Surface and Extracellular Proteome of the Emerging Pathogen Corynebacterium ulcerans
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Isolation of Extracellular Proteins Secreted into the Medium
2.3. In-Solution Tryptic Digest
2.4. NanoLC-MS/MS Analysis
2.5. Isolation of Cell Surface Proteins by Tryptic Shaving
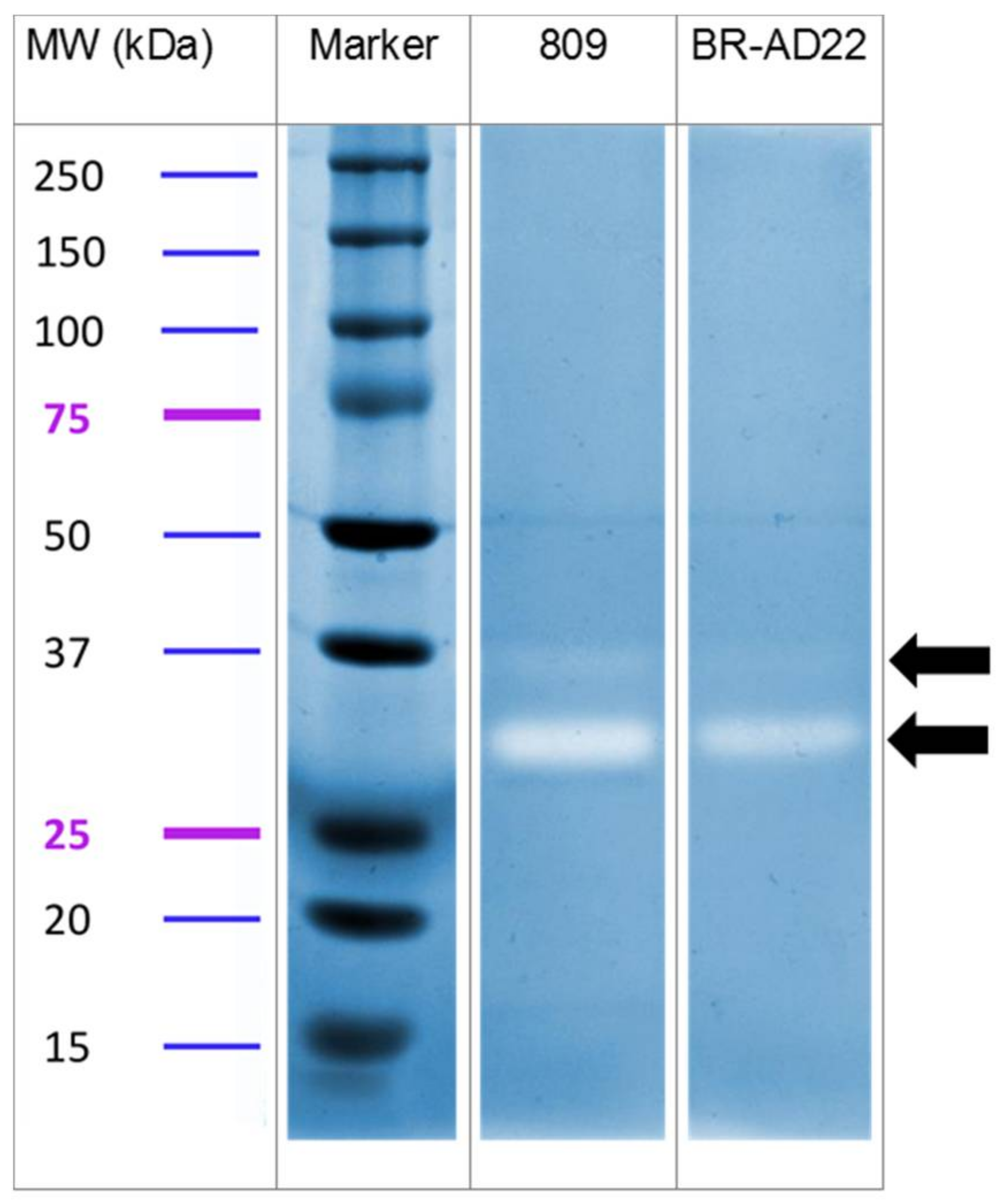
2.6. Zymography
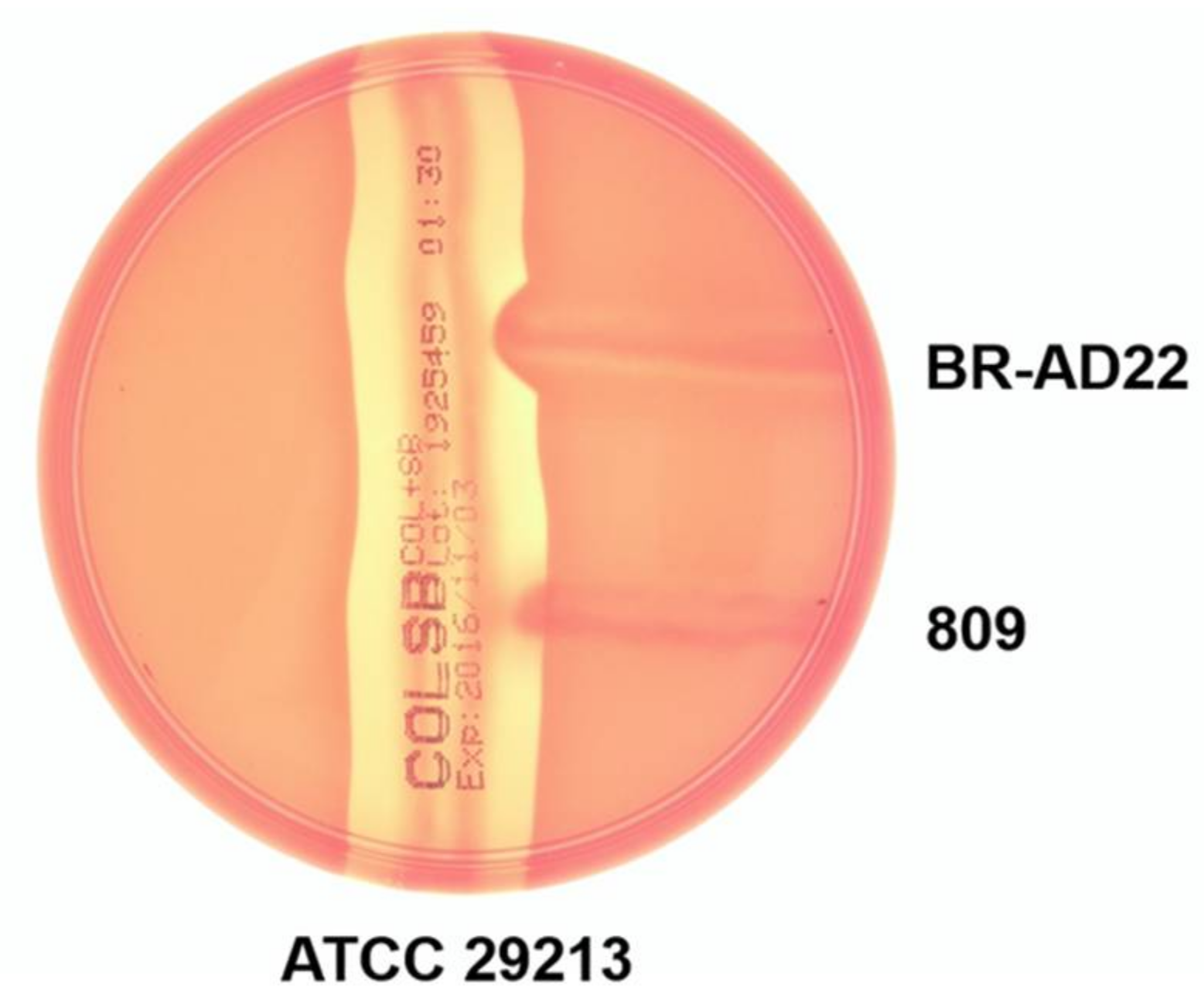
2.7. Inverse CAMP Test
3. Results
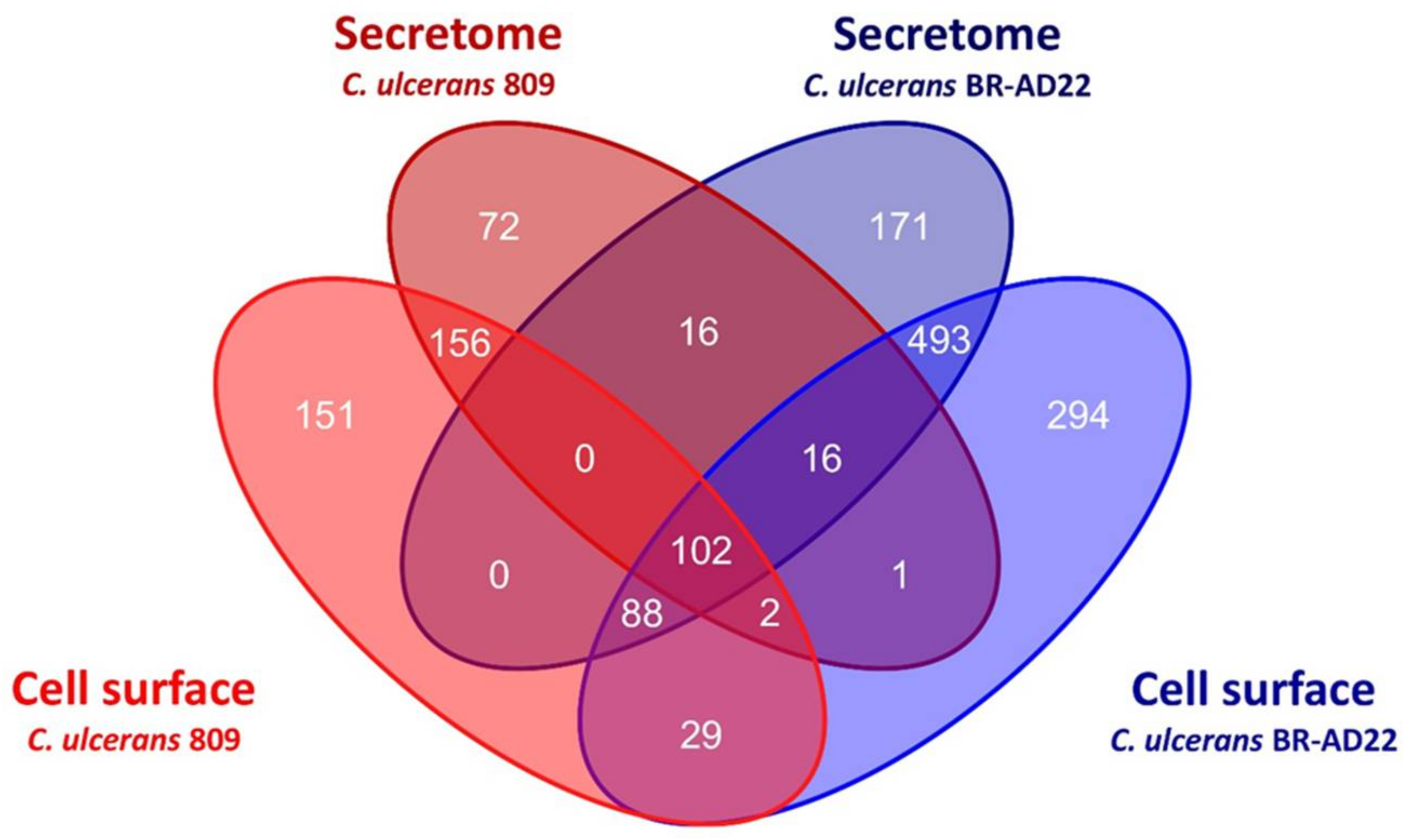
3.1. Analysis of Proteins in Extracellular and Surface Fraction
3.2. Identification of Putative Virulence Factors
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Tauch, A.; Sandbote, J. The family Corynebacteriaceae. In The Prokaryotes; Rosenberg, E., DeLong, E., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 239–277. [Google Scholar]
- Gilbert, R.; Stewart, F.C. Corynebacterium ulcerans: A pathogenic microorganism resembling Corynebacterium diphtheriae. J. Lab. Clin. Med. 1927, 12, 756–761. [Google Scholar]
- Sangal, V.; Hoskisson, P.A. Corynephages: Infections of the infectors. In Diphtheria and Its Etiological Agents; Burkovski, A., Ed.; Springer: Dordrecht, The Netherlands, 2014; pp. 67–82. [Google Scholar]
- Wagner, K.S.; White, J.M.; Crowcroft, N.S.; De Martin, S.; Mann, G.; Efstratiou, A. Diphtheria in the United Kingdom, 1986-2008: The increasing role of Corynebacterium ulcerans. Epidemiol. Infect. 2010, 138, 1519–1530. [Google Scholar] [CrossRef] [PubMed]
- Dias, A.A.; Santos, L.S.; Sabbadini, P.S.; Santos, C.S.; Silva, F.C., Jr.; Napoleão, F.; Nagao, P.E.; Villas-Bôas, M.H.; Hirata, R., Jr.; Guaraldi, A.L. Corynebacterium ulcerans diphtheria: An emerging zoonosis in Brazil and worldwide. Rev. Saude Publica 2011, 45, 1176–1191. [Google Scholar] [CrossRef] [PubMed]
- Burkovski, A. Pathogenesis of Corynebacterium diphtheriae and Corynebacterium ulcerans. In Human Emerging and Re-Emerging Infections; Singh, S.K., Ed.; John Wiley & Sons/Wiley Blackwell Press: Hoboken, NJ, USA, 2016; Volume 2, pp. 697–708. [Google Scholar]
- Hacker, E.; Azevedo Antunes, C.; Mattos-Guaraldi, A.L.; Burkovski, A.; Tauch, A. Corynebacterium ulcerans—An emerging human pathogen. Future Microbiol. 2016, 11, 1191–1208. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.; Teutsch, B.; Heinzinger, S.; Sing, A. Corynebacterium ulcerans—Ein Emerging Pathogen? Daten des Konsiliarlabors für Diphtherie 2011–2016. Epid. Bull. 2018, 8, 83–86. [Google Scholar]
