Molecular and Physiological Study of Candida albicans by Quantitative Proteome Analysis
Abstract
1. Introduction
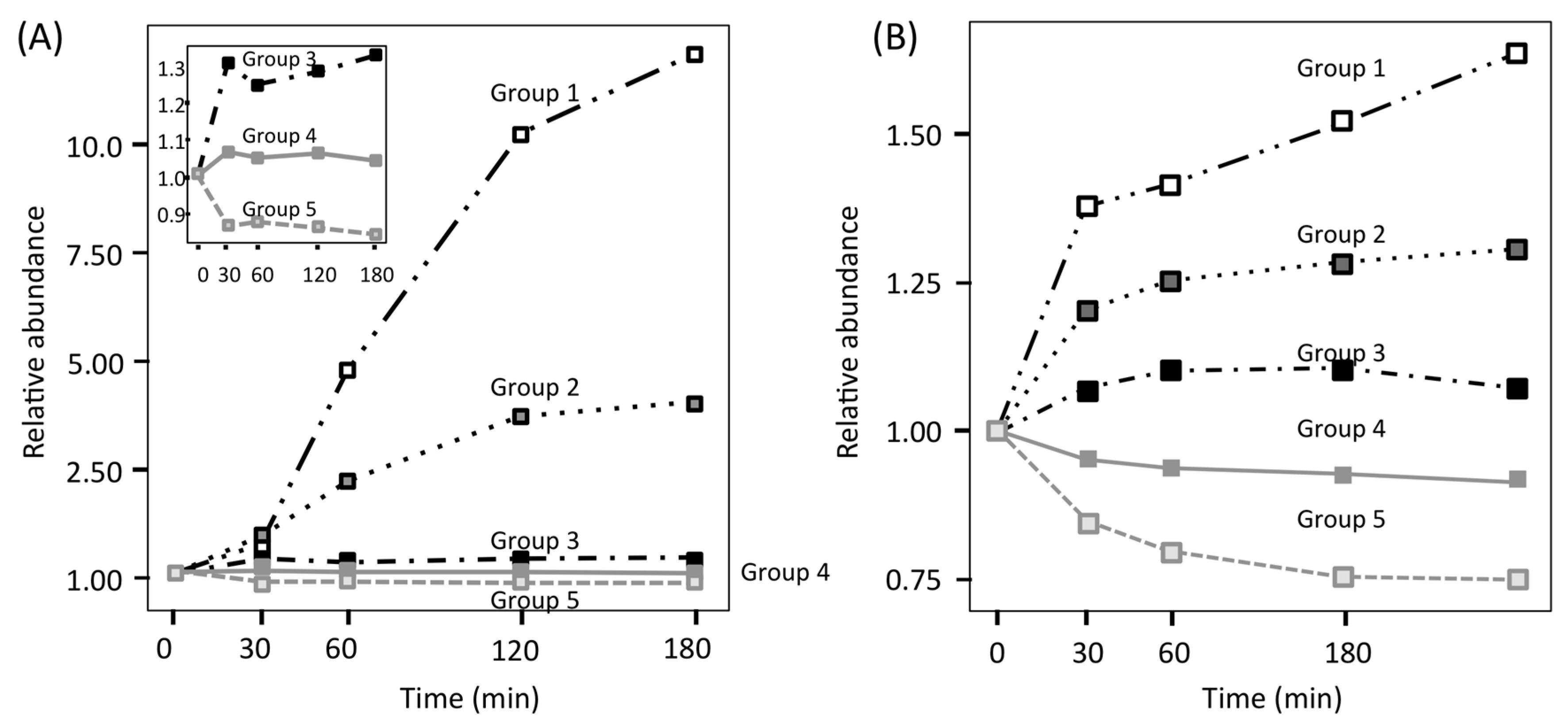
2. Time-Course Proteomics Analysis of C. albicans Adaptation to Serum
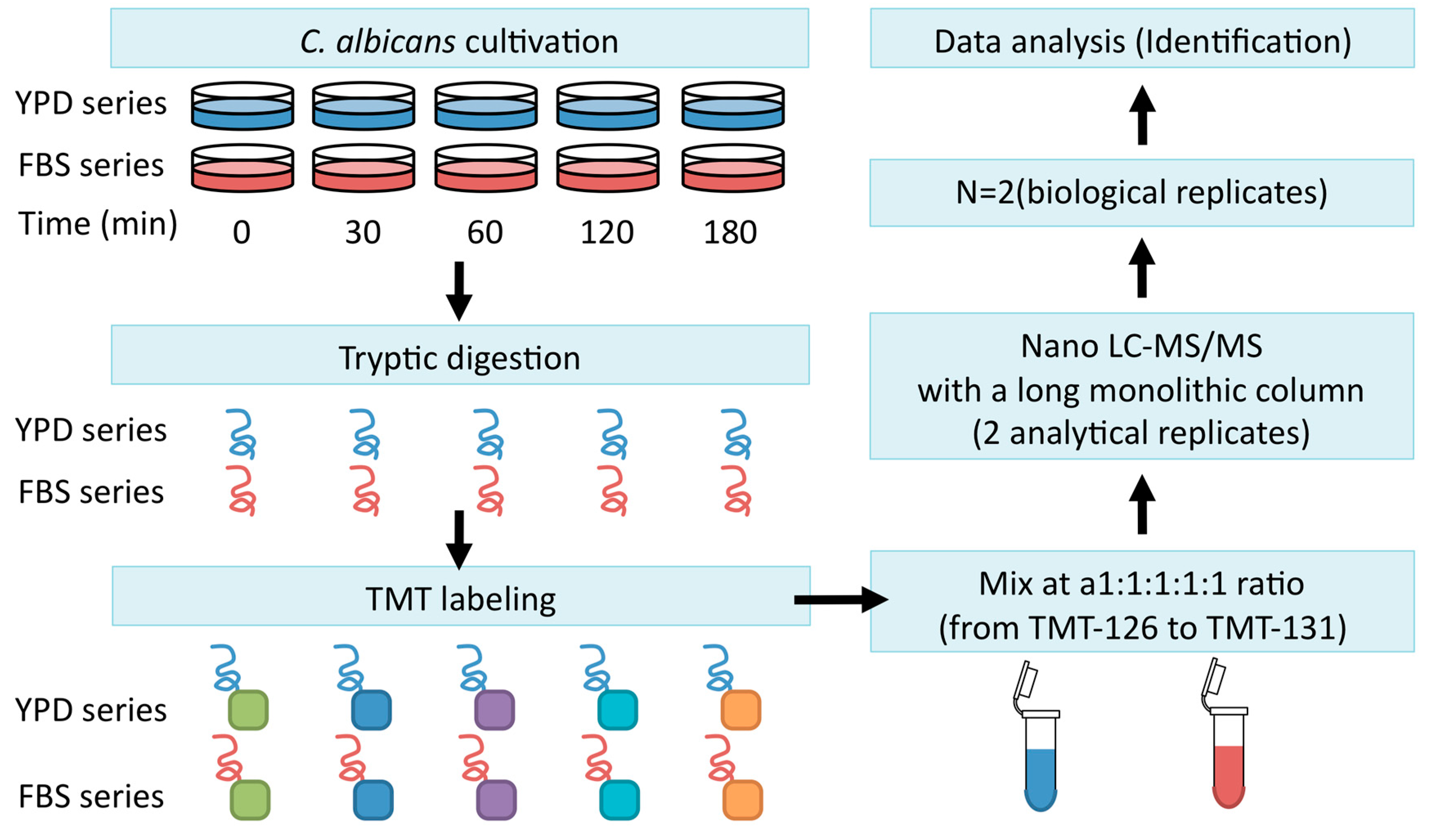
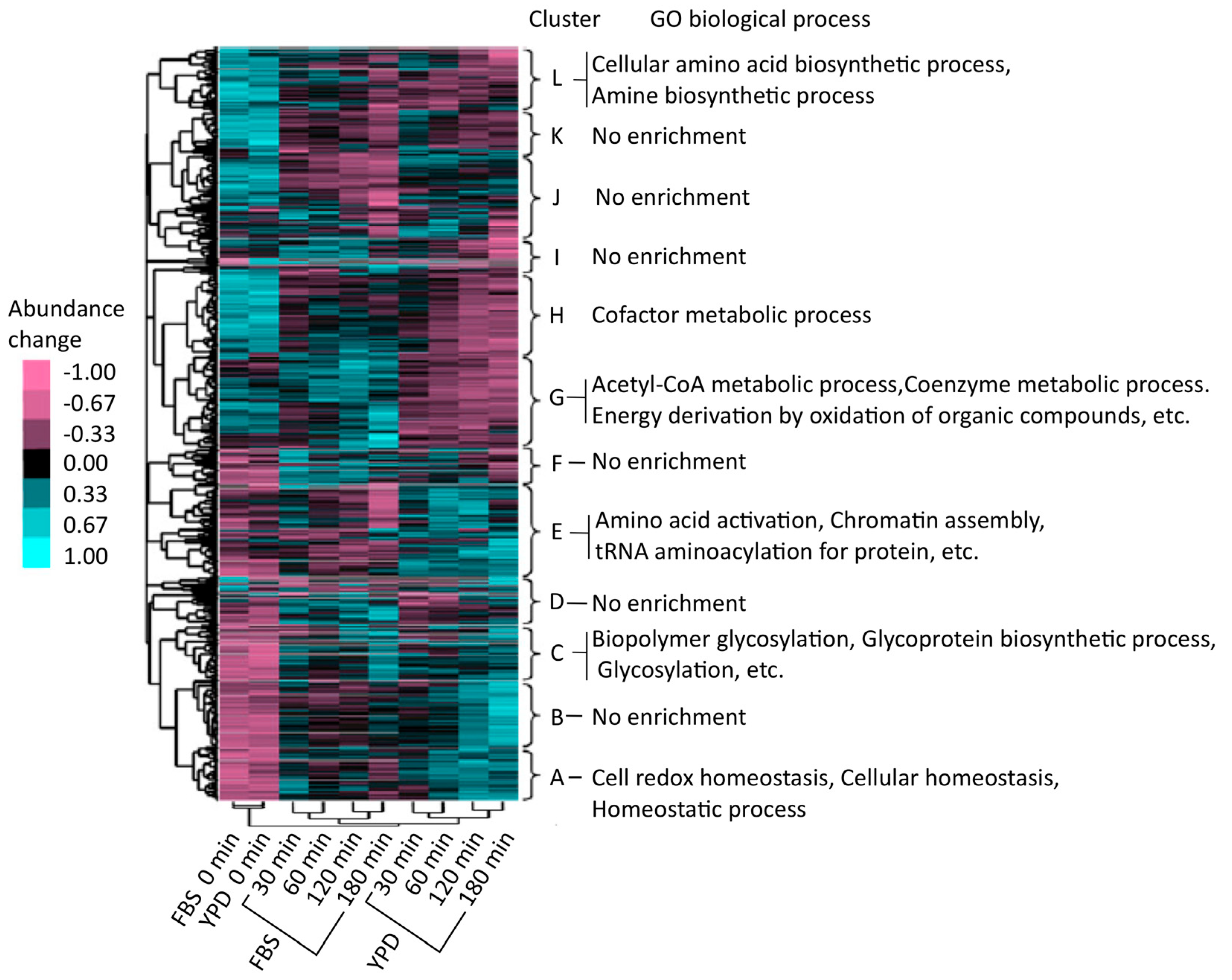
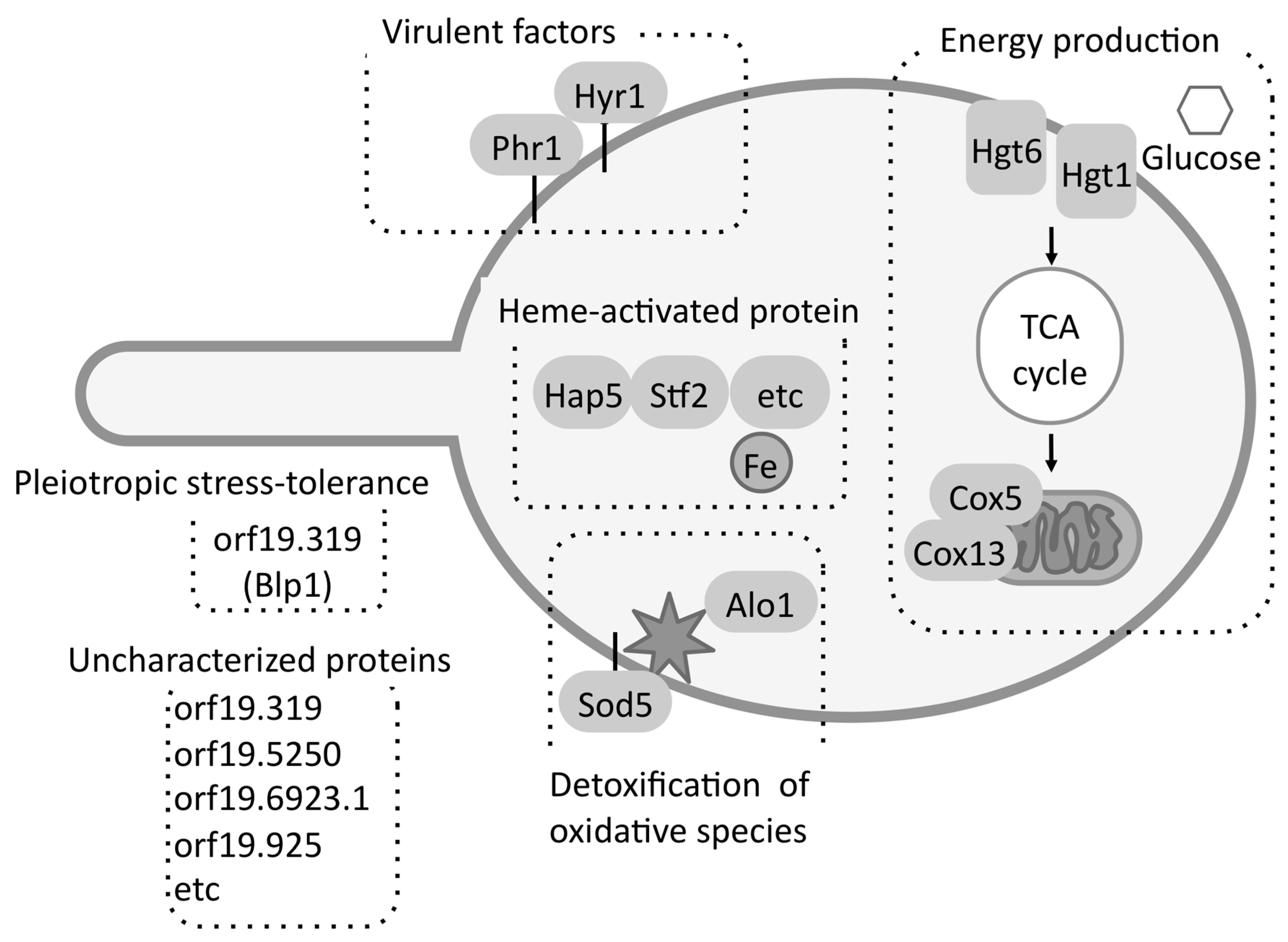
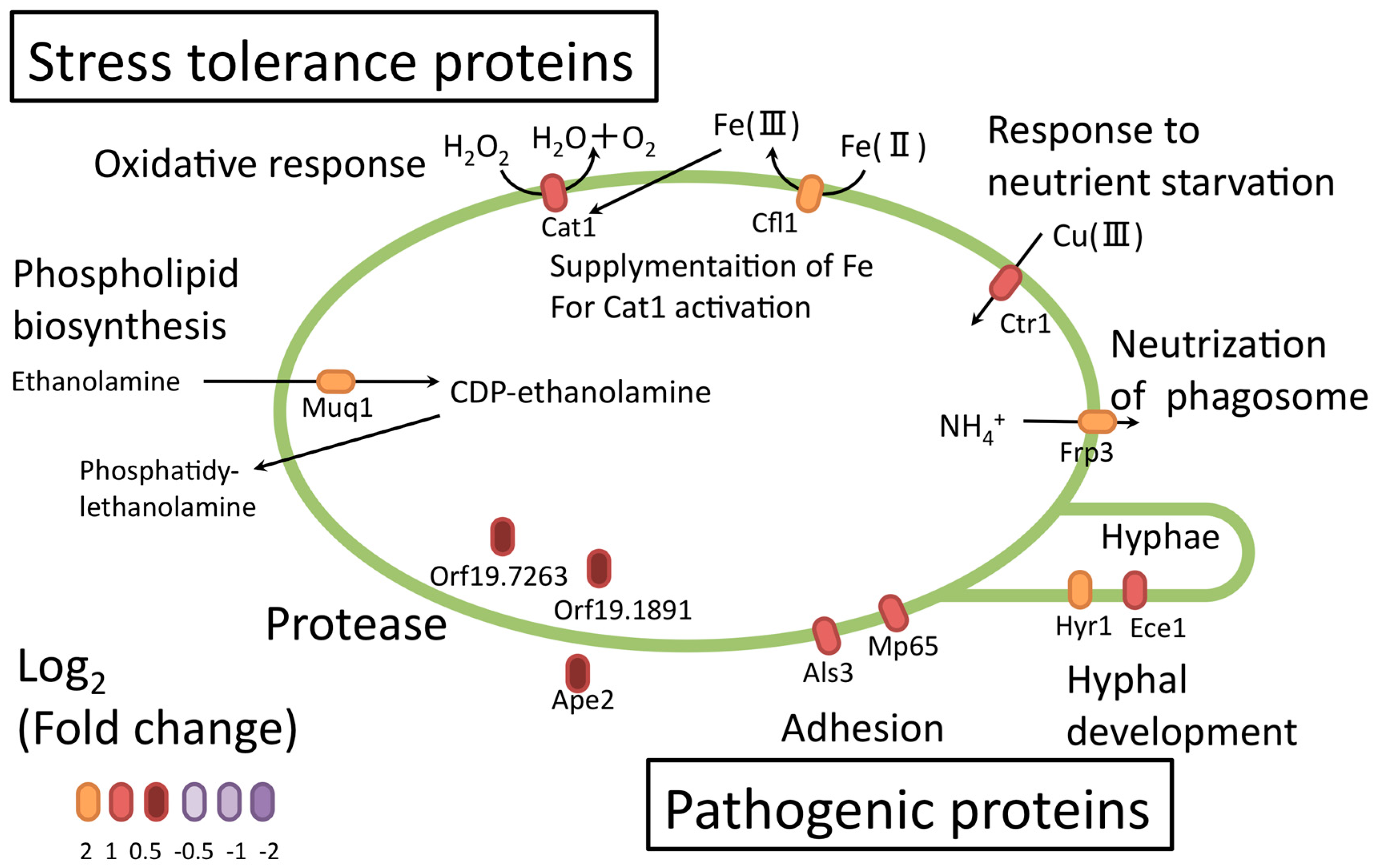
3. Quantitative Time-Course Proteomics Analysis of C. albicans Serum Adaptation
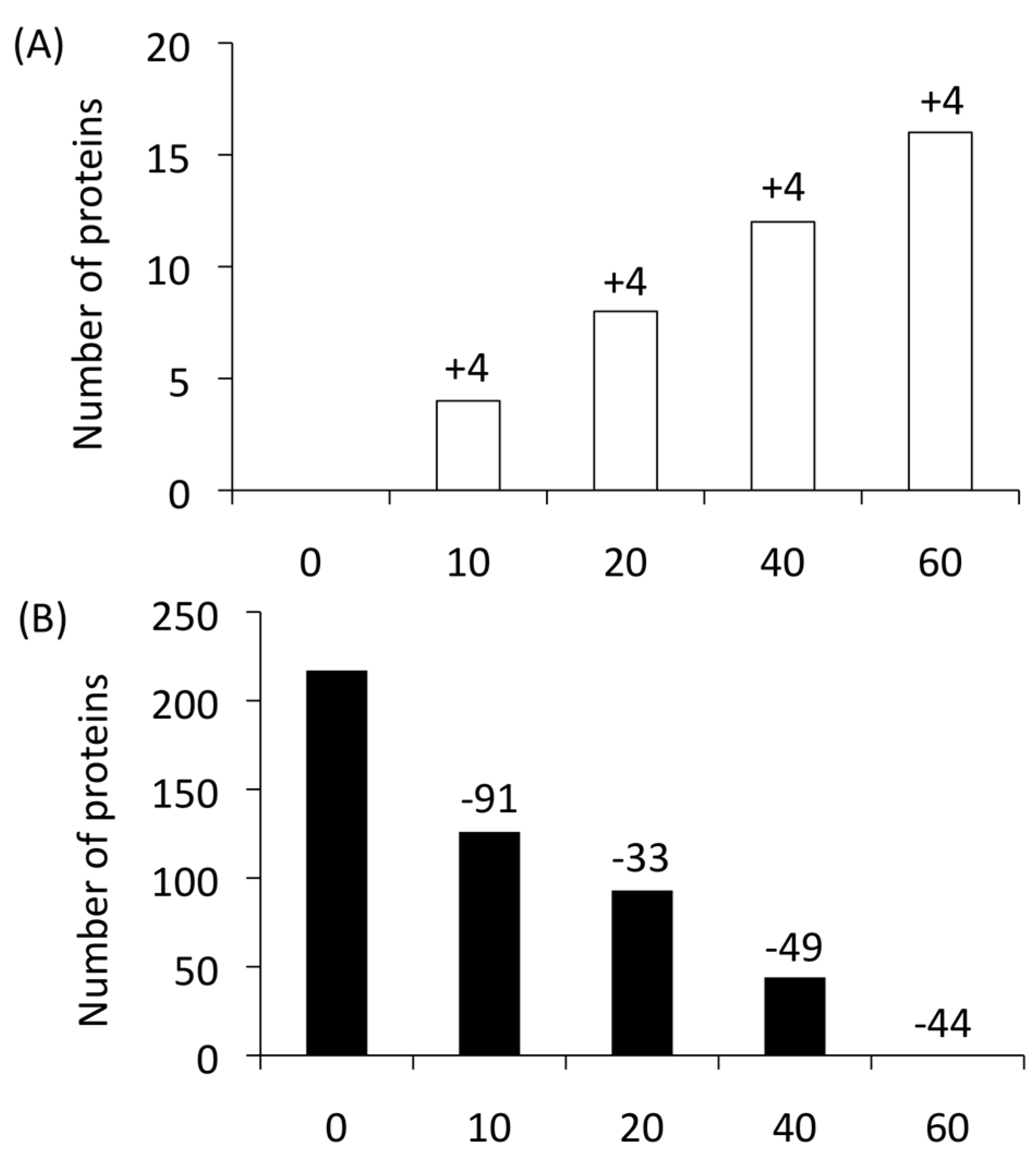
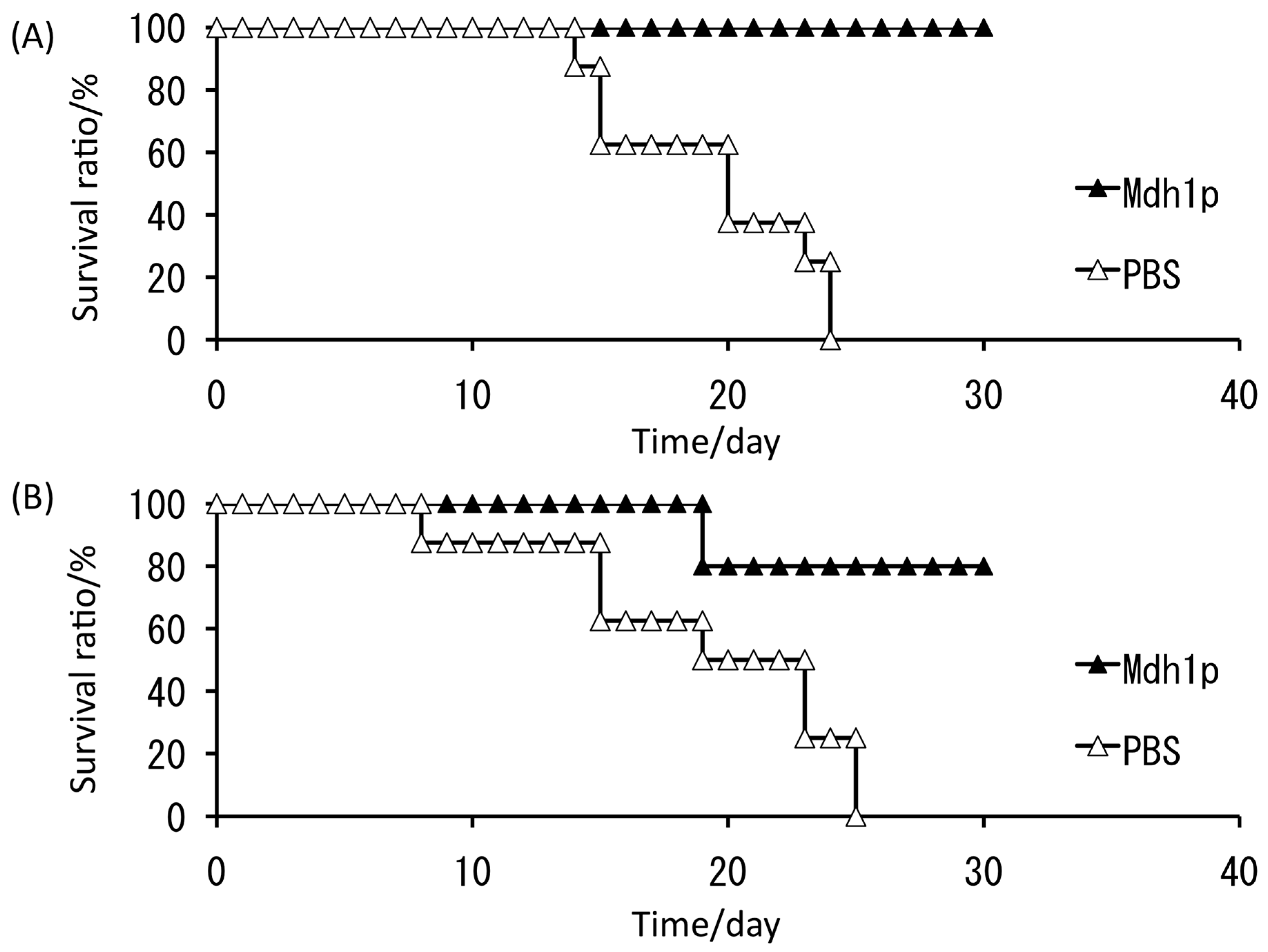
4. Finding an Antigen for a Potential Vaccine
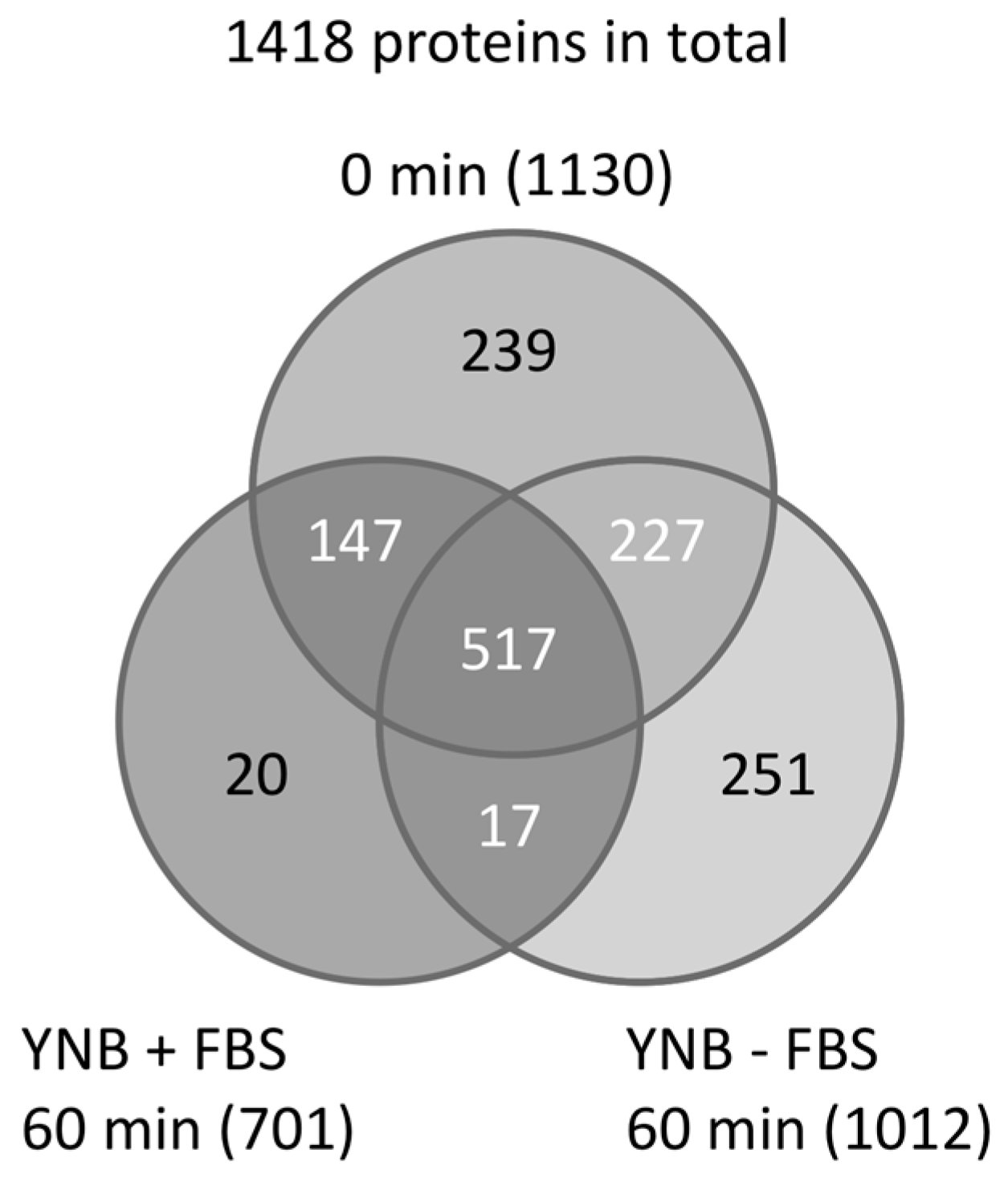
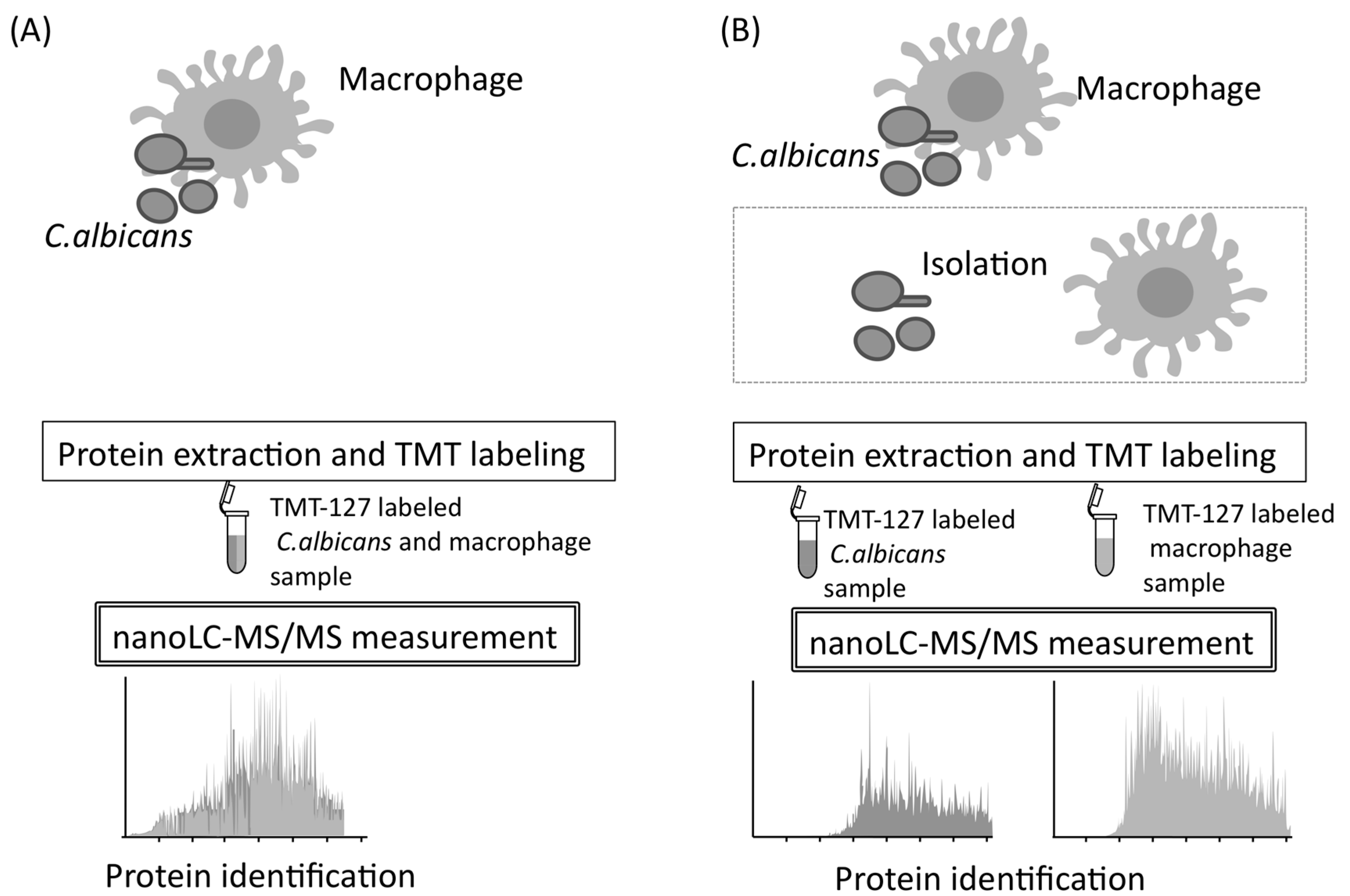
5. Mixed and Quantitative Proteome Analysis
6. Conclusions
Funding
Conflicts of Interest
References
- Verma, A.; Gaffen, S.L.; Swidergall, M. Innate immunity to mucosal Candida infections. J. Fungi (Basel) 2017, 3, 60. [Google Scholar] [CrossRef] [PubMed]
- da Silva Dantas, A.; Lee, K.K.; Raziunaite, I.; Schaefer, K.; Wagener, J.; Yadav, B.; Gow, N.A. Cell biology of Candida albicans-host interactions. Curr. Opin. Microbiol. 2016, 34, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Shibasaki, S.; Karasaki, M.; Ueda, M. Combining proteomic strategies and molecular display technology for development of vaccines against Candida albicans. J. Proteom. Bioinform. 2014, 7, 134–138. [Google Scholar] [CrossRef]
- Del Bono, V.; Giacobbe, D.R. Bloodstream infections in internal medicine. Virulence 2016, 7, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Monteoliva, L.; Martinez-Lopez, R.; Pitarch, A.; Hernaez, M.L.; Serna, A.; Nombela, C.; Albar, J.P.; Gil, C. Quantitative proteome and acidic subproteome profiling of Candida albicans yeast-to-hypha transition. J. Proteome Res. 2011, 10, 502–517. [Google Scholar] [CrossRef] [PubMed]
- Hoehamer, C.F.; Cummings, E.D.; Hilliard, G.M.; Rogers, P.D. Changes in the proteome of Candida albicans in response to azole, polyene, and echinocandin antifungal agents. Antimicrob. Agents Chemother. 2010, 54, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Martínez, J.P.; Blanes, R.; Casanova, M.; Valentín, E.; Murgui, A.; Domínguez, Á. Null mutants of Candida albicans for cell-wall-related genes form fragile biofilms that display an almost identical extracellular matrix proteome. FEMS Yeast Res. 2016, 16, fow075. [Google Scholar] [CrossRef] [PubMed]
- Morisaka, H.; Kirino, A.; Kobayashi, K.; Ueda, M. Two-dimensional protein separation by the HPLC system with a monolithic column. Biosci. Biotechnol. Biochem. 2012, 76, 585–588. [Google Scholar] [CrossRef] [PubMed]
