Isoelectric Point Separations of Peptides and Proteins
Abstract
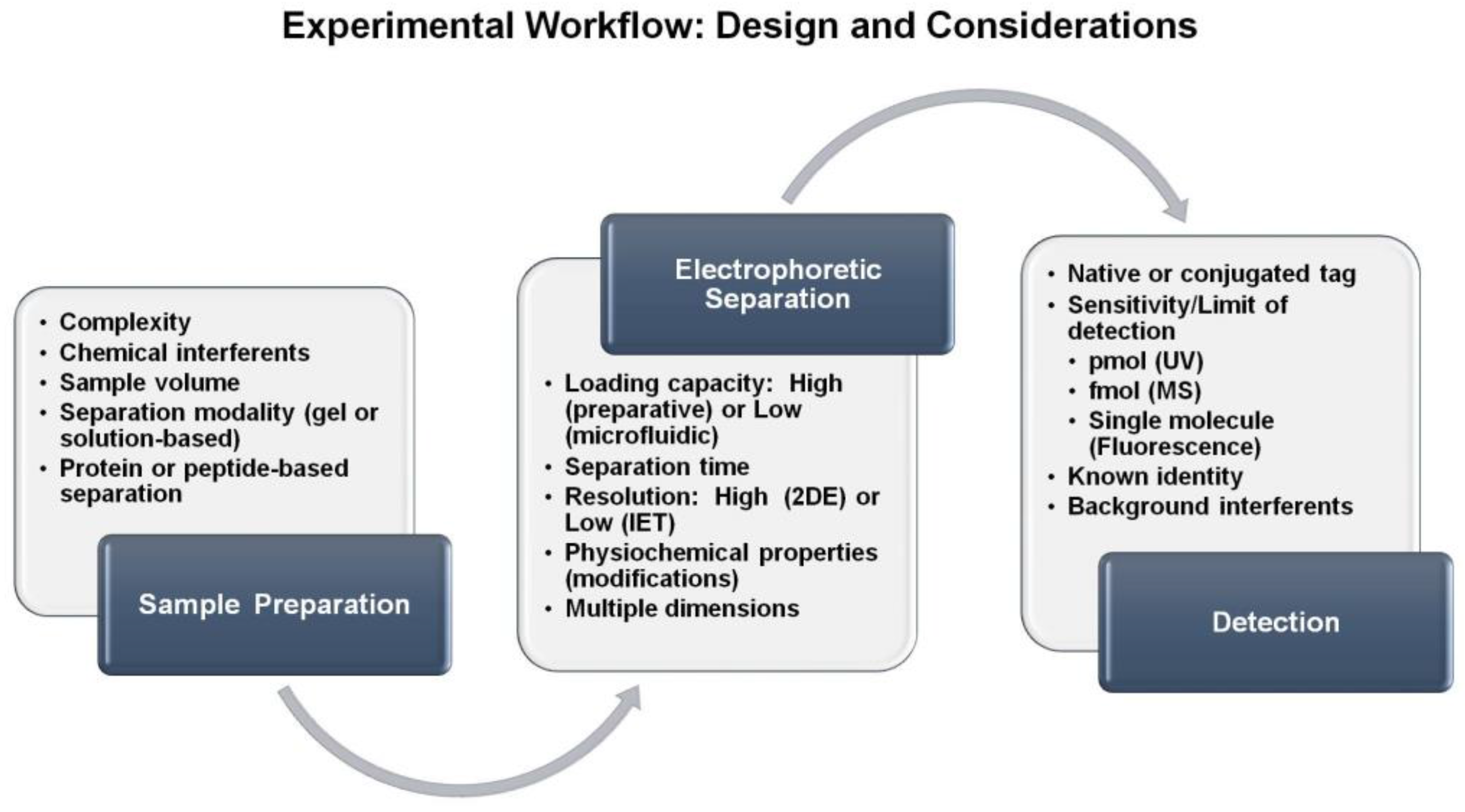
:1. Introduction
2. 2DE and IPG Strips
2.1. Capillary IEF (cIEF)
2.2. Large-Format Devices
2.3. Small Scale Multi-Compartment Electrolyzers (MCE)
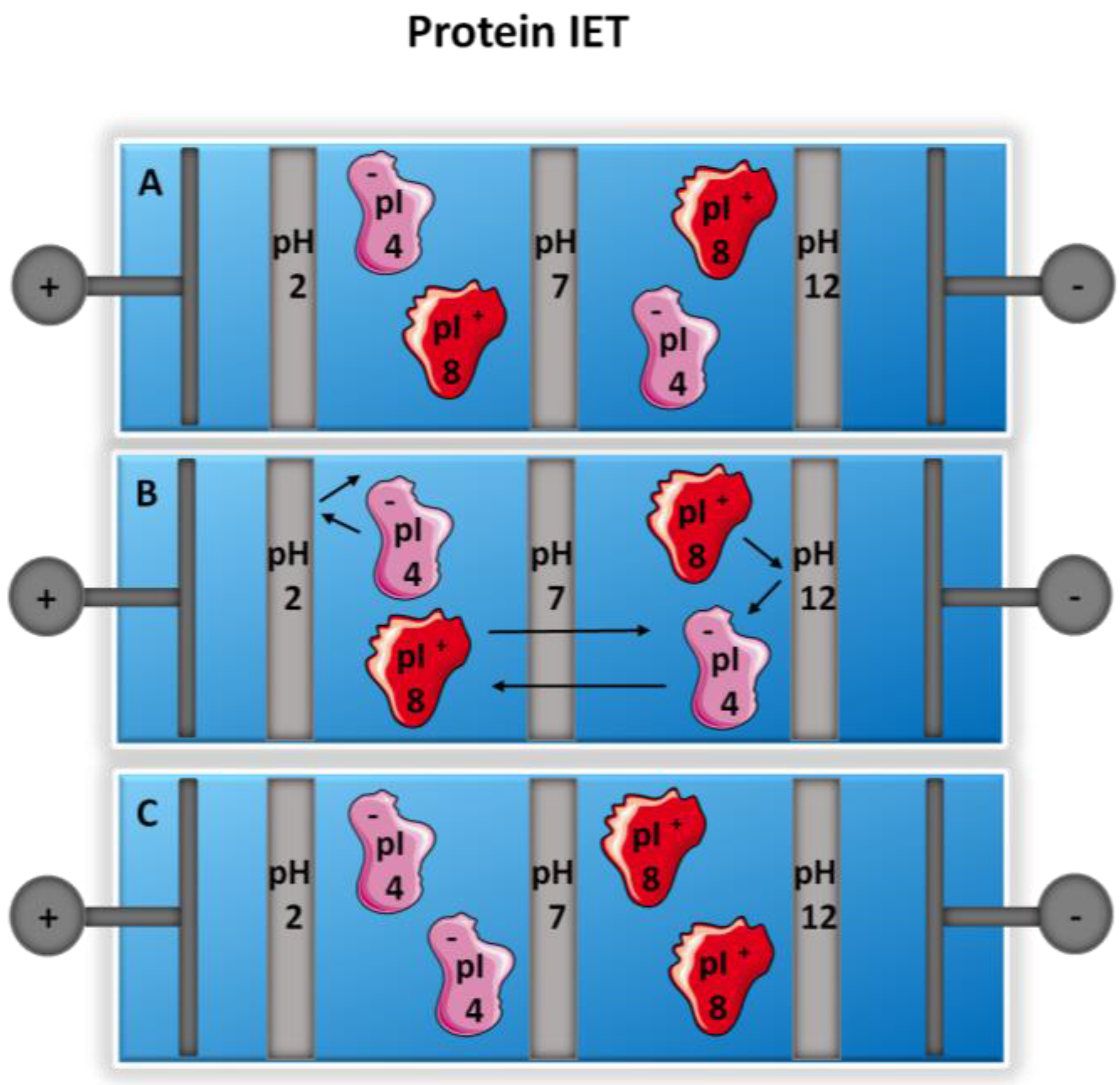
2.4. Isoelectric Trapping—IEF without Carrier Ampholytes
3. Detection of Peptides and Proteins
4. Applications of Isoelectric Focusing and Related Methodologies
4.1. Protein Identification and Post-Translational Modifications
4.2. Separations for Mass Spectrometry Analysis
4.3. Novel Approaches
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| 2DE | Two-dimensional gel electrophoresis |
| cIEF | Capillary isoelectric focusing |
| ESI | Electrospray ionization |
| HiRIEF | High-resolution isoelectric focusing |
| IEF | Isoelectric focusing |
| IET | Isoelectric trapping |
| IPG | Immobilized pH gradients Immobilized pH gradients |
| MALDI | Matrix-assisted laser desorption/ionization |
| MCE | Multi-compartment electrolyzers |
| MSWIFT | Membrane-separated wells for isoelectric focusing and trapping |
| pI | Isoelectric point |
| PVA | Poly(vinyl alcohol) |
References
- Consden, R.; Gordon, A.H.; Martin, A.J. Ionophoresis in Silica Jelly; a method for the separation of amino-acids and peptides. Biochem. J. 1946, 40, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Svensson, H. Isoelectric fractionation, analysis, and characterization of ampholytes in natural pH gradients. I. The differential equation of solute concnetrations at a steady state and its solution for simple cases. Acta Chem. Scand. 1961, 15, 325–341. [Google Scholar] [CrossRef]
- Svensson, H. Isoelectric fractionation, analysis, and characterization of ampholytes in natural pH gradients. II. Buffering capacity and conductance of isoionic ampholytes. Acta Chem. Scand. 1962, 16, 456–466. [Google Scholar] [CrossRef]
- Svensson, H. Isoelectric fractionation, analysis and characterization of ampholytes in natural pH gradient III. Description of apparatus for electrolysis in columns stabilized by denisty gradient and direct gradients and direct determination of isoelectric point. Arch. Biochem. Biophys. 1962, (Suppl. 1), 131–142. [Google Scholar] [CrossRef]
- Vesterberg, O. Synthesis and isoelectric fractionation of carrier ampholytes. Acta Chem. Scand. 1969, 23, 2653–2666. [Google Scholar] [CrossRef]
- Righetti, P.G.; Simo, C.; Sebastiano, R.; Citterio, A. Carrier ampholytes for IEF, on their fortieth anniversary (1967–2007), brought to trial in court: The verdict. Electrophoresis 2007, 28, 3799–3810. [Google Scholar] [CrossRef] [PubMed]
