Understanding the Remodelling of Cell Walls during Brachypodium distachyon Grain Development through a Sub-Cellular Quantitative Proteomic Approach
Abstract
:1. Introduction
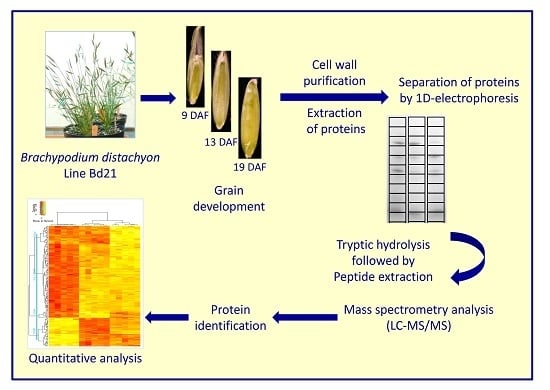
2. Experimental Section
2.1. Plant Material
2.2. Cell Wall Purification and Protein Extraction
2.3. Mass Spectrometry Analysis
2.4. Functional Annotation
2.5. Quantification and Statistical Analysis
2.6. Immunolabelling
2.7. Semi-Quantitative RT-PCR
3. Results
3.1. Extraction of Proteins from Cell Walls of the B. distachyon Grain at Three Developmental Stages
3.2. Proteomic Analysis
3.3. Variations of CWP Abundance during Grain Development
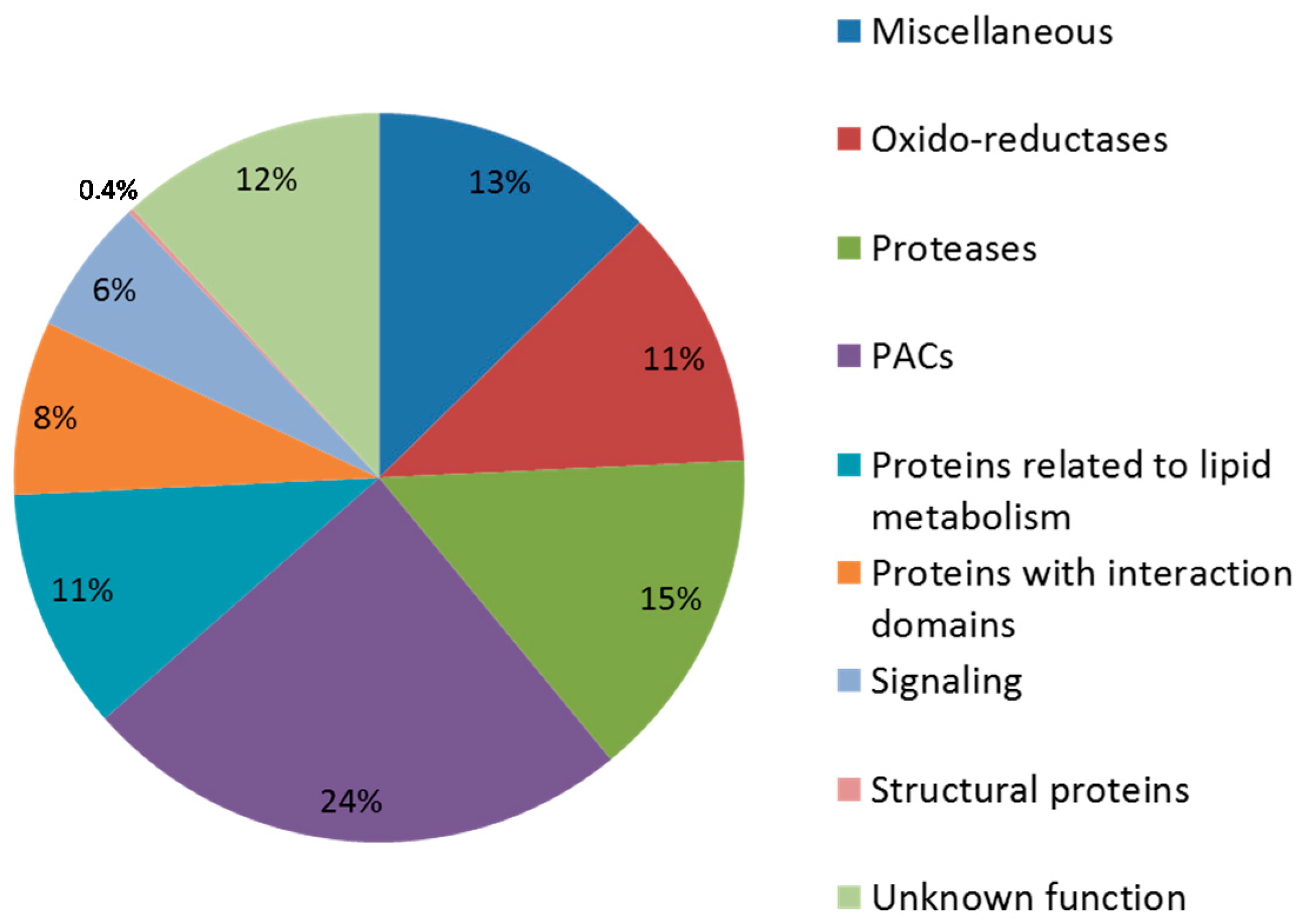
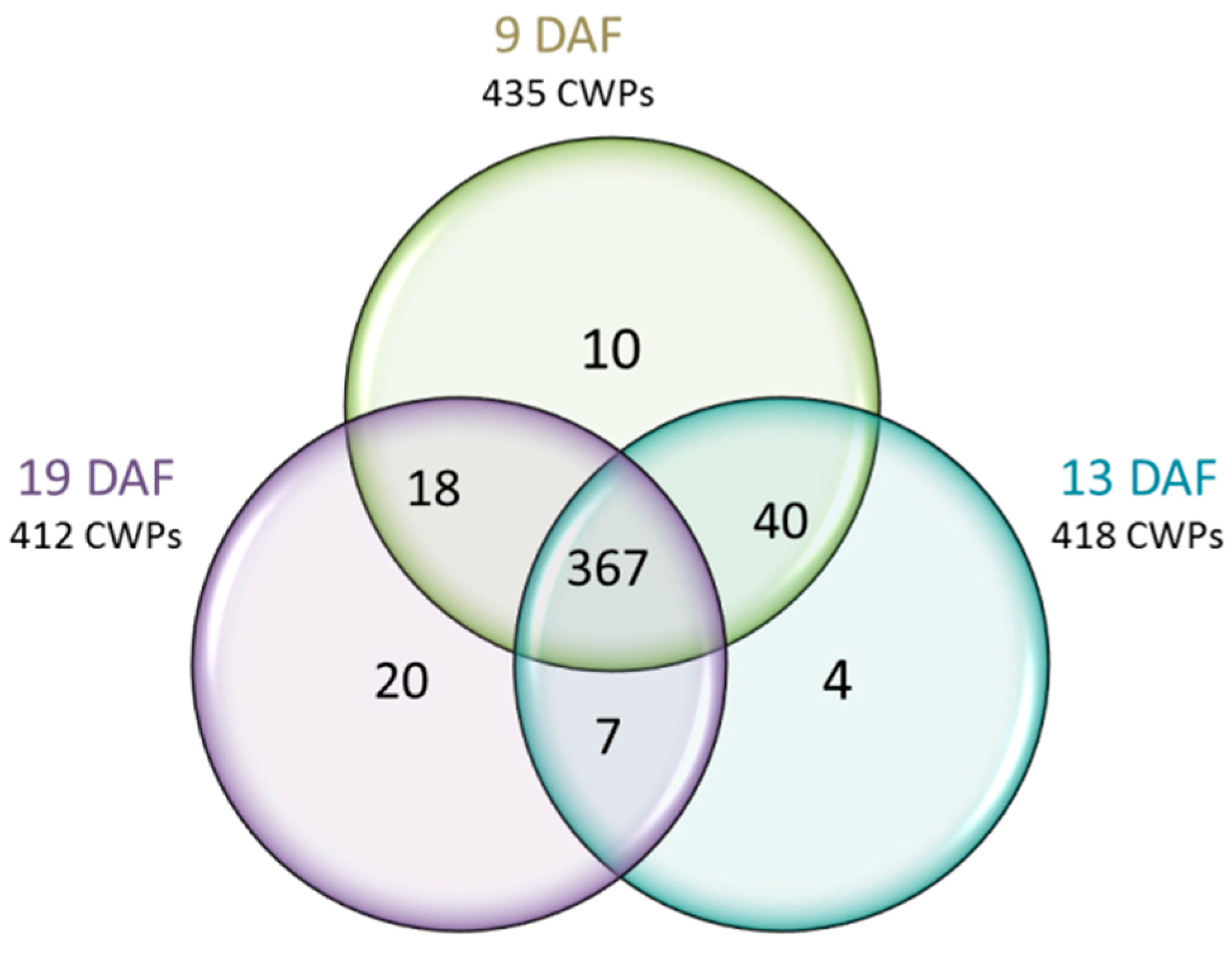
3.3.1. Distribution among the Three Developmental Stages
3.3.2. Quantitative Analysis of CWPs
3.4. Focus on Proteins Acting on Cell Wall Polysaccharides (PACs)
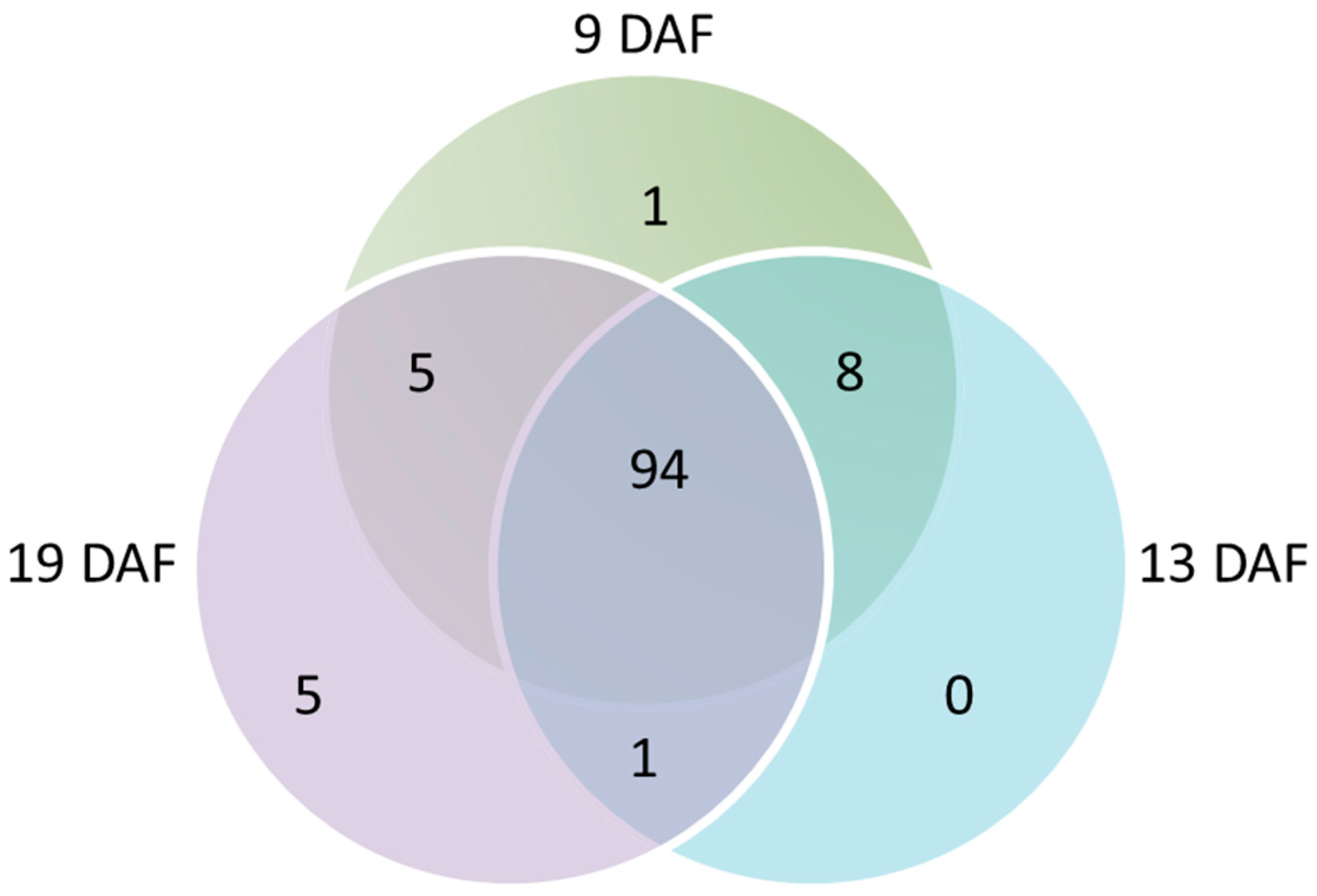
3.4.1. Repartition of PACs According to the Grain Developmental Stages
3.4.2. Quantitative Analysis of the PACs
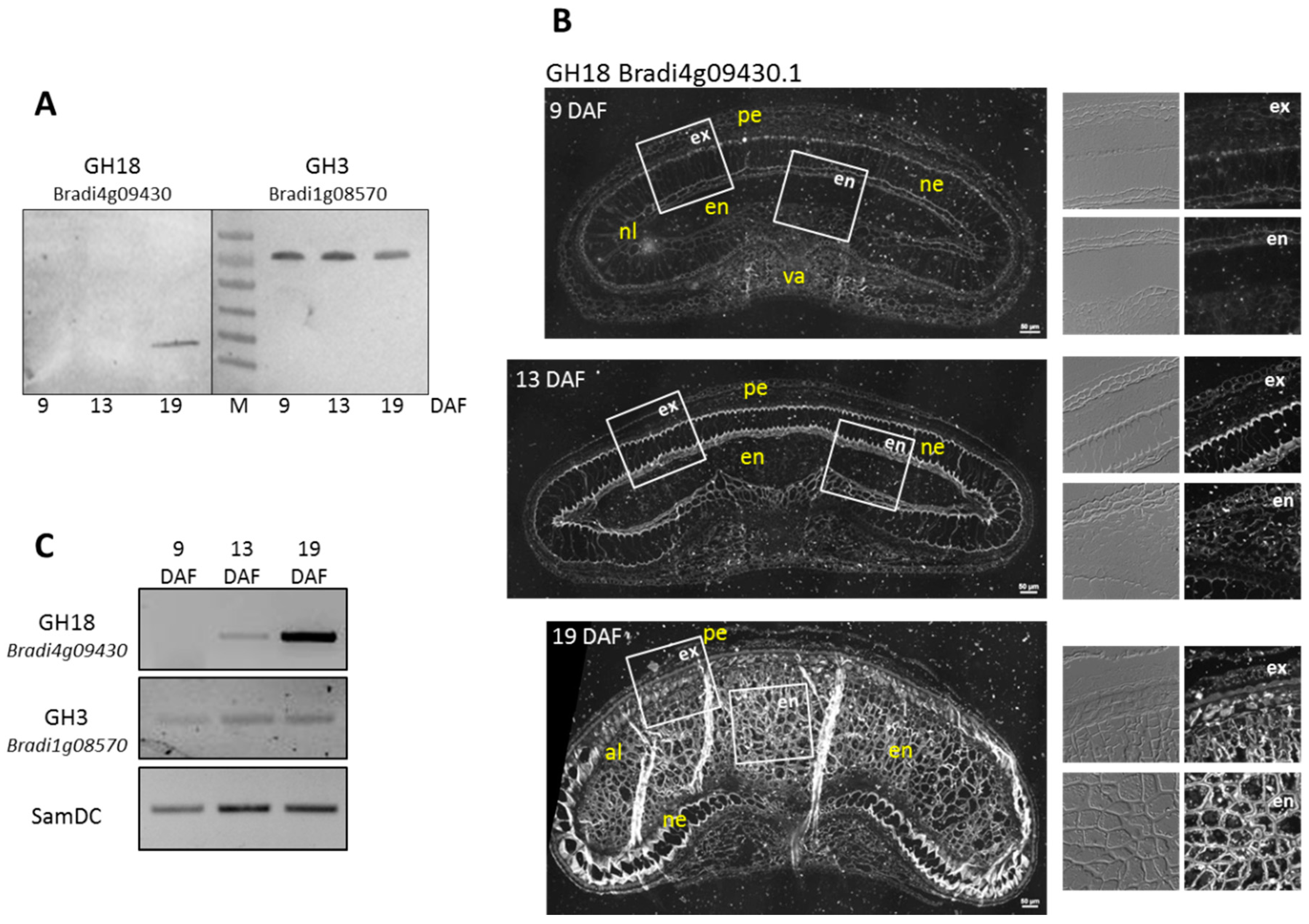
3.4.3. Experimental Validation of the Proteomic Quantitative Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Girin, T.; David, L.C.; Chardin, C.; Sibout, R.; Krapp, A.; Ferrario-Méry, S.; Daniel-Vedele, F. Brachypodium: A promising hub between model species and cereals. J. Exp. Bot. 2014, 65, 5683–5696. [Google Scholar] [CrossRef] [PubMed]
- Draper, J.; Mur, L.A.; Jenkins, G.; Ghosh-Biswas, G.C.; Bablak, P.; Hasterok, R.; Routledge, A.P. Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol. 2001, 127, 1539–1555. [Google Scholar] [CrossRef] [PubMed]
- Initiative, I.B. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 2010, 463, 763–768. [Google Scholar]
- Guillon, F.; Bouchet, B.; Jamme, F.; Robert, P.; Quemener, B.; Barron, C.; Larre, C.; Dumas, P.; Saulnier, L. Brachypodium distachyon grain: Characterization of endosperm cell walls. J. Exp. Bot. 2011, 62, 1001–1015. [Google Scholar] [CrossRef] [PubMed]
