based proteomics techniques and their application in ovarian cancer research 06 Biological Sciences 0601 Biochemistry and Cell Biology 11 Medical and Health Sciences 1112 Oncology and Carcinogenesis. J. Ovarian Res. 2018, 11, 88. [Google Scholar] [CrossRef] [PubMed]
- Geyer, P.E.; Holdt, L.M.; Teupser, D.; Mann, M. Revisiting biomarker discovery by plasma proteomics. Mol. Syst. Biol. 2017, 13, 942. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Chang, H.M.; Wang, A.; Song, J.; Zhang, X.; Guo, J.; Leung, P.C.K.; Lian, F. Identification of potential metabolic biomarkers of polycystic ovary syndrome in follicular fluid by SWATH mass spectrometry. Reprod. Biol. Endocrinol. 2019, 17, 45. [Google Scholar] [CrossRef]
- Tessitore, A.; Gaggiano, A.; Cicciarelli, G.; Verzella, D.; Capece, D.; Fischietti, M.; Zazzeroni, F.; Alesse, E. Serum biomarkers identification by mass spectrometry in high-mortality tumors. Int. J. Proteom. 2013, 1–51. [Google Scholar] [CrossRef] [PubMed]
- Asare-Werehene, M.; Communal, L.; Carmona, E.; Le, T.; Provencher, D.; Mes-Masson, A.M.; Tsang, B.K. Pre-operative Circulating Plasma Gelsolin Predicts Residual Disease and Detects Early Stage Ovarian Cancer. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Hüttenhain, R.; Choi, M.; de la Fuente, L.M.; Oehl, K.; Chang, C.Y.; Zimmermann, A.K.; Malander, S.; Olsson, H.; Surinova, S.; Clough, T.; et al. A targeted mass spectrometry strategy for developing proteomic biomarkers: A case study of epithelial ovarian cancer. Mol. Cell. Proteom. 2019, 18, 1836–1850. [Google Scholar] [CrossRef]
- Hristova, V.A.; Chan, D.W. Cancer biomarker discovery and translation: Proteomics and beyond. Expert Rev. Proteom. 2019, 16, 93–103. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, T.; Zhang, Z.; Payne, S.H.; Zhang, B.B.B.B.; McDermott, J.E.; Zhou, J.-Y.Y.; Petyuk, V.A.; Chen, L.; Ray, D.; et al. Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer. Cell 2016, 166, 755–765. [Google Scholar] [CrossRef]
- Macklin, A.; Khan, S.; Kislinger, T. Recent advances in mass spectrometry based clinical proteomics: Applications to cancer research. Clin. Proteom. 2020, 17, 1–25. [Google Scholar] [CrossRef]
- Kristjansdottir, B.; Levan, K.; Partheen, K.; Carlsohn, E.; Sundfeldt, K. Potential tumor biomarkers identified in ovarian cyst fluid by quantitative proteomic analysis, itraq. Clin. Proteom. 2013, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Hu, Y.; Sun, S.; Chen, L.; Schnaubelt, M.; Clark, D.; Ao, M.; Zhang, Z.; Chan, D.; Qian, J.; et al. Glycoproteomics-based signatures for tumor subtyping and clinical outcome prediction of high-grade serous ovarian cancer. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Zervantonakis, I.K.; Iavarone, C.; Chen, H.Y.; Selfors, L.M.; Palakurthi, S.; Liu, J.F.; Drapkin, R.; Matulonis, U.; Leverson, J.D.; Sampath, D.; et al. Systems analysis of apoptotic priming in ovarian cancer identifies vulnerabilities and predictors of drug response. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Scott, C.L.; Becker, M.A.; Haluska, P.; Samimi, G. Patient-Derived Xenograft Models to Improve Targeted Therapy in Epithelial Ovarian Cancer Treatment. Front. Oncol. 2013, 3, 295. [Google Scholar] [CrossRef] [PubMed]
- Cruz, I.N.; Coley, H.M.; Kramer, H.B.; Madhuri, T.K.; Safuwan, N.A.M.; Angelino, A.R.; Yang, M. Proteomics analysis of ovarian cancer cell lines and tissues reveals drug resistance-associated proteins. Cancer Genom. Proteom. 2017, 14, 35–52. [Google Scholar] [CrossRef] [PubMed]