- Righetti, P.G.; Candiano, G. Recent advances in electrophoretic techniques for the characterization of protein biomolecules: A poker of aces. J. Chromatogr. A 2011, 1218, 8727–8737. [Google Scholar] [CrossRef] [PubMed]
- Bjellqvist, B.; Ek, K.; Righetti, P.G.; Gianazza, E.; Gorg, A.; Westermeier, R.; Postel, W. Isoelectric focusing in immobilized pH gradients: Principle, methodology and some applications. J. Biochem. Biophys. Methods 1982, 6, 317–339. [Google Scholar] [CrossRef]
- Stoyanov, A. Ief-based multidimensional applications in proteomics: Toward higher resolution. Electrophoresis 2012, 33, 3281–3290. [Google Scholar] [CrossRef] [PubMed]
- O’Farrell, P.H. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 1975, 250, 4007–4021. [Google Scholar] [PubMed]
- O’Farrell, P.H.; O’Farrell, P.Z. Two-dimensional polyacrylamide gel electrophoretic fractionation. Methods Cell Biol. 1977, 16, 407–420. [Google Scholar] [PubMed]
- O’Farrell, P.Z.; Goodman, H.M.; O’Farrell, P.H. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell 1977, 12, 1133–1141. [Google Scholar] [CrossRef]
- Ek, K.; Bjellqvist, B.; Righetti, P.G. Preparative isoelectric focusing in immobilized pH gradients. I. General principles and methodology. J. Biochem. Biophys. Methods 1983, 8, 135–155. [Google Scholar] [CrossRef]
- Stoyanov, A.V.; Righetti, P.G. pH changes in immobiline gels due to low-molecular mass ion adsorption and conditions for salt front formation during electrophoretic desorption. Electrophoresis 1997, 18, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Gorg, A.; Postel, W.; Westermeier, R. Ultrathin-layer isoelectric focusing in polyacrylamide gels on cellophane. Anal. Biochem. 1978, 89, 60–70. [Google Scholar] [CrossRef]
- Gorg, A.; Postel, W.; Westermeier, R.; Gianazza, E.; Righetti, P.G. Gel gradient electrophoresis, isoelectric focusing and two-dimensional techniques in horizontal, ultrathin polyacrylamide layers. J. Biochem. Biophys. Methods 1980, 3, 273–284. [Google Scholar] [CrossRef]
- Westermeier, R. Method 10—Ief in Immobilized pH Gradients, 4th ed.; Wiley: Hoboken, NJ, USA, 2005; p. 16. [Google Scholar]
- Westermeier, R. 2D gel-based proteomics: There’s life in the old dog yet. Arch. Physiol. Biochem. 2016, 122, 236–237. [Google Scholar] [CrossRef] [PubMed]
- Westermeier, R. Looking at proteins from two dimensions: A review on five decades of 2D electrophoresis. Arch. Physiol. Biochem. 2014, 120, 168–172. [Google Scholar] [CrossRef] [PubMed]
- Curreem, S.O.; Watt, R.M.; Lau, S.K.; Woo, P.C. Two-dimensional gel electrophoresis in bacterial proteomics. Protein Cell 2012, 3, 346–363. [Google Scholar] [CrossRef] [PubMed]