- Zakikhany, K.; Efstratiou, A. Diphtheria in Europe: Current problems and new challenges. Future Microbiol. 2012, 7, 595–607. [Google Scholar] [CrossRef] [PubMed]
- Meinel, D.M.; Margos, G.; Konrad, R.; Krebs, S.; Blum, H.; Sing, A. Next generation sequencing analysis of nine Corynebacterium ulcerans isolates reveals zoonotic transmission and a novel putative diphtheria toxin-encoding pathogenicity island. Genome Med. 2014, 6, 113. [Google Scholar] [CrossRef] [PubMed]
- Meinel, D.M.; Konrad, R.; Berger, A.; König, C.; Schmidt-Wieland, T.; Hogardt, M.; Bischoff, H.; Ackermann, N.; Hörmansdorfer, S.; Krebs, S.; et al. Zoonotic transmission of toxigenic Corynebacterium ulcerans strain, Germany, 2012. Emerg. Infect. Dis. 2015, 21, 356–358. [Google Scholar] [CrossRef] [PubMed]
- Trost, E.; Al-Dilaimi, A.; Papavasiliou, P.; Schneider, J.; Viehoever, P.; Burkovski, A.; de Castro Soares, S.; Silva Almeida, S.; Alves Dorella, F.; Miyoshi, A.; et al. Comparative analysis of two complete Corynebacterium ulcerans genomes and detection of candidate virulence factors. BMC Genom. 2011, 12, 383. [Google Scholar] [CrossRef] [PubMed]
- Dias, A.A.; Silva, F.C.; Pereira, G.A.; Souza, M.C.; Camello, T.C.; Damasceno, J.A.; Pacheco, L.G.; Miyoshi, A.; Azevedo, V.A.; Hirata, R., Jr.; et al. Corynebacterium ulcerans isolated from an asymptomatic dog kept in an animal shelter in the metropolitan area of Rio de Janeiro, Brazil. Vector Borne Zoonotic Dis. 2010, 10, 743–748. [Google Scholar] [CrossRef] [PubMed]
- Mattos-Guaraldi, A.L.; Sampaio, J.L.; Santos, C.S.; Pimenta, F.P.; Pereira, G.A.; Pacheco, L.G.; Miyoshi, A.; Azevedo, V.; Moreira, L.O.; Gutierrez, F.L.; et al. First detection of Corynebacterium ulcerans producing a diphtheria-like toxin in a case of human with pulmonary infection in the Rio de Janeiro metropolitan area, Brazil. Mem. Inst. Oswaldo Cruz 2008, 103, 396–400. [Google Scholar] [CrossRef] [PubMed]
- Dias, A.A.; Silva, F.C., Jr.; Santos, L.S.; Ribeiro-Carvalho, M.M.; Sabbadini, P.S.; Santos, C.S.; Filardy, A.A.; Miyoshi, A.; Azevedo, V.A.; Hirata, R., Jr.; et al. Strain-dependent arthritogenic potential of the zoonotic pathogen Corynebacterium ulcerans. Vet. Microbiol. 2011, 153, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Hacker, E.; Ott, L.; Hasselt, K.; Mattos-Guaraldi, A.L.; Tauch, A.; Burkovski, A. Colonization of human epithelial cell lines by Corynebacterium ulcerans from human and animal sources. Microbiology 2015, 161, 1582–1591. [Google Scholar] [CrossRef] [PubMed]