- Guillon, F.; Larre, C.; Petipas, F.; Berger, A.; Moussawi, J.; Rogniaux, H.; Santoni, A.; Saulnier, L.; Jamme, F.; Miquel, M.; et al. A comprehensive overview of grain development in Brachypodium distachyon variety Bd21. J. Exp. Bot. 2012, 63, 739–755. [Google Scholar] [CrossRef] [PubMed]
- Larre, C.; Penninck, S.; Bouchet, B.; Lollier, V.; Tranquet, O.; Denery-Papini, S.; Guillon, F.; Rogniaux, H. Brachypodium distachyon grain: Identification and subcellular localization of storage proteins. J. Exp. Bot. 2010, 61, 1771–1783. [Google Scholar] [CrossRef] [PubMed]
- Trafford, K.; Haleux, P.; Henderson, M.; Parker, M.; Shirley, N.; Tucker, M.; Fincher, G.; Burton, R. Grain development in brachypodium and other grasses: Possible interactions between cell expansion, starch deposition, and cell-wall synthesis. J. Exp. Bot. 2013, 64, 5033–5047. [Google Scholar] [CrossRef] [PubMed]
- Opanowicz, M.; Hands, P.; Betts, D.; Parker, M.L.; Toole, G.A.; Mills, E.N.; Doonan, J.H.; Drea, S. Endosperm development in Brachypodium distachyon. J. Exp. Bot. 2011, 62, 735–748. [Google Scholar] [CrossRef] [PubMed]
- Saulnier, L.; Robert, P.; Grintchenko, M.; Jamme, F.; Bouchet, B.; Guillon, F. Wheat endosperm cell walls: Spatial heterogeneity of polysaccharide structure and composition using micro-scale enzymatic fingerprinting and ft-ir microspectroscopy. J. Cereal Sci. 2009, 50, 312–317. [Google Scholar] [CrossRef]
- Knox, J.P. Revealing the structural and functional diversity of plant cell walls. Curr. Opin. Plant Biol. 2008, 11, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Christensen, U.; Alonso-Simon, A.; Scheller, H.V.; Willats, W.G.; Harholt, J. Characterization of the primary cell walls of seedlings of Brachypodium distachyon—A potential model plant for temperate grasses. Phytochemistry 2010, 71, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Tyler, L.; Bragg, J.N.; Wu, J.; Yang, X.; Tuskan, G.A.; Vogel, J.P. Annotation and comparative analysis of the glycoside hydrolase genes in Brachypodium distachyon. BMC Genom. 