- Motokawa, M.; Kobayashi, H.; Ishizuka, N.; Minakuchi, H.; Nakanishi, K.; Jinnai, H.; Hosoya, K.; Ikegami, T.; Tanaka, N. Monolithic silica columns with various skeleton sizes and through-pore sizes for capillary liquid chromatography. J. Chromatogr. A 2002, 961, 53–63. [Google Scholar] [CrossRef]
- Shibasaki, S.; Karasaki, M.; Aburaya, S.; Morisaka, H.; Takeda, Y.; Aoki, W.; Kitano, S.; Kitano, M.; Ueda, M.; Sano, H.; et al. A comparative proteomics study of a synovial cell line stimulated with TNF-α. FEBS Open Bio 2016, 6, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Buu, L.M.; Chen, Y.C. Sap6, a secreted aspartyl proteinase, participates in maintenance the cell surface integrity of Candida albicans. J. Biomed. Sci. 2013, 30, 101. [Google Scholar] [CrossRef] [PubMed]
- Staniszewska, M.; Bondaryk, M.; Malewski, T.; Kurzatkowski, W. Quantitative expression of Candida albicans aspartyl proteinase genes SAP7, SAP8, SAP9, SAP10 in human serum in vitro. Pol. J. Microbiol. 2014, 63, 15–20. [Google Scholar] [PubMed]
- Aoki, W.; Kitahara, N.; Miura, N.; Morisaka, H.; Yamamoto, Y.; Kuroda, K.; Ueda, M. Comprehensive characterization of secreted aspartic proteases encoded by a virulence gene family in Candida albicans. J. Biochem. 2011, 150, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Wang, T.; Yan, Y.; Shi, G.; Cheng, H.; Wu, D.; Wang, C. Matrine reduces yeast-to-hypha transition and resistance of a fluconazole-resistant strain of Candida albicans. J. Appl. Microbiol. 2014, 117, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Shareck, J.; Belhumeur, P. Modulation of morphogenesis in Candida albicans by various small molecules. Eukaryot. Cell 2011, 10, 1004–1012. [Google Scholar] [CrossRef] [PubMed]
- Aoki, W.; Kitahara, N.; Miura, N.; Morisaka, H.; Kuroda, K.; Ueda, M. Profiling of adhesive properties of the agglutinin-like sequence (ALS) protein family, a virulent attribute of Candida albicans. FEMS Immunol. Med. Microbiol. 2012, 65, 121–124. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Sherman, M.C.; Lysak, N.; Filonenko, A.; Richards, H.; Sobonya, R.E.; Klotz, S.A.; Lipke, P.N. Peptide detection of fungal functional amyloids in infected tissue. PLoS ONE 2014, 9, e86067. [Google Scholar] [CrossRef] [PubMed]
- Hoyer, L.L.; Cota, E. Candida albicans agglutinin-like sequence (Als) family vignettes: A review of Als protein structure and function. Front. Microbiol. 2016, 15, 280. [Google Scholar] [CrossRef] [PubMed]