- Hacker, E.; Ott, L.; Schulze-Luehrmann, J.; Lührmann, A.; Wiesmann, V.; Wittenberg, T.; Burkovski, A. The killing of macrophages by Corynebacterium ulcerans. Virulence 2016, 7, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Simpson-Louredo, L.; Ramos, J.N.; Peixoto, R.S.; Santos, L.S.; Antunes, C.A.; Ladeira, E.M.; Santos, C.S.; Vieira, V.V.; Boas, M.H.; Hirata, R., Jr.; et al. Corynebacterium ulcerans isolates from humans and dogs: Fibrinogen, fibronectin and collagen-binding, antimicrobial and PFGE profiles. Anton. Leeuwenhoek 2014, 105, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Hansmeier, N.; Chao, T.C.; Kalinowski, J.; Pühler, A.; Tauch, A. Mapping and comprehensive analysis of the extracellular and cell surface proteome of the human pathogen Corynebacterium diphtheriae. Proteomics 2006, 6, 2465–2476. [Google Scholar] [CrossRef] [PubMed]
- Hansmeier, N.; Chao, T.C.; Daschkey, S.; Müsken, M.; Kalinowski, J.; Pühler, A.; Tauch, A. A comprehensive proteome map of the lipid-requiring nosocomial pathogen Corynebacterium jeikeium K411. Proteomics 2007, 7, 1076–1096. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, L.G.; Slade, S.E.; Seyffert, N.; Santos, A.R.; Castro, T.L.; Silva, W.M.; Santos, A.V.; Santos, S.G.; Farias, L.M.; Carvalho, M.A.; et al. A combined approach for comparative exoproteome analysis of Corynebacterium pseudotuberculosis. BMC Microbiol. 2011, 11, 12. [Google Scholar] [CrossRef] [PubMed]
- Poetsch, A.; Haußmann, U.; Burkovski, A. Proteomics of corynebacteria: From biotechnology workhorses to pathogens. Proteomics 2011, 11, 3244–3255. [Google Scholar] [CrossRef] [PubMed]
- Rees, M.A.; Kleifeld, O.; Crellin, P.K.; Ho, B.; Stinear, T.P.; Smith, A.I.; Coppel, R.L. Proteomic characterization of a natural host-pathogen interaction: Repertoire of in vivo expressed bacterial and host surface-associated proteins. J. Proteome Res. 2015, 14, 120–132. [Google Scholar] [CrossRef] [PubMed]
- Silva, W.M.; Seyffert, N.; Ciprandi, A.; Santos, A.V.; Castro, T.L.; Pacheco, L.G.; Barh, D.; Le Loir, Y.; Pimenta, A.M.; Miyoshi, A.; et al. Differential exoproteome analysis of two Corynebacterium pseudotuberculosis biovar ovis strains isolated from goat (1002) and sheep (C231). Curr. Microbiol. 2013, 67, 460–465. [Google Scholar] [CrossRef] [PubMed]