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Albenne, C.; Canut, H.; Jamet, E. Plant cell wall proteomics: The leadership of Arabidopsis thaliana. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Albenne, C.; Canut, H.; Hoffmann, L.; Jamet, E. Plant cell wall proteins: A large body of data, but what about runaways? Proteomes 2014, 2, 224–242. [Google Scholar] [CrossRef]
- Douche, T.; Clemente, H.; Burlat, V.; Roujol, D.; Valot, B.; Zivy, M.; Pont-Lezica, R.; Jamet, E. Brachypodium distachyon as a model plant toward improved biofuel crops: Search for secreted proteins involved in biogenesis and disassembly of cell wall polymers. Proteomics 2013, 13, 2438–2454. [Google Scholar] [CrossRef] [PubMed]
- Francin-Allami, M.; Merah, K.; Albenne, C.; Rogniaux, H.; Pavlovic, M.; Lollier, V.; Sibout, R.; Guillon, F.; Jamet, E.; Larré, C. Cell wall proteomic of Brachypodium distachyon grains: A focus on cell wall remodeling proteins. Proteomics 2015, 15, 2296–2306. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Chen, G.X.; Lv, D.W.; Li, X.H.; Yan, Y.M. N-linked glycoproteome profiling of seedling leaf in Brachypodium distachyon L. J. Proteome Res. 2015, 14, 1727–1738. [Google Scholar] [CrossRef] [PubMed]
- Franková, L.; Fry, S.C. Biochemistry and physiological roles of enzymes that 'cut and paste' plant cell-wall polysaccharides. J. Exp. Bot. 2013, 64, 3519–3550. [Google Scholar] [CrossRef] [PubMed]
- Minic, Z. Physiological roles of plant glycoside hydrolases. Planta 2008, 227, 723–740. [Google Scholar] [CrossRef] [PubMed]
