Omics Approaches for the Study of Adaptive Immunity to Staphylococcus aureus and the Selection of Vaccine Candidates
Abstract
:1. Introduction
- the in vivo behavior of S. aureus;
- the antibody and T-cell response against S. aureus; and
- vaccination development.
2. Challenges in Deciphering the Adaptive Immune Response to S. aureus
- the diversity and complexity of S. aureus host interactions;
- the impressive genetic variability of the species S. aureus;
- a deficit in infection-relevant in vitro and in vivo models; and
- the high variability of the anti-staphylococcal immune responses.
3. Omics Technologies in S. aureus Research
3.1. Genomics
3.2. Transcriptomics
3.3. Proteomics
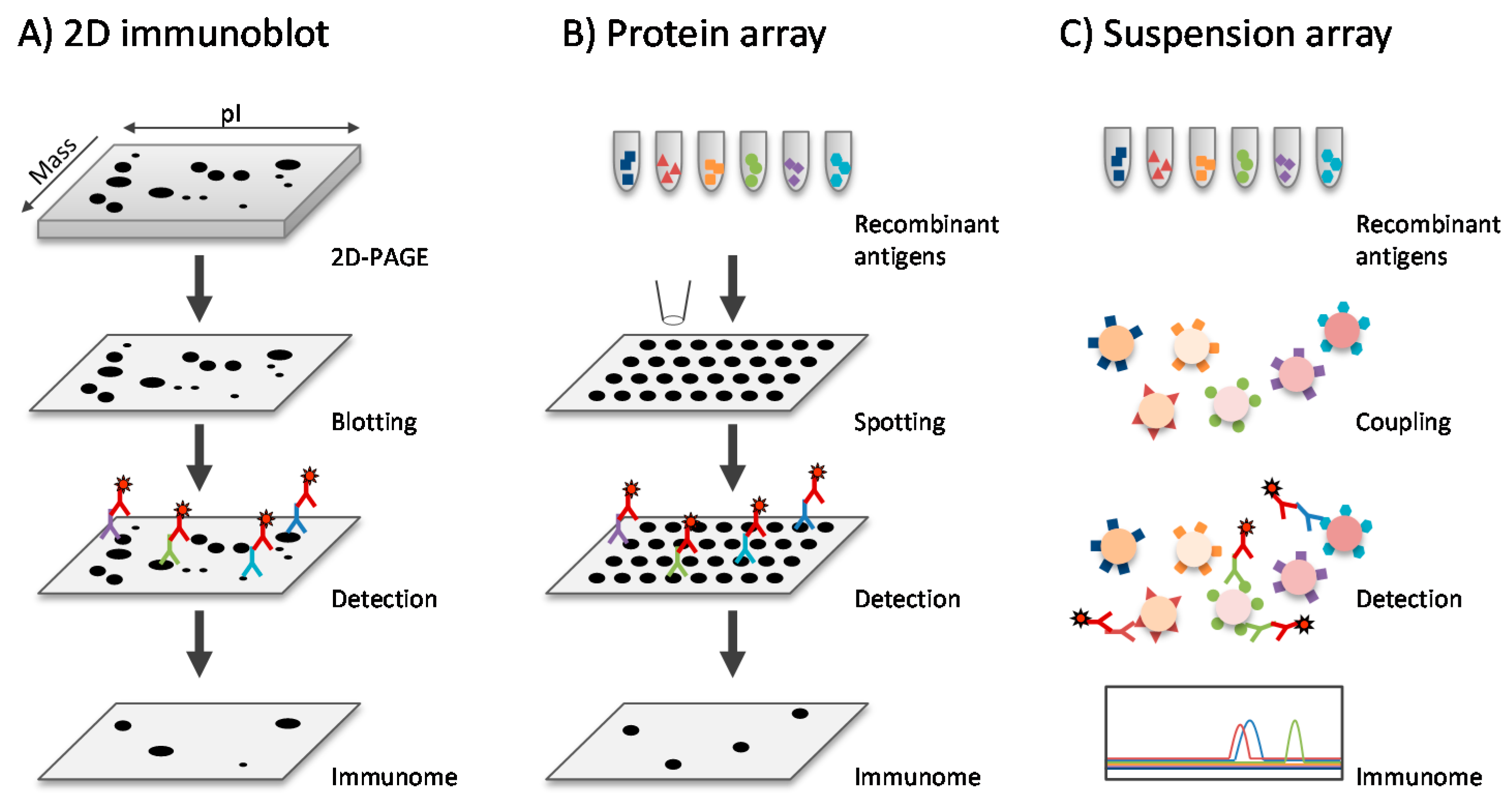
3.4. Immunoproteomics
3.4.1. Antibody Profiles in Healthy Individuals
3.4.2. Antibody Profiles during Infection
3.4.3. T-cell Responses to S. aureus
3.5. Metabolomics
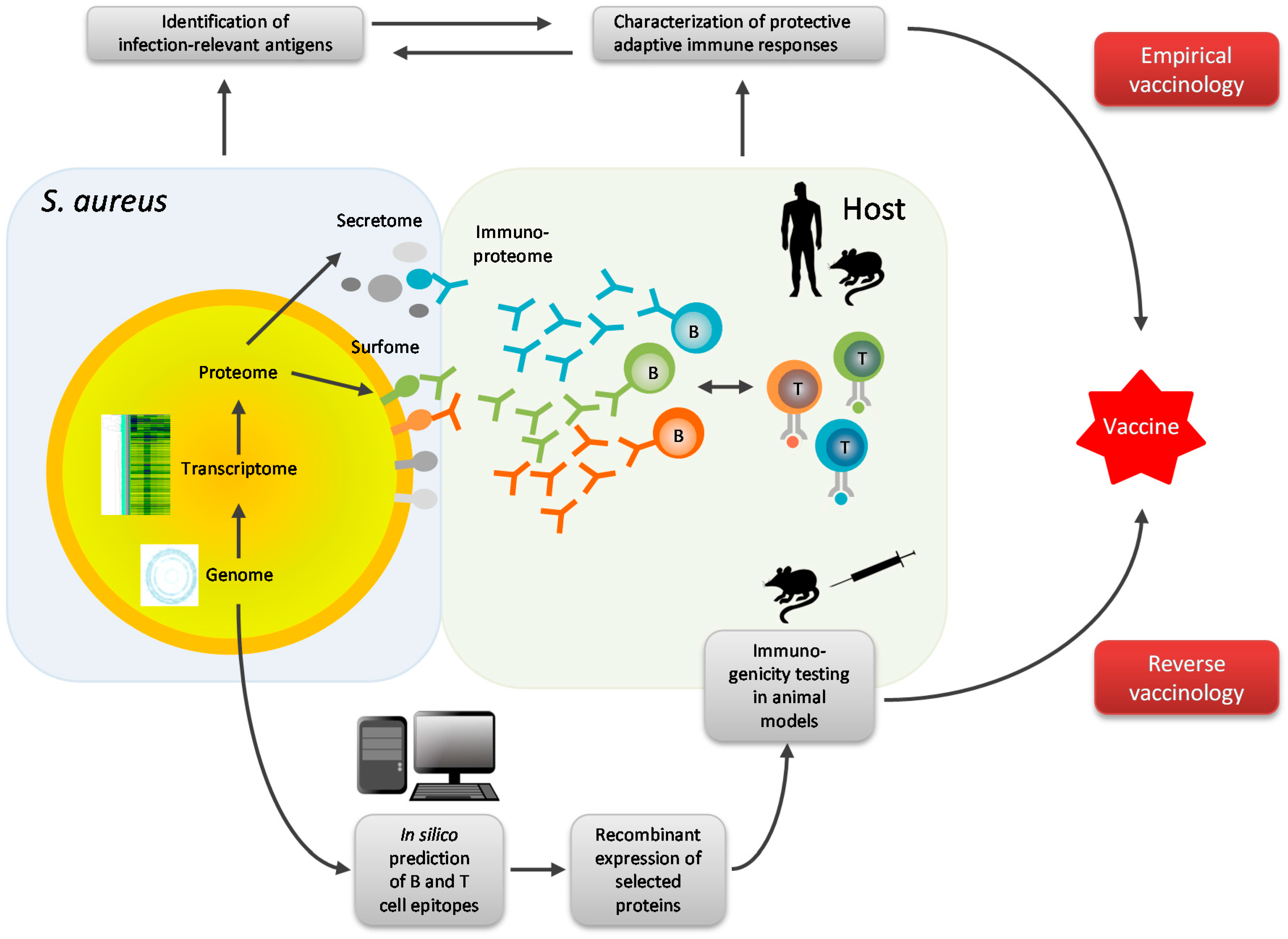
4. Omics Technologies in S. aureus Vaccine Development
5. Future Directions of Research
- (1)
- Improvement of important aspects of pre-analytics, such as the rapid enrichment of the pathogen from infected cells and tissues, as well as further increases of the sensitivity of detection methods, should ultimately permit the analysis of pathogen-host interactions directly ex vivo.
- (2)
- Interpretation of the “big data” generated by omics techniques relies on sophisticated bioinformatics and depends on the inter-disciplinary dialogue as well as on innovation and technical optimization in the field of computational statistics and bioinformatics.
- (3)
- Prospective clinical trials in well-defined patient cohorts will remain key to finding answers to the burning questions at hand. Such clinical studies should simultaneously consider the pathogen and the immune response, collecting bacterial strains as well as patient sera and immune cells.