- Silva, W.M.; Seyffert, N.; Santos, A.V.; Castro, T.L.; Pacheco, L.G.; Santos, A.R.; Ciprandi, A.; Dorella, F.A.; Andrade, H.M.; Barh, D.; et al. Identification of 11 new exoproteins in Corynebacterium pseudotuberculosis by comparative analysis of the exoproteome. Microb. Pathog. 2013, 61–62, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Silva, W.M.; Carvalho, R.D.; Soares, S.C.; Bastos, I.F.; Folador, E.L.; Souza, G.H.; Le Loir, Y.; Miyoshi, A.; Silva, A.; Azevedo, V. Label-free proteomic analysis to confirm the predicted proteome of Corynebacterium pseudotuberculosis under nitrosative stress mediated by nitric oxide. BMC Genom. 2014, 15, 1065. [Google Scholar] [CrossRef] [PubMed]
- Hiery, E.; Poetsch, A.; Moosbauer, T.; Amin, B.; Hofmann, J.; Burkovski, A. A proteomic study of Clavibacter michiganensis subsp. michiganensis culture supernatants. Proteomes 2015, 3, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Tjalsma, H.; Lambooy, L.; Hermans, P.W.; Swinkels, D.W. Shedding & shaving: Disclosure of proteomic expressions on a bacterial face. Proteomics 2008, 8, 1415–1428. [Google Scholar] [PubMed]
- Löwer, M.; Weydig, C.; Metzler, D.; Reuter, A.; Starzinski-Powitz, A.; Wessler, S.; Schneider, G. Prediction of extracellular proteases of the human pathogen Helicobacter pylori reveals proteolytic activity of the Hp1018/19 protein HtrA. PLoS ONE 2008, 3, e3510. [Google Scholar]
- Suvajdzic, L.; Potkonjak, A.; Milanov, D.; Lako, B.; Kocic, B.; Milic, N.; Cabarkapa, I. A proposal of a diagnostic protocol for isolation of Corynebacterium ulcerans from cow’s milk. Acta Sci. Vet. 2012, 40, 1039. [Google Scholar]
- Burkovski, A. Cell envelope of corynebacteria: Structure and influence on pathogenicity. ISRN Microbiol. 2013, 935736. [Google Scholar] [CrossRef] [PubMed]
- Tauch, A.; Burkovski, A. Molecular armory or niche factors: Virulence determinants of Corynebacterium species. FEMS Microbiol. Lett. 2015, 362, 1–6. [Google Scholar]
- Burkovski, A. The role of corynomycolic acids in Corynebacterium-host interaction. Anton. Leeuwenhoek 2018. [Google Scholar] [CrossRef] [PubMed]
- Fels, U.; Gevaert, K.; Van Damme, P. Proteogenomics in aid of host-pathogen interaction: A bacterial perspective. Proteomes 2017, 5, 26. [Google Scholar] [CrossRef] [PubMed]
| Accession No. | Description | Cellular Localisation |
|---|---|---|