- Herbers, K.; Lorences, E.P.; Barrachina, C.; Sonnewald, U. Functional characterisation of Nicotiana tabacum xyloglucan endotransglycosylase (NtXET-1): Generation of transgenic tobacco plants and changes in cell wall xyloglucan. Planta 2001, 212, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.B.; Lu, S.M.; Zhang, J.F.; Liu, S.; Lu, Y.T. A xyloglucan endotransglucosylase/hydrolase involves in growth of primary root and alters the deposition of cellulose in arabidopsis. Planta 2007, 226, 1547–1560. [Google Scholar] [CrossRef] [PubMed]
- Francoz, E.; Ranocha, P.; Nguyen-Kim, H.; Jamet, E.; Burlat, V.; Dunand, C. Roles of cell wall peroxidases in plant development. Phytochemistry 2015, 112, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Greiner, S. Growth control by cell wall pectins. Protoplasma 2012, 249 (Suppl. S2), S169–S175. [Google Scholar] [CrossRef] [PubMed]
- Feiz, L.; Irshad, M.; Pont-Lezica, R.F.; Canut, H.; Jamet, E. Evaluation of cell wall preparations for proteomics: A new procedure for purifying cell walls from arabidopsis hypocotyls. Plant Methods 2006, 2. [Google Scholar] [CrossRef] [PubMed]
- Irshad, M.; Canut, H.; Borderies, G.; Pont-Lezica, R.; Jamet, E. A new picture of cell wall protein dynamics in elongating cells of Arabidopsis thaliana: Confirmed actors and newcomers. BMC Plant Biol. 2008, 8. [Google Scholar] [CrossRef] [PubMed]
- Suliman, M.; Chateigner-Boutin, A.; Francin-Allami, M.; Partier, A.; Bouchet, B.; Salse, J.; Pont, C.; Marion, J.; Rogniaux, H.; Tessier, D.; et al. Identification of glycosyltransferases involved in cell wall synthesis of wheat endosperm. J. Proteom. 2013, 78, 508–521. [Google Scholar] [CrossRef] [PubMed]
- Clemente, H.S.; Pont-Lezica, R.; Jamet, E. Bioinformatics as a tool for assessing the quality of sub-cellular proteomic strategies and inferring functions of proteins: Plant cell wall proteomics as a test case. Bioinform. Biol. Insights 2009, 3, 15–28. [Google Scholar] [PubMed]
- San Clemente, H.; Jamet, E. WALLProtDB, a database resource for plant cell wall proteomics. Plant Methods 2015, 11. [Google Scholar] [CrossRef] [PubMed]