- Mayer, K.; Albrecht, S.; Schaller, A. Targeted analysis of protein phosphorylation by 2D electrophoresis. Methods Mol. Biol. 2015, 1306, 167–176. [Google Scholar] [PubMed]
- Maccarrone, G.; Filiou, M.D. Protein profiling and phosphoprotein analysis by isoelectric focusing. Methods Mol. Biol. 2015, 1295, 293–303. [Google Scholar] [PubMed]
- Mauro, S.; Colignon, B.; Dieu, M.; Delaive, E.; Raes, M. Three-dimensional electrophoresis for quantitative profiling of complex proteomes. Methods Mol. Biol. 2015, 1295, 427–440. [Google Scholar] [PubMed]
- De Jong, C.A.; Risley, J.; Lee, A.K.; Zhao, S.S.; Chen, D.D. Separation of recombinant therapeutic proteins using capillary gel electrophoresis and capillary isoelectric focusing. Methods Mol. Biol. 2016, 1466, 137–149. [Google Scholar] [PubMed]
- Mokaddem, M.; d’Orlye, F.; Varenne, A. Online capillary isoelectric focusing-electrospray ionization mass spectrometry (CIEF-ESI MS) in glycerol-water media for the separation and characterization of hydrophilic and hydrophobic proteins. Methods Mol. Biol. 2016, 1466, 57–66. [Google Scholar] [PubMed]
- Tang, H.Y.; Speicher, D.W. Complex proteome prefractionation using microscale solution isoelectrofocusing. Expert Rev. Proteom. 2005, 2, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Ma, J.; Zhu, G.; Shan, Y.; Liang, Z.; Zhang, L.; Zhang, Y. Integration of capillary isoelectric focusing with monolithic immobilized pH gradient, immobilized trypsin microreactor and capillary zone electrophoresis for on-line protein analysis. J. Sep. Sci. 2010, 33, 3194–3200. [Google Scholar] [CrossRef] [PubMed]
- Kasicka, V. Recent developments in CE and CEC of peptides (2009–2011). Electrophoresis 2012, 33, 48–73. [Google Scholar] [CrossRef] [PubMed]
- Righetti, P.G.; Wenisch, E.; Faupel, M. Preparative protein-purification in a multi-compartment electrolyzer with immobiline membranes. J. Chromatogr. 1989, 475, 293–309. [Google Scholar] [CrossRef]
- Righetti, P.G.; Wenisch, E.; Jungbauer, A.; Katinger, H.; Faupel, M. Preparative purification of human monoclonal-antibody isoforms in a multicompartment electrolyzer with immobiline membranes. J. Chromatogr. 1990, 500, 681–696. [Google Scholar] [CrossRef]
- Egen, N.B.; Thromann, W.; Twitty, G.E.; Bier, M. A new preparative isoelectric focusing apparatus. In Electrophoresis; Hirai, H., Ed.; de Gruyter: Berlin, Germany, 1984; pp. 547–549. [Google Scholar]
- Shen, H.; Li, X.; Bieberich, C.J.; Frey, D.D. Reducing sample complexity in proteomics by chromatofocusing with simple buffer mixtures. Methods Mol. Biol. 2008, 424, 187–203. [Google Scholar] [PubMed]
- Sluyterman, L.A.A.; Elgersma, O. Chromatofocusing: Isoelectric focusing on ion-exchange columns. J. Chromatogr. 1978, 150, 17–30. [Google Scholar] [CrossRef]