- Aoki, W.; Ueda, T.; Tatsukami, Y.; Kitahara, N.; Morisaka, H.; Kuroda, K.; Ueda, M. Time-course proteomic profile of Candida albicans during adaptation to a fetal serum. Pathog. Dis. 2013, 67, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.L.; Lee, R.T.; Fang, H.M.; Wang, Y.M.; Li, R.; Zou, H.; Zhu, Y.; Wang, Y. Bacterial peptidoglycan triggers Candida albicans hyphal growth by directly activating the adenylyl cyclase Cyr1p. Cell Host Microbe 2008, 4, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Dalle, F.; Wächtler, B.; L’Ollivier, C.; Holland, G.; Bannert, N.; Wilson, D.; Labruère, C.; Bonnin, A.; Hube, B. Cellular interactions of Candida albicans with human oral epithelial cells and enterocytes. Cell. Microbiol. 2010, 12, 248–271. [Google Scholar] [CrossRef] [PubMed]
- Morisaka, H.; Matsui, K.; Tatsukami, Y.; Kuroda, K.; Miyake, H.; Tamaru, Y.; Ueda, M. Profile of native cellulosomal proteins of Clostridium cellulovorans adapted to various carbon sources. AMB Express 2012, 2, 37. [Google Scholar] [CrossRef] [PubMed]
- Desai, P.R.; Thakur, A.; Ganguli, D.; Paul, S.; Morschhäuser, J.; Bachhawat, A.K. Glutathione utilization by Candida albicans requires a functional glutathione degradation (DUG) pathway and OPT7, an unusual member of the oligopeptide transporter family. J. Biol Chem. 2011, 286, 41183–41194. [Google Scholar] [CrossRef] [PubMed]
- Davis, D.A.; Bruno, V.M.; Loza, L.; Filler, S.G.; Mitchell, A.P. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics 2002, 162, 1573–1581. [Google Scholar] [PubMed]
- Aoki, W.; Tatsukami, Y.; Kitahara, N.; Matsui, K.; Morisaka, H.; Kuroda, K.; Ueda, M. Elucidation of potentially virulent factors of Candida albicans during serum adaptation by using quantitative time-course proteomics. J. Proteom. 2013, 91, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, M.B.; Costanzo, M.C.; Skrzypek, M.S.; Binkley, G.; Lane, C.; Miyasato, S.R.; Sherlock, G. The Candida Genome Database (CGD), a community resource for Candida albicans gene and protein information. Nucleic Acids Res. 2005, 33, 358–363. [Google Scholar] [CrossRef] [PubMed]
- de Hoon, M.J.; Imoto, S.; Nolan, J.; Miyano, S. Open source clustering software. Bioinformatics 2004, 20, 1453–1454. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007, 8, R183. [Google Scholar] [CrossRef] [PubMed]
- Cabezón, V.; Llama-Palacios, A.; Nombela, C.; Monteoliva, L.; Gil, C. Analysis of Candida albicans plasma membrane proteome. Proteomics 2009, 9, 4770–4786. [Google Scholar] [CrossRef] [PubMed]