| G0CTA8; embl-cds:AEG81534 | 50S ribosomal protein L35 | ribosome |
| G0CTV7; embl-cds:AEG81724 | ABC-type transport system involved in Fe-S cluster assembly ATP-binding protein | membrane |
| G0CU06; embl-cds:AEG81772 | Elongation factor EF-P | cytoplasm |
| G0CU14; embl-cds:AEG81780 | Putative secreted protein | |
| G0CU20; embl-cds:AEG80548 | Uncharacterized protein | |
| G0CU44; embl-cds:AEG81799 | Putative secreted protein | |
| G0CU89; embl-cds:AEG81844 | DtxR-family transcription regulator | cytoplasm |
| G0CU95; embl-cds:AEG81850 | Alkyl hydroperoxide reductase | cytoplasm |
| G0CUC6; embl-cds:AEG80578 | Uncharacterized protein | |
| G0CUG4; embl-cds:AEG80613 | Uncharacterized protein | |
| G0CUM1; embl-cds:AEG81896 | Polyribonucleotide nucleotidyltransferase | cytoplasm |
| G0CUR3; embl-cds:AEG81937 | Elongation factor Ts | cytoplasm |
| G0CUW8; embl-cds:AEG80687 | Putative secreted protein | |
| G0CV31; embl-cds:AEG81974 | Uncharacterized protein | |
| G0CV41; embl-cds:AEG81983 | Putative secreted protein | |
| G0CV58; embl-cds:AEG82000 | Putative secreted protein | |
| G0CV74; embl-cds:AEG82016 | Putative secreted protein | |
| G0CV86; embl-cds:AEG80721 | Pyridoxal 5′-phosphate synthase subunit PdxS | cytoplasm |
| G0CVA1; embl-cds:AEG80736 | Nucleoid-associated protein CULC22_00193 | cytoplasm |
| G0CVC7; embl-cds:AEG80762 | Aspartokinase | cytoplasm |
| G0CVK8; embl-cds:AEG82067 | Branched-chain-amino-acid aminotransferase | cytoplasm |
| G0CVR2; embl-cds:AEG80808 | Cold shock-like protein A | cytoplasm |
| G0CVS1; embl-cds:AEG80815 | Putative secreted protein | |
| G0CVT0; embl-cds:AEG80823 | Dihydrolipoyl dehydrogenase | cytoplasm |
| G0CVT9; embl-cds:AEG80832 | Putative secreted protein | |
| G0CVU4; embl-cds:AEG80837 | Uncharacterized protein | |
| G0CVU8; embl-cds:AEG80841 | 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase | cytoplasm |
| G0CVX3; embl-cds:AEG80866 | Thiol-disulfide isomerase/thioredoxin | cytoplasm |
| G0CW00; embl-cds:AEG82124 | Glutamine--fructose-6-phosphate aminotransferase (isomerizing) | cytoplasm |
| G0CW81; embl-cds:AEG80892 | 50S ribosomal protein L11 | ribosome |
| G0CW82; embl-cds:AEG80893 | 50S ribosomal protein L1 | ribosome |
| G0CW87; embl-cds:AEG80898 | 50S ribosomal protein L7/L12 | ribosome |
| G0CW95; embl-cds:AEG80906 | DNA-directed RNA polymerase subunit beta | cytoplasm |
| G0CWB3; embl-cds:AEG80924 | Elongation factor G | cytoplasm |