- Valot, B.; Langella, O.; Nano, E.; Zivy, M. Masschroq: A versatile tool for mass spectrometry quantification. Proteomics 2011, 11, 3572–3577. [Google Scholar] [CrossRef] [PubMed]
- Jamet, E.; Albenne, C.; Boudart, G.; Irshad, M.; Canut, H.; Pont-Lezica, R. Recent advances in plant cell wall proteomics. Proteomics 2008, 8, 893–908. [Google Scholar] [CrossRef] [PubMed]
- Jashni, M.K.; Mehrabi, R.; Collemare, J.; Mesarich, C.H.; de Wit, P.J. The battle in the apoplast: Further insights into the roles of proteases and their inhibitors in plant-pathogen interactions. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Calderan-Rodrigues, M.J.; Jamet, E.; Douché, T.; Bonassi, M.B.; Cataldi, T.R.; Fonseca, J.G.; San Clemente, H.; Pont-Lezica, R.; Labate, C.A. Cell wall proteome of sugarcane stems: Comparison of a destructive and a non-destructive extraction method showed differences in glycoside hydrolases and peroxidases. BMC Plant Biol. 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-Kim, H.; Clemente, H.S.; Balliau, T.; Zivy, M.; Dunand, C.; Albenne, C.; Jamet, E. Arabidopsis thaliana root cell wall proteomics: Increasing the proteome coverage using a combinatorial peptide ligand library and description of unexpected hyp in peroxidase amino acid sequences. Proteomics 2016, 3, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Bokhari, S.A.; Dong, C.J.; Liu, J.Y. Comparative proteomics analysis of the root apoplasts of rice seedlings in response to hydrogen peroxide. PLoS ONE 2011, 6, e16723. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Giboulot, A.; Zivy, M.; Valot, B.; Jamet, E.; Albenne, C. Combining various strategies to increase the coverage of the plant cell wall glycoproteome. Phytochemistry 2011, 72, 1109–1123. [Google Scholar] [CrossRef] [PubMed]
- Fry, S.C. Primary cell wall metabolism: Tracking the careers of wall polymers in living plant cells. New Phytol. 2004, 161, 641–675. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Saulnier, L.; Guillon, F.; Chateigner-Boutin, A.-L. Cell wall deposition and metabolism in wheat grain. J. Cereal Sci. 2012, 56, 91–108. [Google Scholar] [CrossRef]