- Fradin, C.; De Groot, P.; MacCallum, D.; Schaller, M.; Klis, F.; Odds, F.C.; Hube, B. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol. Microbiol. 2005, 56, 397–415. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, A.; Grant, C.M. Oxidant-specific regulation of protein synthesis in Candida albicans. Fungal Genet. Biol. 2014, 67, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Kasuga, T.; Sachs, M.S.; Glass, N.L. Transcriptional profiling of cross pathway control in Neurospora crassa and comparative analysis of the Gcn4 and CPC1 regulons. Eukaryot. Cell 2007, 6, 1018–1029. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, A.; Grant, C.M. A single inhibitory upstream open reading frame (uORF) is sufficient to regulate Candida albicans GCN4 translation in response to amino acid starvation conditions. RNA 2014, 20, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Han, T.L.; Cannon, R.D.; Villas-Bôas, S.G. The metabolic basis of Candida albicans morphogenesis and quorum sensing. Fungal Genet. Biol. 2011, 48, 747–763. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Stead, D.; Walker, J.; Selway, L.; Smith, D.A.; Brown, A.J.; Quinn, J. A proteomic analysis of the salt, cadmium and peroxide stress responses in Candida albicans and the role of the Hog1 stress-activated MAPK in regulating the stress-induced proteome. Proteomics 2009, 9, 4686–4703. [Google Scholar] [CrossRef] [PubMed]
- Sandai, D.; Yin, Z.; Selway, L.; Stead, D.; Walker, J.; Leach, M.D.; Bohovych, I.; Ene, I.V.; Kastora, S.; Budge, S.; et al. The evolutionary rewiring of ubiquitination targets has reprogrammed the regulation of carbon assimilation in the pathogenic yeast Candida albicans. mBio 2012, 3, e00495-12. [Google Scholar] [CrossRef] [PubMed]
- Edfors, F.; Danielsson, F.; Hallström, BM.; Käll, L.; Lundberg, E.; Pontén, F.; Forsström, B.; Uhlén, M. Gene-specific correlation of RNA and protein levels in human cells and tissues. Mol. Syst. Biol. 2016, 12, 883. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.M.; Kauffman, S.J.; Hauser, M.; Huang, L.; Lin, M.; Sillaots, S.; Jiang, B.; Xu, D.; Roemer, T. Pathway analysis of Candida albicans survival and virulence determinants in a murine infection model. Proc. Natl. Acad. Sci. USA 2010, 107, 22044–22049. [Google Scholar] [CrossRef] [PubMed]
- De Bernardis, F.; Mühlschlegel, F.A.; Cassone, A.; Fonzi, W.A. The pH of the host niche controls gene expression in and virulence of Candida albicans. Infect. Immun. 1998, 66, 3317–3325. [Google Scholar] [PubMed]