- McDermott, J.E.; Arshad, O.A.; Petyuk, V.A.; Fu, Y.; Gritsenko, M.A.; Clauss, T.R.; Moore, R.J.; Schepmoes, A.A.; Zhao, R.; Monroe, M.E.; et al. Proteogenomic Characterization of Ovarian HGSC Implicates Mitotic Kinases, Replication Stress in Observed Chromosomal Instability. Cell Reports Med. 2020, 1, 100004. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Pan, J.; Shah, P.; Ao, M.; Thomas, S.N.; Liu, Y.; Chen, L.; Schnaubelt, M.; Clark, D.J.; Rodriguez, H.; et al. Integrated Proteomic and Glycoproteomic Characterization of Human High-Grade Serous Ovarian Carcinoma. Cell Rep. 2020, 33, 108276. [Google Scholar] [CrossRef]
- Proteomic Data Commons. Available online: https://pdc.cancer.gov/pdc/browse/filters/study_name:Prospective Ovarian JHU Proteome%7CProspective Ovarian PNNL Phosphoproteome Lumos%7CProspective Ovarian PNNL Proteome Qeplus (accessed on 15 January 2021).
- Tong, M.; Yu, C.; Zhan, D.; Zhang, M.; Zhen, B.; Zhu, W.; Wang, Y.; Wu, C.; He, F.; Qin, J.; et al. Molecular subtyping of cancer and nomination of kinase candidates for inhibition with phosphoproteomics: Reanalysis of CPTAC ovarian cancer. EBioMedicine 2019, 40, 305–317. [Google Scholar] [CrossRef]
- Parekh, R.B.; Rohlff, C. Post-translational modification of proteins and the discovery of new medicine. Curr. Opin. Biotechnol. 1997, 8, 718–723. [Google Scholar] [CrossRef]
- Walsh, C.T.; Garneau-Tsodikova, S.; Gatto, G.J. Protein Posttranslational Modifications: The Chemistry of Proteome Diversifications. Angew. Chem. Int. Ed. 2005, 44, 7342–7372. [Google Scholar] [CrossRef]
- Olsen, J.V.; Mann, M. Status of large-scale analysis of posttranslational modifications by mass spectrometry. Mol. Cell. Proteom. 2013, 12, 3444–3452. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Chen, L.; Zhang, B.; Song, Q.; Yu, Y.; Moore, C.; Wang, T.L.; Shih, I.M.; Zhang, H.; Chan, D.W.; et al. Proteome-wide tyrosine phosphorylation analysis reveals dysregulated signaling pathways in ovarian tumors. Mol. Cell. Proteom. 2019, 18, 448–460. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, M.E.; Moen, A.; Davidson, B.; Falnes, P. Hsp70 (HSPA1) lysine methylation status as a potential prognostic factor in metastatic high-grade serous carcinoma. PLoS ONE 2015, 10, e0140168. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, M.E.; Moen, A.; Falnes, P. Correspondence: On the enzymology and significance of HSPA1 lysine methylation. Nat. Commun. 2016, 7, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Cloutier, P.; Lavallée-Adam, M.; Faubert, D.; Blanchette, M.; Coulombe, B. A Newly Uncovered Group of Distantly Related Lysine Methyltransferases Preferentially Interact with Molecular Chaperones to Regulate Their Activity. PLoS Genet. 2013, 9, e1003210. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, M.E.; Moen, A.; Bousset, L.; Egge-Jacobsen, W.; Kernstock, S.; Melki, R.; Falnes, P. Identification and characterization of a novel human methyltransferase modulating Hsp70 protein function through lysine methylation. J. Biol. Chem. 2013, 288, 27752–27763. [Google Scholar] [CrossRef] [PubMed]
- Truman, A.W. Cracking the Chaperone Code: Cellular Roles for Hsp70 Phosphorylation. Trends Biochem. Sci. 2017, 42, 932–935. [Google Scholar] [CrossRef]