| G0CWB4; embl-cds:AEG80925 | Elongation factor Tu | cytoplasm |
| G0CWC3; embl-cds:AEG80934 | 30S ribosomal protein S10 | ribosome |
| G0CWC4; embl-cds:AEG80935 | 50S ribosomal protein L3 | ribosome |
| G0CWC5; embl-cds:AEG80936 | 50S ribosomal protein L4 | ribosome |
| G0CWC6; embl-cds:AEG80937 | 50S ribosomal protein L23 | ribosome |
| G0CWC7; embl-cds:AEG80938 | 50S ribosomal protein L2 | ribosome |
| G0CWC9; embl-cds:AEG80940 | 50S ribosomal protein L22 | ribosome |
| G0CWD0; embl-cds:AEG80941 | 30S ribosomal protein S3 | ribosome |
| G0CWD1; embl-cds:AEG80942 | 50S ribosomal protein L16 | ribosome |
| G0CWD8; embl-cds:AEG80949 | 50S ribosomal protein L14 | ribosome |
| G0CWE7; embl-cds:AEG80958 | 50S ribosomal protein L18 | ribosome |
| G0CWL5; embl-cds:AEG82198 | 50S ribosomal protein L27 | ribosome |
| G0CWL6; embl-cds:AEG82199 | 50S ribosomal protein L21 | ribosome |
| G0CWM9; embl-cds:AEG82211 | ATP-dependent Clp protease proteolytic subunit | cytoplasm |
| G0CWN2; embl-cds:AEG80960 | 50S ribosomal protein L30 | ribosome |
| G0CWN3; embl-cds:AEG80961 | 50S ribosomal protein L15 | ribosome |
| G0CWN5; embl-cds:AEG80963 | Maltotriose-binding protein | membrane |
| G0CWQ1; embl-cds:AEG80980 | DNA-directed RNA polymerase subunit alpha | cytoplasm |
| G0CWS7; embl-cds:AEG81005 | 10 kDa chaperonin | cytoplasm |
| G0CWU3; embl-cds:AEG81019 | Putative secreted protein | |
| G0CX97; embl-cds:AEG81093 | LytR-family transcription regulator | cytoplasm |
| G0CXC9; embl-cds:AEG81124 | Putative secreted protein | |
| G0CXG7; embl-cds:AEG82307 | Succinyl-CoA:Coenzyme A transferase | redox chain |
| G0CXJ0; embl-cds:AEG82330 | Phosphoribosylformylglycinamidine synthase subunit PurS | cytoplasm |
| G0CXK8; embl-cds:AEG81127 | Uncharacterized protein | |
| G0CXM1; embl-cds:AEG81140 | Uncharacterized protein | |
| G0CXP5; embl-cds:AEG81163 | Carbonic anhydrase | cytoplasm |
| G0CXQ6; embl-cds:AEG81174 | Resuscitation-promoting factor | extracellular |
| G0CXQ9; embl-cds:AEG81178 | Putative secreted protein | |
| G0CXR6; embl-cds:AEG81185 | Citrate synthase | cytoplasm |
| G0CXR9; embl-cds:AEG81188 | Superoxide dismutase [Cu-Zn] | cell envelope |
| G0CY06; embl-cds:AEG82362 | Putative secreted protein | |
| G0CY28; embl-cds:AEG81208 | Uncharacterized protein | |
| G0CY42; embl-cds:AEG81222 | Uncharacterized protein | |