- Zuo, X.; Speicher, D.W. A method for global analysis of complex proteomes using sample prefractionation by solution isoelectrofocusing prior to two-dimensional electrophoresis. Anal. Biochem. 2000, 284, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Zuo, X.; Speicher, D.W. Comprehensive analysis of complex proteomes using microscale solution isoelectrofocusing prior to narrow pH range two-dimensional electrophoresis. Proteomics 2002, 2, 58–68. [Google Scholar] [CrossRef]
- An, Y.; Fu, Z.; Gutierrez, P.; Fenselau, C. Solution isoelectric focusing for peptide analysis: Comparative investigation of an insoluble nuclear protein fraction. J. Proteome Res. 2005, 4, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Tran, J.C.; Doucette, A.A. Rapid and effective focusing in a carrier ampholyte solution isoelectric focusing system: A proteome prefractionation tool. J. Proteome Res. 2008, 7, 1761–1766. [Google Scholar] [CrossRef] [PubMed]
- Tran, J.C.; Wall, M.J.; Doucette, A.A. Evaluation of a solution isoelectric focusing protocol as an alternative to ion exchange chromatography for charge-based proteome prefractionation. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2009, 877, 807–813. [Google Scholar] [CrossRef] [PubMed]
- Ros, A.; Faupel, M.; Mees, H.; Oostrum, J.; Ferrigno, R.; Reymond, F.; Michel, P.; Rossier, J.S.; Girault, H.H. Protein purification by off-gel electrophoresis. Proteomics 2002, 2, 151–156. [Google Scholar] [CrossRef]
- Michel, P.E.; Reymond, F.; Arnaud, I.L.; Josserand, J.; Girault, H.H.; Rossier, J.S. Protein fractionation in a multicompartment device using off-gel (TM) isoelectric focusing. Electrophoresis 2003, 24, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Moreda-Pineiro, A.; Garcia-Otero, N.; Bermejo-Barrera, P. A review on preparative and semi-preparative offgel electrophoresis for multidimensional protein/peptide assessment. Anal. Chim. Acta 2014, 836, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Magdeldin, S.; Elguoshy, A.; Yoshida, Y.; Hirao, Y.; Xu, B.; Zhang, Y.; Yamamoto, K.; Takimoto, H.; Fujinaka, H.; Kinoshita, N.; et al. Complementary protein and peptide offgel fractionation for high-throughput proteomic analysis. Anal. Chem. 2015, 87, 8481–8488. [Google Scholar] [CrossRef] [PubMed]
- Kasicka, V. Recent developments in capillary and microchip electroseparations of peptides (2013-middle 2015). Electrophoresis 2016, 37, 162–188. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, S.; Kasicka, V. Analysis of proteins and peptides by electromigration methods in microchips. J. Sep. Sci. 2016. [Google Scholar] [CrossRef]