- Yamada-Okabe, T.; Yamada-Okabe, H. Characterization of the CaNAG3, CaNAG4, and CaNAG6 genes of the pathogenic fungus Candida albicans: Possible involvement of these genes in the susceptibilities of cytotoxic agents. FEMS Microbiol. Lett. 2002, 212, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Huh, W.K.; Kim, S.T.; Kim, H.; Jeong, G.; Kang, S.O. Deficiency of D-erythroascorbic acid attenuates hyphal growth and virulence of Candida albicans. Infect. Immun. 2001, 69, 3939–3946. [Google Scholar] [CrossRef] [PubMed]
- Martchenko, M.; Alarco, A.M.; Harcus, D.; Whiteway, M. Superoxide dismutases in Candida albicans: Transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene. Mol. Biol. Cell 2004, 15, 456–467. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, R.; Kodgire, S.; Santhakumari, B.; Patil, R.; Kulkarni, M.; Zore, G. Serum responsive proteome reveals correlation between oxidative phosphorylation and morphogenesis in Candida albicans ATCC10231. J. Proteom. 2018, 185, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Heilmann, C.J.; Sorgo, A.G.; Siliakus, A.R.; Dekker, H.L.; Brul, S.; de Koster, C.G.; de Koning, L.J.; Klis, F.M. Hyphal induction in the human fungal pathogen Candida albicans reveals a characteristic wall protein profile. Microbiology 2011, 157, 2297–2307. [Google Scholar] [CrossRef] [PubMed]
- Tylicki, A.; Ziolkowska, G.; Bolkun, A.; Siemieniuk, M.; Czerniecki, J.; Nowakiewicz, A. Comparative study of the activity and kinetic properties of malate dehydrogenase and pyruvate decarboxylase from Candida albicans, Malassezia pachydermatis, and Saccharomyces cerevisiae. Can. J. Microbiol. 2008, 54, 734–741. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Arenas, E.; Molero, G.; Nombela, C.; Diez-Orejas, R.; Gil, C. Low virulent strains of Candida, albicans: Unravelling the antigens for a future vaccine. Proteomics 2004, 4, 3007–3020. [Google Scholar] [CrossRef] [PubMed]
- Shibasaki, S.; Aoki, W.; Nomura, T.; Karasaki, M.; Sewaki, T.; Ueda, M. Evaluation of Mdh1 protein as an antigenic candidate for a vaccine against candidiasis. Biocontrol Sci. 2014, 19, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Orsi, C.F.; Borghi, E.; Colombari, B.; Neglia, R.G.; Quaglino, D.; Ardizzoni, A.; Morace, G.; Blasi, E. Impact of Candida albicans hyphal wall protein 1 (HWP1) genotype on biofilm production and fungal susceptibility to microglial cells. Microb. Pathog. 