| G0CY56; embl-cds:AEG81236 | 50S ribosomal protein L28 | ribosome |
| G0CY58; embl-cds:AEG81238 | 50S ribosomal protein L32 | ribosome |
| G0CY59; embl-cds:AEG81239 | Two-component system transcriptional regulatory protein | cytoplasm |
| G0CYA4; embl-cds:AEG81281 | 50S ribosomal protein L25 | ribosome |
| G0CYH1; embl-cds:AEG82442 | Purine phosphoribosyltransferase | cytoplasm |
| G0CYH2; embl-cds:AEG82443 | Putative membrane protein | membrane |
| G0CYK0; embl-cds:AEG81292 | Enolase | extracellular; cytoplasm; cell surface |
| G0CYK9; embl-cds:AEG81301 | Transcription elongation factor GreA | cytoplasm |
| G0CYM4; embl-cds:AEG81317 | Fumarate hydratase class II | cytoplasm |
| G0CYQ3; embl-cds:AEG81345 | Putative secreted protein | |
| G0CYQ7; embl-cds:AEG81349 | Uncharacterized protein | |
| G0CYS8; embl-cds:AEG82468 | Fructose-bisphosphate aldolase | cytoplasm |
| G0CYV5; embl-cds:AEG82498 | Alcohol dehydrogenase | cytoplasm |
| G0CYV6; embl-cds:AEG82499 | Aldehyde dehydrogenase | cytoplasm |
| G0CYW2; embl-cds:AEG82505 | Chaperone protein DnaK | cytoplasm |
| G0CYY1; embl-cds:AEG82523 | Urease accessory protein UreG | cytoplasm |
| G0CYY4; embl-cds:AEG82526 | Urease subunit alpha | cytoplasm |
| G0CYY5; embl-cds:AEG82527 | Urease subunit beta | cytoplasm |
| G0CZ22; embl-cds:AEG81380 | 2-oxoglutarate dehydrogenase E1 component | membrane |
| G0CZ53; embl-cds:AEG81411 | Uncharacterized protein | |
| G0CZ62; embl-cds:AEG81420 | Peptide chain release factor 1 | cytoplasm |
| G0CZ71; embl-cds:AEG81429 | ATP synthase subunit alpha | membrane |
| G0CZ73; embl-cds:AEG81431 | ATP synthase subunit beta | membrane |
| G0CZ86; embl-cds:AEG81444 | Electron transfer flavoprotein beta subunit | redox chain |
| G0CZ94; embl-cds:AEG82552 | Phosphoenolpyruvate carboxykinase (GTP) | cytoplasm |
| G0CZA6; embl-cds:AEG82564 | Putative secreted protein | |
| G0CZA9; embl-cds:AEG82567 | Trehalose corynomycolyl transferase | cytoplasm |
| G0CZB9; embl-cds:AEG82576 | UDP-galactopyranose mutase | cytoplasm |
| G0CZC6; embl-cds:AEG82583 | Serine-tRNA ligase | cytoplasm |
| G0CZC8; embl-cds:AEG82585 | Putative secreted protein | |
| G0CZG8; embl-cds:AEG82621 | Uncharacterized protein | |
| G0CZP6; embl-cds:AEG81523 | 30S ribosomal protein S1 | ribosome |
| G0CZQ1; embl-cds:AEG81528 | Uncharacterized protein | cytoplasm |
| G0CZR5; embl-cds:AEG82638 | 30S ribosomal protein S6 | ribosome |