- Branan, N. Microfluidic IEF without carrier molecules. Anal. Chem. 2006, 78, 3488. [Google Scholar] [CrossRef] [PubMed]
- Macounova, K.; Cabrera, C.R.; Holl, M.R.; Yager, P. Generation of natural pH gradients in microfluidic channels for use in isoelectric focusing. Anal. Chem. 2000, 72, 3745–3751. [Google Scholar] [CrossRef] [PubMed]
- Righetti, P.G.; Sebastiano, R.; Citterio, A. Capillary electrophoresis and isoelectric focusing in peptide and protein analysis. Proteomics 2013, 13, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yang, S.; Lee, C.S.; DeVoe, D.L. Polyacrylamide gel plugs enabling 2-D microfluidic protein separations via isoelectric focusing and multiplexed sodium dodecyl sulfate gel electrophoresis. Electrophoresis 2008, 29, 2241–2250. [Google Scholar] [CrossRef] [PubMed]
- Vlckova, M.; Kalman, F.; Schwarz, M.A. Pharmaceutical applications of isoelectric focusing on microchip with imaged UV detection. J. Chromatogr. A 2008, 1181, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Mikkonen, S.; Jacksen, J.; Roeraade, J.; Thormann, W.; Emmer, A. Microfluidic isoelectric focusing of amyloid beta peptides followed by micropillar-matrix-assisted laser desorption ionization-mass spectrometry. Anal. Chem. 2016, 88, 10044–10051. [Google Scholar] [CrossRef] [PubMed]
- Chiari, M.; Micheletti, C.; Nesi, M.; Fazio, M.; Righetti, P.G. Towards new formulations for polyacrylamide matrices—N-acryloylaminoethoxyethanol, a novel monomer combining high hydrophilicity with extreme hydrolytic stability. Electrophoresis 1994, 15, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Lalwani, S.; Vigh, G. A family of high-buffering capacity diamino sulfate isoelectric buffers for ph-biased isoelectric trapping separations. Electrophoresis 2005, 26, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Lalwani, S.; Tutu, E.; Vigh, G. Synthesis and characterization of quaternary ammonium dicarboxylic acid isoelectric buffers and their use in pH-biased isoelectric trapping separations. Electrophoresis 2005, 26, 2047–2055. [Google Scholar] [CrossRef] [PubMed]
- Lim, P.; North, R.; Vigh, G. Rapid isoelectric trapping in a micropreparative-scale multicompartment electrolyzer. Electrophoresis 2007, 28, 1851–1859. [Google Scholar] [CrossRef] [PubMed]
- Cologna, S.M.; Russell, W.K.; Lim, P.J.; Vigh, G.; Russell, D.H. Combining isoelectric point-based fractionation, liquid chromatography and mass spectrometry to improve peptide detection and protein identification. J. Am. Soc. Mass Spectrom. 2010, 21, 1612–1619. [Google Scholar] [CrossRef] [PubMed]