2014, 69–70, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Garcia, A.; Gallot, N.; Abad, A.; Mendoza, L.; Rementeria, A.; Hernando, F.L. Molecular fractionation and characterization of a Candida albicans fraction that increases tumor cell adhesion to hepatic endothelium. Appl. Microbiol. Biotechnol. 2011, 92, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Cutler, J.E. Vaccine and monoclonal antibody that enhance mouse resistance to candidiasis. Clin. Vaccine Immunol. 2011, 18, 1656–1667. [Google Scholar] [CrossRef] [PubMed]
- Shibasaki, S.; Ueda, M. Oral vaccine development by molecular display methods using microbial cells. Methods Mol. Biol. 2016, 1404, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Shibasaki, S.; Ueda, M. Preparation of an oral vaccine by proteome analysis and molecular display technology. Methods Mol. Biol. 2017, 1625, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Shibasaki, S.; Aoki, W.; Nomura, T.; Miyoshi, A.; Tafuku, S.; Sewaki, T.; Ueda, M. An oral vaccine against candidiasis generated by a yeast molecular display system. Pathog. Dis. 2013, 69, 262–628. [Google Scholar] [CrossRef] [PubMed]
- Shibasaki, S.; Karasaki, M.; Tafuku, S.; Aoki, W.; Sewaki, T.; Ueda, M. Oral immunization against candidiasis using lactobacillus casei displaying enolase 1 from Candida albicans. Sci. Pharm. 2014, 82, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Miramon, P.; Kasper, L.; Hube, B. Thriving within the host: Candida spp. interactions with phagocytic cells. Med. Microbiol. Immunol. 2013, 202, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Galli, S.J.; Borregaard, N.; Wynn, T.A. Phenotypic and functional plasticity of, cells of innate immunity: Macrophages, mast cells and neutrophils. Nat. Immunol. 2011, 12, 1035–1044. [Google Scholar] [CrossRef] [PubMed]
- Martin-Manso, G.; Navarathna, D.H.; Galli, S.; Soto-Pantoja, D.R.; Kuznetsova, S.A.; Tsokos, M.; Roberts, D.D. Endogenous thrombospondin-1 regulates leukocyte recruitment and activation and accelerates death from systemic candidiasis. PLoS ONE 2012, 7, e48775. [Google Scholar] [CrossRef] [PubMed]
- Seider, K.; Heyken, A.; Luttich, A.; Miramon, P.; Hube, B. Interaction of pathogenic, yeasts with phagocytes: Survival, persistence and escape. Curr. Opin. Microbiol. 2010, 13, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Brunke, S.; Seider, K.; Fischer, D.; Jacobsen, I.D.; Kasper, L.; Jablonowski, N.; Wartenberg, A.; Bader, O.; Enache-Angoulvant, A.; Schaller, M.; et al. One small step for a yeast—Microevolution within macrophages renders Candida glabrata hypervirulent due to a single point mutation. PLoS Pathog. 2014, 10, e1004478. [Google Scholar] [CrossRef] [PubMed]