- Cairns, J.R.K.; Esen, A. Β-glucosidases. Cell. Mol. Life Sci. 2010, 67, 3389–3405. [Google Scholar] [CrossRef] [PubMed]
- Lammens, W.; Le Roy, K.; Schroeven, L.; Van Laere, A.; Rabijns, A.; Van den Ende, W. Structural insights into glycoside hydrolase family 32 and 68 enzymes: Functional implications. J. Exp. Bot. 2009, 60, 727–740. [Google Scholar] [CrossRef] [PubMed]
- Hirose, T.; Takano, M.; Terao, T. Cell wall invertase in developing rice caryopsis: Molecular cloning of oscin1 and analysis of its expression in relation to its role in grain filling. Plant Cell Physiol. 2002, 452–459. [Google Scholar] [CrossRef]
- Vogel, J. Unique aspects of the grass cell wall. Curr. Opin. Plant Biol. 2008, 11, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Somerville, C.; Anderson, C.T. POLYGALACTURONASE INVOLVED IN EXPANSION1 functions in cell elongation and flower development in Arabidopsis. Plant Cell 2014, 26, 1018–1035. [Google Scholar] [CrossRef] [PubMed]
- Chateigner-Boutin, A.L.; Bouchet, B.; Alvarado, C.; Bakan, B.; Guillon, F. The wheat grain contains pectic domains exhibiting specific spatial and development-associated distribution. PLoS ONE 2014, 9, e89620. [Google Scholar]
- Palmer, R.; Cornuault, V.; Marcus, S.E.; Knox, J.P.; Shewry, P.R.; Tosi, P. Comparative in situ analyses of cell wall matrix polysaccharide dynamics in developing rice and wheat grain. Planta 2015, 241, 669–685. [Google Scholar] [CrossRef] [PubMed]
- Chateigner-Boutin, A.L.; Suliman, M.; Bouchet, B.; Alvarado, C.; Lollier, V.; Rogniaux, H.; Guillon, F.; Larré, C. Endomembrane proteomics reveals putative enzymes involved in cell wall metabolism in wheat grain outer layers. J. Exp. Bot. 2015, 66, 2649–2658. [Google Scholar] [CrossRef] [PubMed]