- Cologna, S.M.; Williams, B.J.; Russell, W.K.; Pai, P.J.; Vigh, G.; Russell, D.H. Studies of histidine as a suitable isoelectric buffer for tryptic digestion and isoelectric trapping fractionation followed by capillary electrophoresis-mass spectrometry for proteomic analysis. Anal. Chem. 2011, 83, 8108–8114. [Google Scholar] [CrossRef] [PubMed]
- Pai, P.J.; Cologna, S.M.; Russell, W.K.; Vigh, G.; Russell, D.H. Efficient electrophoretic method to remove neutral additives from protein solutions followed by mass spectrometry analysis. Anal. Chem. 2011, 83, 2814–2818. [Google Scholar] [CrossRef] [PubMed]
- Lim, P.J.; Vigh, G. Concentration and fractionation by isoelectric trapping in a micropreparative-scale multicompartmental electrolyzer having orthogonal pH gradients. Part 1. Electrophoresis 2011, 32, 1647–1653. [Google Scholar] [PubMed]
- Lim, P.J.; Vigh, G. Concentration and fractionation by isoelectric trapping in a micropreparative-scale multicompartmental electrolyzer having orthogonal pH gradients. Part 2. Electrophoresis 2011, 32, 1654–1658. [Google Scholar] [PubMed]
- North, R.Y.; Vigh, G. Preparative-scale isoelectric trapping by recursive electrophoresis in a compartmentalized system having orthogonal primary and secondary pH gradients. Part 1—Construction and standard operation. Electrophoresis 2011, 32, 2797–2804. [Google Scholar] [CrossRef] [PubMed]
- North, R.Y.; Vigh, G. Preparative-scale isoelectric trapping by recursive electrophoresis in a compartmentalized system having orthogonal primary and secondary pH gradients. Part 2—Operation in cascade mode. Electrophoresis 2011, 32, 2805–2808. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Campana, A.M.; Taverna, M.; Fabre, H. LIF detection of peptides and proteins in CE. Electrophoresis 2007, 28, 208–232. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Tragas, C.; Wu, J.; Mao, Q.; Pawliszyn, J. Recent developments in capillary isoelectric focusing with whole-column imaging detection. Electrophoresis 1998, 19, 2290–2295. [Google Scholar] [CrossRef] [PubMed]
- Gencoglu, A.; Minerick, A.R. Electrochemical detection techniques in micro- and nanofluidic devices. Microfluid. Nanofluid. 2014, 17, 781–807. [Google Scholar] [CrossRef]
- Berkelman, T.; Bandhakavi, S.; Paulus, A. In-gel peptide ief sample preparation for LC/MS analysis. Methods Mol. Biol. 2015, 1295, 369–379. [Google Scholar] [PubMed]
- Lengqvist, J.; Eriksson, H.; Gry, M.; Uhlen, K.; Bjorklund, C.; Bjellqvist, B.; Jakobsson, P.J.; Lehtio, J. Observed peptide pI and retention time shifts as a result of post-translational modifications in multidimensional separations using narrow-range IPG-IEF. Amino Acids 2011, 40, 697–711. [Google Scholar] [CrossRef] [PubMed]