- Kitahara, N.; Morisaka, H.; Aoki, W.; Takeda, Y.; Shibasaki, S.; Kuroda, K.; Ueda, M. Description of the interaction between Candida albicans and macrophages by mixed and quantitative proteome analysis without isolation. AMB Express 2015, 5, 127. [Google Scholar] [CrossRef] [PubMed]
- Geraldino, T.H.; de Vito, E.; Custódio, L.A.; Conchon-Costa, I.; Gaziri, L.C.; Felipe, I.; Loyola, W.; Bonifácio, K.L. Increased tumour necrosis factor-alpha production, higher mannose receptor activity and ability to kill Candida by concanavalin-A-activated macrophages. FEMS Immunol. Med. Microbiol. 2010, 59, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Netea, M.G.; van Tits, L.J.; Curfs, J.H.; Amiot, F.; Meis, J.F.; van der Meer, J.W.; Kullberg, B.J. Increased susceptibility of TNF-alpha lymphotoxin-alpha double knockout mice to systemic candidiasis through impaired recruitment of neutrophils and phagocytosis of Candida albicans. J. Immunol. 1999, 163, 1498–1505. [Google Scholar] [PubMed]
- Phan, Q.T.; Myers, C.L.; Fu, Y.; Sheppard, D.C.; Yeaman, M.R.; Welch, W.H.; Ibrahim, A.S.; Edwards, J.E., Jr.; Filler, S.G. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 2007, 5, e64. [Google Scholar] [CrossRef] [PubMed]
- Cleary, I.A.; Reinhard, S.M.; Miller, C.L.; Murdoch, C.; Thornhill, M.H.; Lazzell, A.L.; Monteagudo, C.; Thomas, D.P.; Saville, S.P. Candida albicans adhesin Als3p is dispensable for virulence in the mouse model of disseminated candidiasis. Microbiology 2011, 157, 1806–1815. [Google Scholar] [CrossRef] [PubMed]
- Staniszewska, M.; Bondaryk, M.; Żukowski, K.; Chudy, M. Quantification of the APE2 gene expression level in Candida albicans clinical isolates from patients with diagnosed fungal infections. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1429–1435. [Google Scholar] [CrossRef] [PubMed]
- Reales-Calderon, J.A.; Sylvester, M.; Strijbis, K.; Jensen, O.N.; Nombela, C.; Molero, G.; Gil, C. Candida albicans induces pro-inflammatory and anti-apoptotic signals in macrophages as revealed by quantitative proteomics and phosphoproteomics. J. Proteom. 2013, 91, 106–135. [Google Scholar] [CrossRef] [PubMed]
- Ibata-Ombetta, S.; Idziorek, T.; Trinel, PA.; Poulain, D.; Jouault, T. Candida albicans phospholipomannan promotes survival of phagocytosed yeasts through modulation of bad phosphorylation and macrophage apoptosis. J. Biol. Chem. 2003, 278, 13086–13093. [Google Scholar] [CrossRef] [PubMed]