- Porter, C.M.; Truman, A.W.; Truttmann, M.C. Post-translational modifications of Hsp70 family proteins: Expanding the chaperone code. J. Biol. Chem. 2020, 295, 10689–10708. [Google Scholar]
- Francavilla, C.; Lupia, M.; Tsafou, K.; Villa, A.; Kowalczyk, K.; Rakownikow Jersie-Christensen, R.; Bertalot, G.; Confalonieri, S.; Brunak, S.; Jensen, L.J.; et al. Phosphoproteomics of Primary Cells Reveals Druggable Kinase Signatures in Ovarian Cancer. Cell Rep. 2017, 18, 3242–3256. [Google Scholar] [CrossRef]
- Januchowski, R.; Sterzyńska, K.; Zawierucha, P.; Ruciński, M.; Świerczewska, M.; Partyka, M.; Bednarek-Rajewska, K.; Brazert, M.; Nowicki, M.; Zabel, M.; et al. Microarray-based detection and expression analysis of new genes associated with drug resistance in ovarian cancer cell lines. Oncotarget 2017, 8, 49944–49958. [Google Scholar] [CrossRef]
- Cheng, L.; Zhang, K.; Qing, Y.; Li, D.; Cui, M.; Jin, P.; Xu, T. Proteomic and lipidomic analysis of exosomes derived from ovarian cancer cells and ovarian surface epithelial cells. J. Ovarian Res. 2020, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Du Bois, A. A Randomized Clinical Trial of Cisplatin/Paclitaxel Versus Carboplatin/Paclitaxel as First-Line Treatment of Ovarian Cancer. CancerSpectrum Knowl. Environ. 2003, 95, 1320–1329. [Google Scholar] [CrossRef]
- Yu, K.H.; Levine, D.A.; Zhang, H.; Chan, D.W.; Zhang, Z.; Snyder, M. Predicting ovarian cancer patients’ clinical response to platinum-based chemotherapy by their tumor proteomic signatures. J. Proteome Res. 2016, 15, 2455–2465. [Google Scholar] [CrossRef]
- Wu, W.; Wang, Q.; Yin, F.; Yang, Z.; Zhang, W.; Gabra, H.; Li, L. Identification of proteomic and metabolic signatures associated with chemoresistance of human epithelial ovarian cancer. Int. J. Oncol. 2016, 49, 1651–1665. [Google Scholar] [CrossRef] [PubMed]
- Ediriweera, M.K.; Tennekoon, K.H.; Samarakoon, S.R. Role of the PI3K/AKT/mTOR signaling pathway in ovarian cancer: Biological and therapeutic significance. Semin. Cancer Biol. 2019, 59, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Mabuchi, S.; Kuroda, H.; Takahashi, R.; Sasano, T. The PI3K/AKT/mTOR pathway as a therapeutic target in ovarian cancer. Gynecol. Oncol. 2015, 137, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Li, H.X.; Zeng, J.F.; Shen, K. PI3K/AKT/mTOR signaling pathway as a therapeutic target for ovarian cancer. Arch. Gynecol. Obstet. 2014, 290, 1067–1078. [Google Scholar] [CrossRef]
- Wu, R.; Hu, T.C.; Rehemtulla, A.; Fearon, E.R.; Cho, K.R. Preclinical testing of PI3K/AKT/mTOR signaling inhibitors in a mouse model of ovarian endometrioid adenocarcinoma. Clin. Cancer Res. 2011, 17, 7359–7372. [Google Scholar] [CrossRef]
- Hudson, L.G.; Zeineldin, R.; Silberberg, M.; Stack, M.S. Activated Epidermal Growth Factor Receptor in Ovarian Cancer I. Background: The EGF Receptor. Cancer Treat. Res. 2009, 149, 203–226. [Google Scholar] [CrossRef]
- Sheng, Q.; Liu, J. The therapeutic potential of targeting the EGFR family in epithelial ovarian cancer. Br. J. Cancer 2011, 104, 1241–1245. [Google Scholar] [CrossRef]
- Blume-Jensen, P.; Hunter, T. Oncogenic kinase signalling. Nature 2001, 411, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Marx, V. Method of the Year: Spatially resolved transcriptomics. Nat. Methods 2021, 18, 9–14. [Google Scholar] [CrossRef]
- Marx, V. A dream of single-cell proteomics. Nat. Methods 2019, 16, 809–812. [Google Scholar] [CrossRef] [PubMed]