- Gou, J.Y.; Miller, L.M.; Hou, G.; Yu, X.H.; Chen, X.Y.; Liu, C.J. Acetylesterase-mediated deacetylation of pectin impairs cell elongation, pollen germination, and plant reproduction. Plant Cell 2012, 24, 50–65. [Google Scholar] [CrossRef] [PubMed]
- Marowa, P.; Ding, A.; Kong, Y. Expansins: Roles in plant growth and potential applications in crop improvement. Plant Cell Rep. 2016, 35, 949–965. [Google Scholar] [CrossRef] [PubMed]
- Lizana, X.C.; Riegel, R.; Gomez, L.D.; Herrera, J.; Isla, A.; McQueen-Mason, S.J.; Calderini, D.F. Expansins expression is associated with grain size dynamics in wheat (Triticum aestivum L.). J. Exp. Bot. 2010, 61, 1147–1157. [Google Scholar] [CrossRef] [PubMed]
- Passardi, F.; Penel, C.; Dunand, C. Performing the paradoxical: How plant peroxidases modify the cell wall. Trends Plant Sci. 2004, 9, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Chen, Y.; Yin, S.; Zhang, M.; Wang, W. Over-expression of TaEXPB23, a wheat expansin gene, improves oxidative stress tolerance in transgenic tobacco plants. J. Plant Physiol. 2014, 173, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.E.O.; Gomes, V.M. Role of plant lipid transfer proteins in plant cell physiology—A concise review. Peptides 2007, 28, 1144–1153. [Google Scholar] [CrossRef] [PubMed]
- Yeats, T.H.; Rose, J.K. The biochemistry and biology of extracellular plant lipid-transfer proteins (LTPs). Protein Sci. 2008, 17, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Vissenberg, K.; Fry, S.C.; Pauly, M.; Höfte, H.; Verbelen, J.P. XTH acts at the microfibril-matrix interface during cell elongation. J. Exp. Bot. 2005, 56, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Strohmeier, M.; Hrmova, M.; Fischer, M.; Harvey, A.J.; Fincher, G.B.; Pleiss, J. Molecular modeling of family GH16 glycoside hydrolases: Potential roles for xyloglucan transglucosylases/hydrolases in cell wall modification in the poaceae. Protein Sci. 2004, 13, 3200–3213. [Google Scholar] [CrossRef] [PubMed]