- Branca, R.M.; Orre, L.M.; Johansson, H.J.; Granholm, V.; Huss, M.; Perez-Bercoff, A.; Forshed, J.; Kall, L.; Lehtio, J. Hirief LC-MS enables deep proteome coverage and unbiased proteogenomics. Nat. Methods 2014, 11, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Choi, N.; Lim, B.; Kim, T.W.; Song, S.; Kim, Y.P. Sequential phosphorylation analysis using dye-tethered peptides and microfluidic isoelectric focusing electrophoresis. Biosens. Bioelectron. 2015, 73, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.K.; Harrata, A.K.; Lee, C.S. Monitoring protein refolding induced by disulfide formation using capillary isoelectric focusing electrospray ionization mass spectrometry. Anal. Chem. 1998, 70, 2044–2049. [Google Scholar] [CrossRef] [PubMed]
- Tragas, C.; Pawliszyn, J. On-line coupling of high performance gel filtration chromatography with imaged capillary isoelectric focusing using a membrane interface. Electrophoresis 2000, 21, 227–237. [Google Scholar] [CrossRef]
- Chen, J.Z.; Lee, C.S.; Shen, Y.F.; Smith, R.D.; Baehrecke, E.H. Integration of capillary isoelectric focusing with capillary reversed-phase liquid chromatography for two-dimensional proteomics separation. Electrophoresis 2002, 23, 3143–3148. [Google Scholar] [CrossRef]
- Yang, C.; Zhang, L.Y.; Liu, H.C.; Zhang, W.B.; Zhang, Y.K. Two-dimensional capillary electrophoresis involving capillary isoelectric focusing and capillary zone electrophoresis. J. Chromatogr. A 2003, 1018, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Dickerson, J.A.; Ramsay, L.M.; Dada, O.O.; Cermak, N.; Dovichi, N.J. Two-dimensional capillary electrophoresis: Capillary isoelectric focusing and capillary zone electrophoresis with laser-induced fluorescence detection. Electrophoresis 2010, 31, 2650–2654. [Google Scholar] [CrossRef] [PubMed]
- Washburn, M.P.; Wolters, D.; Yates, J.R., 3rd. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001, 19, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Millioni, R.; Franchin, C.; Tessari, P.; Polati, R.; Cecconi, D.; Arrigoni, G. Pros and cons of peptide isolectric focusing in shotgun proteomics. J. Chromatogr. A 2013, 1293, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Geiser, L.; Vaezzadeh, A.R.; Deshusses, J.M.; Hochstrasser, D.F. Shotgun proteomics: A qualitative approach applying isoelectric focusing on immobilized pH gradient and LC-MS/MS. Methods Mol. Biol. 2011, 681, 449–458. [Google Scholar] [PubMed]