- Coscia, F.; Watters, K.M.; Curtis, M.; Eckert, M.A.; Chiang, C.Y.; Tyanova, S.; Montag, A.; Lastra, R.R.; Lengyel, E.; Mann, M. Integrative proteomic profiling of ovarian cancer cell lines reveals precursor cell associated proteins and functional status. Nat. Commun. 2016, 7, 1–14. [Google Scholar] [CrossRef]
- Domcke, S.; Sinha, R.; Levine, D.A.; Sander, C.; Schultz, N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun. 2013, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Borrebaeck, C.A.K. Precision diagnostics: Moving towards protein biomarker signatures of clinical utility in cancer. Nat. Rev. Cancer 2017, 17, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, J.; Li, J.; Zhang, H.; Guo, S.; Yan, M.; Zhu, Z.; Lan, B.; Ding, Y.; Xu, M.; et al. Identification of serum biomarkers for gastric cancer diagnosis using a human proteome microarray. Mol. Cell. Proteom. 2016, 15, 614–623. [Google Scholar] [CrossRef]
- Gunawardana, C.G.; Memari, N.; Diamandis, E.P. Identifying novel autoantibody signatures in ovarian cancer using high-density protein microarrays. Clin. Biochem. 2009, 42, 426–429. [Google Scholar] [CrossRef]
- Hudson, M.E.; Pozdnyakova, I.; Haines, K.; Mor, G.; Snyder, M. Identification of differentially expressed proteins in ovarian cancer using high-density protein microarrays. Proc. Natl. Acad. Sci. USA 2007, 104, 17494–17499. [Google Scholar] [CrossRef]
- Borrebaeck, C.A.K.; Wingren, C. Design of high-density antibody microarrays for disease proteomics: Key technological issues. J. Proteom. 2009, 72, 928–935. [Google Scholar] [CrossRef]
- Mellby, L.D.; Nyberg, A.P.; Johansen, J.S.; Wingren, C.; Nordestgaard, B.G.; Bojesen, S.E.; Mitchell, B.L.; Sheppard, B.C.; Sears, R.C.; Borrebaeck, C.A.K. Serum biomarker signature-based liquid biopsy for diagnosis of early-stage pancreatic cancer. J. Clin. Oncol. 2018, 36, 2887–2894. [Google Scholar] [CrossRef] [PubMed]
- Ioannidis, J.P.A.; Panagiotou, O.A. Comparison of effect sizes associated with biomarkers reported in highly cited individual articles and in subsequent meta-analyses. JAMA J. Am. Med. Assoc. 2011, 305, 2200–2210. [Google Scholar] [CrossRef] [PubMed]
- Geyer, P.E.; Voytik, E.; Treit, P.V.; Doll, S.; Kleinhempel, A.; Niu, L.; Müller, J.B.; Buchholtz, M.; Bader, J.M.; Teupser, D.; et al. Plasma Proteome Profiling to detect and avoid sample-related biases in biomarker studies. EMBO Mol. Med. 2019, 11, e10427. [Google Scholar] [CrossRef]
- Jacobs, I.; Oram, D.; Fairbanks, J.; Turner, J.; Frost, C.; Grudzinskas, J.G. A risk of malignancy index incorporating CA 125, ultrasound and menopausal status for the accurate preoperative diagnosis of ovarian cancer. BJOG An. Int. J. Obstet. Gynaecol. 1990, 97, 922–929. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.L.; Lu, Z.; Bast, R.C. The role of biomarkers in the management of epithelial ovarian cancer. Expert Rev. Mol. Diagn. 2017, 17, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Visintin, I.; Alvero, A.B.; Mor, G. Development and Validation of a Protein-based Signature for the Detection of Ovarian Cancer. Clin. Lab. Med. 2009, 29, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Bristow, R.E.; Smith, A.; Zhang, Z.; Chan, D.W.; Crutcher, G.; Fung, E.T.; Munroe, D.G. Ovarian malignancy risk stratification of the adnexal mass using a multivariate index assay. Gynecol. Oncol. 2013, 128, 252–259. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).