- Hrmova, M.; Farkas, V.; Lahnstein, J.; Fincher, G.B. A barley xyloglucan xyloglucosyl transferase covalently links xyloglucan, cellulosic substrates, and (1,3;1,4)-β-d-glucans. J. Biol. Chem. 2007, 282, 12951–12962. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Burton, R.A.; Hrmova, M.; Fincher, G.B. Barley arabinoxylan arabinofuranohydrolases: Purification, characterization and determination of primary structures from cdna clones. Biochem. J. 2001, 356, 181–189. [Google Scholar] [CrossRef] [PubMed]


© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Francin-Allami, M.; Lollier, V.; Pavlovic, M.; San Clemente, H.; Rogniaux, H.; Jamet, E.; Guillon, F.; Larré, C. Understanding the Remodelling of Cell Walls during Brachypodium distachyon Grain Development through a Sub-Cellular Quantitative Proteomic Approach. Proteomes 2016, 4, 21. https://doi.org/10.3390/proteomes4030021
Francin-Allami M, Lollier V, Pavlovic M, San Clemente H, Rogniaux H, Jamet E, Guillon F, Larré C. Understanding the Remodelling of Cell Walls during Brachypodium distachyon Grain Development through a Sub-Cellular Quantitative Proteomic Approach. Proteomes. 2016; 4(3):21. https://doi.org/10.3390/proteomes4030021
Chicago/Turabian StyleFrancin-Allami, Mathilde, Virginie Lollier, Marija Pavlovic, Hélène San Clemente, Hélène Rogniaux, Elisabeth Jamet, Fabienne Guillon, and Colette Larré. 2016. "Understanding the Remodelling of Cell Walls during Brachypodium distachyon Grain Development through a Sub-Cellular Quantitative Proteomic Approach" Proteomes 4, no. 3: 21. https://doi.org/10.3390/proteomes4030021
APA StyleFrancin-Allami, M., Lollier, V., Pavlovic, M., San Clemente, H., Rogniaux, H., Jamet, E., Guillon, F., & Larré, C. (2016). Understanding the Remodelling of Cell Walls during Brachypodium distachyon Grain Development through a Sub-Cellular Quantitative Proteomic Approach. Proteomes, 4(3), 21. https://doi.org/10.3390/proteomes4030021