| Name (NCBI) | Gene | Uniprot Identifier | Localization of Protein | LPxTG Motif | ||||
|---|---|---|---|---|---|---|---|---|
| Supernatant | Cell Surface | |||||||
| 809 | BR-AD22 | 809 | BR-AD22 | 809 | BR-AD22 | |||
| Putative ribosome binding protein | rbp | AEG80717 | - | - | - | - | - | none |
| Corynebacterial protease CP40 precursor | cpp | AEG82501 | AEG84830 | + | + | - | - | none |
| Phospholipase D | pld | AEG80581 | AEG82759 | - | + | - | - | none |
| Surface-anchored protein, fimbrial subunit | spaF | AEG82476 | AEG84808 | + | + | + | + | LPKTG |
| Surface-anchored protein, fimbrial subunit | spaE | AEG82477 | AEG84809 | - | - | - | - | LPLTG |
| Surface-anchored protein, fimbrial subunit | spaD | AEG82479 | AEG84811 | + | + | - | - | LPMTG |
| Surface-anchored protein, fimbrial subunit | spaC | AEG82506 | AEG84835 | + | + | + | + | LPLTG |
| Surface-anchored protein, fimbrial subunit | spaB | AEG82507 | AEG84836 | + | + | - | - | LARTG |
| Resuscitation-promoting factor interacting protein | rpfI | AEG81666 | AEG83858 | + | + | + | + | none |
| Cell wall-associated hydrolase | cwlH | AEG82053 | AEG84247 | - | + | - | + | none |
| Sialidase precursor | nanH | AEG80974 | AEG83155 | + | + | + | + | none |
| Venom serine protease KN13 | vsp1 | AEG81049 | AEG83233 | + | + | + | + | none |
| Venom serine protease 2A | vsp2 | AEG82491 | - | - | - | - | - | none |
| Trypsin-like serine protease | tspA | AEG82376 | AEG84712 | + | + | - | - | none |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bittel, M.; Gastiger, S.; Amin, B.; Hofmann, J.; Burkovski, A. Surface and Extracellular Proteome of the Emerging Pathogen Corynebacterium ulcerans. Proteomes 2018, 6, 18. https://doi.org/10.3390/proteomes6020018
Bittel M, Gastiger S, Amin B, Hofmann J, Burkovski A. Surface and Extracellular Proteome of the Emerging Pathogen Corynebacterium ulcerans. Proteomes. 2018; 6(2):18. https://doi.org/10.3390/proteomes6020018
Chicago/Turabian StyleBittel, Miriam, Susanne Gastiger, Bushra Amin, Jörg Hofmann, and Andreas Burkovski. 2018. "Surface and Extracellular Proteome of the Emerging Pathogen Corynebacterium ulcerans" Proteomes 6, no. 2: 18. https://doi.org/10.3390/proteomes6020018
APA StyleBittel, M., Gastiger, S., Amin, B., Hofmann, J., & Burkovski, A. (2018). Surface and Extracellular Proteome of the Emerging Pathogen Corynebacterium ulcerans. Proteomes, 6(2), 18. https://doi.org/10.3390/proteomes6020018