- Gasparoto, T.H.; Gaziri, L.C.; Burger, E.; de Almeida, R.S.; Felipe, I. Apoptosis of phagocytic cells induced by Candida albicans and production of IL-10. FEMS Immunol. Med. Microbiol. 2004, 42, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Heidler, J.; Al-Furoukh, N.; Kukat, C.; Salwig, I.; Ingelmann, M.E.; Seibel, P.; Krüger, M.; Holtz, J.; Wittig, I.; Braun, T.; et al. Nitric oxide-associated protein 1 (NOA1) is necessary for oxygen-depend-ent regulation of mitochondrial respiratory complexes. J. Biol. Chem. 2011, 286, 32086–32093. [Google Scholar] [CrossRef] [PubMed]
- Sasi, B.K.; Sonawane, P.J.; Gupta, V.; Sahu, B.S.; Mahapatra, N.R. Coordinated transcriptional regulation of Hspa1a gene by multiple transcription factors: Crucial roles for HSF-1, NF-Y, NF-kappaB, and CREB. J. Mol. Biol. 2014, 426, 116–135. [Google Scholar] [CrossRef] [PubMed]
- Axsen, W.S.; Styer, C.M.; Solnick, JV. Inhibition of heat shock protein expression by Helicobacter pylori. Microb. Pathog. 2009, 47, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Aoki, W.; Kitahara, N.; Fujita, A.; Shibasaki, S.; Morisaka, H.; Kuroda, K.; Ueda, M. Detection of Candida albicans by using a designed fluorescence-quenched peptide. J. Biosci Bioeng. 2013, 116, 573–575. [Google Scholar] [CrossRef] [PubMed]
- Aoki, W.; Kitahara, N.; Miura, N.; Morisaka, H.; Kuroda, K.; Ueda, M. Design of a novel antimicrobial peptide activated by virulent proteases. Chem. Biol. Drug Des. 2012, 80, 725–733. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shibasaki, S.; Karasaki, M.; Aoki, W.; Ueda, M. Molecular and Physiological Study of Candida albicans by Quantitative Proteome Analysis. Proteomes 2018, 6, 34. https://doi.org/10.3390/proteomes6030034
Shibasaki S, Karasaki M, Aoki W, Ueda M. Molecular and Physiological Study of Candida albicans by Quantitative Proteome Analysis. Proteomes. 2018; 6(3):34. https://doi.org/10.3390/proteomes6030034
Chicago/Turabian StyleShibasaki, Seiji, Miki Karasaki, Wataru Aoki, and Mitsuyoshi Ueda. 2018. "Molecular and Physiological Study of Candida albicans by Quantitative Proteome Analysis" Proteomes 6, no. 3: 34. https://doi.org/10.3390/proteomes6030034
APA StyleShibasaki, S., Karasaki, M., Aoki, W., & Ueda, M. (2018). Molecular and Physiological Study of Candida albicans by Quantitative Proteome Analysis. Proteomes, 6(3), 34. https://doi.org/10.3390/proteomes6030034