- Kohrs, F.; Heyer, R.; Magnussen, A.; Benndorf, D.; Muth, T.; Behne, A.; Rapp, E.; Kausmann, R.; Heiermann, M.; Klocke, M.; et al. Sample prefractionation with liquid isoelectric focusing enables in depth microbial metaproteome analysis of mesophilic and thermophilic biogas plants. Anaerobe 2014, 29, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Stein, D.R.; Hu, X.; McCorrister, S.J.; Westmacott, G.R.; Plummer, F.A.; Ball, T.B.; Carpenter, M.S. High pH reversed-phase chromatography as a superior fractionation scheme compared to off-gel isoelectric focusing for complex proteome analysis. Proteomics 2013, 13, 2956–2966. [Google Scholar] [CrossRef] [PubMed]
- Lengqvist, J.; Uhlen, K.; Lehtio, J. Itraq compatibility of peptide immobilized pH gradient isoelectric focusing. Proteomics 2007, 7, 1746–1752. [Google Scholar] [CrossRef] [PubMed]
- Herzog, C.; Poehler, E.; Peretzki, A.J.; Borisov, S.M.; Aigner, D.; Mayr, T.; Nagl, S. Continuous on-chip fluorescence labelling, free-flow isoelectric focusing and marker-free isoelectric point determination of proteins and peptides. Lab. Chip 2016, 16, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhu, Z.; Yu, H.; Lu, J.J.; Liu, S. Simple means for fractionating protein based on isoelectric point without ampholyte. Anal. Chem. 2016, 88, 9293–9299. [Google Scholar] [CrossRef] [PubMed]
- Brod, E.; Ben-Yosef, V.S.; Bandhakavi, S.; Sivan, U. Charge-based separation of proteins and peptides by electrically induced dynamic pH profiles. J. Chromatogr. A 2016, 1431, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.G.; Shang, Z.; Yan, J.; Li, S.; Li, G.Q.; Liu, R.Z.; Qing, Y.; Fan, L.Y.; Xiao, H.; Cao, C.X. A tunable isoelectric focusing via moving reaction boundary for two-dimensional gel electrophoresis and proteomics. Talanta 2015, 137, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Ma, J.; Wu, S.; Yuan, H.; Zhang, L.; Liang, Z.; Zhang, Y. Integrated platform of capillary isoelectric focusing, trypsin immobilized enzyme microreactor and nanoreversed-phase liquid chromatography with mass spectrometry for online protein profiling. Electrophoresis 2011, 32, 2848–2856. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pergande, M.R.; Cologna, S.M. Isoelectric Point Separations of Peptides and Proteins. Proteomes 2017, 5, 4. https://doi.org/10.3390/proteomes5010004
Pergande MR, Cologna SM. Isoelectric Point Separations of Peptides and Proteins. Proteomes. 2017; 5(1):4. https://doi.org/10.3390/proteomes5010004
Chicago/Turabian StylePergande, Melissa R., and Stephanie M. Cologna. 2017. "Isoelectric Point Separations of Peptides and Proteins" Proteomes 5, no. 1: 4. https://doi.org/10.3390/proteomes5010004
APA StylePergande, M. R., & Cologna, S. M. (2017). Isoelectric Point Separations of Peptides and Proteins. Proteomes, 5(1), 4. https://doi.org/10.3390/proteomes5010004