- (4)
- Disease specific transcriptome and proteome profiles are required to explain the broad spectrum of diseases caused by the versatile species S. aureus and to develop targeted counter-measures.
- (5)
- Advanced omics technologies should be applied to study the adaptive immune response to S. aureus. Mapping of antigen, antibody and T-cell repertoires may reveal correlates of protection on which vaccination strategies can then be based.
- (6)
- The multi-omics-approach is very much focused on genes and proteins. Non-protein molecules, however, may be equally important in S. aureus-host interaction.
Acknowledgments
Conflicts of Interest
References
- Wertheim, H.F.; Melles, D.C.; Vos, M.C.; van Leeuwen, W.; van Belkum, A.; Verbrugh, H.A.; Nouwen, J.L. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005, 5, 751–762. [Google Scholar] [CrossRef]
- Lowy, F.D. Staphylococcus aureus Infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Hidron, A.I.; Edwards, J.R.; Patel, J.; Horan, T.C.; Sievert, D.M.; Pollock, D.A.; Fridkin, S.K.; National Healthcare Safety Network Team; Participating National Healthcare Safety Network Facilities. NHSN annual update: Antimicrobial-resistant pathogens associated with healthcare-associated infections: Annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect. Control Hosp. Epidemiol. 2008, 29, 996–1011. [Google Scholar] [CrossRef] [PubMed]
- Klevens, R.M.; Morrison, M.A.; Nadle, J.; Petit, S.; Gershman, K.; Ray, S.; Harrison, L.H.; Lynfield, R.; Dumyati, G.; Townes, J.M.; et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 2007, 298, 1763–1771. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.N. Microbial etiologies of hospital-acquired bacterial pneumonia and ventilator-associated bacterial pneumonia. Clin. Infect. Dis. 2010, 51 (Suppl. 1), S81–S87. [Google Scholar] [CrossRef] [PubMed]
- David, M.Z.; Daum, R.S. Community-associated methicillin-resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 2010, 23, 616–687. [Google Scholar] [CrossRef] [PubMed]
- De Kraker, M.E.; Wolkewitz, M.; Davey, P.G.; Koller, W.; Berger, J.; Nagler, J.; Icket, C.; Kalenic, S.; Horvatic, J.; Seifert, H.; et al. Clinical impact of antimicrobial resistance in European hospitals: Excess mortality and length of hospital stay related to methicillin-resistant Staphylococcus aureus bloodstream infections. Antimicrob. Agents Chemother. 2011, 55, 1598–1605. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, S.E.; Sakoulas, G.; Perencevich, E.N.; Schwaber, M.J.; Karchmer, A.W.; Carmeli, Y. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: A meta-analysis. Clin. Infect. Dis. 2003, 36, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Boucher, H.W.; Corey, G.R. Epidemiology of Methicillin-Resistant Staphylococcus aureus. Clin. Infect. Dis. 2008, 46, S344–S349. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Daum, R.S.; Spellberg, B. Progress Toward a Staphylococcus aureus Vaccine. Clin. Infect. Dis. 2012, 54, 560–567. [Google Scholar] [CrossRef] [PubMed]
- Proctor, R.A. Challenges for a universal Staphylococcus aureus vaccine. Clin. Infect. Dis. 2012, 54, 1179–1186. [Google Scholar] [CrossRef] [PubMed]
- Fowler, V.G., Jr.; Proctor, R.A. Where does a Staphylococcus aureus vaccine stand? Clin. Microbiol. Infect. 2014, 20 (Suppl. 5), 66–75. [Google Scholar] [CrossRef] [PubMed]
- Fleischmann, R.D.; Adams, M.D.; White, O.; Clayton, R.A.; Kirkness, E.F.; Kerlavage, A.R.; Bult, C.J.; Tomb, J.F.; Dougherty, B.A.; Merrick, J.M.; et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 1995, 269, 496–512. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A.; et al. The sequence of the human genome. Science 2001, 291, 1304–1351. [Google Scholar] [CrossRef] [PubMed]
- Van Belkum, A.; Verkaik, N.J.; de Vogel, C.P.; Boelens, H.A.; Verveer, J.; Nouwen, J.L.; Verbrugh, H.A.; Wertheim, H.F. Reclassification of Staphylococcus aureus nasal carriage types. J. Infect. Dis. 2009, 15, 1820–1826. [Google Scholar] [CrossRef] [PubMed]
- Wertheim, H.F.; Vos, M.C.; Ott, A.; van Belkum, A.; Voss, A.; Kluytmans, J.A.; van Keulen, P.H.; Vandenbroucke-Grauls, C.M.; Meester, M.H.; Verbrugh, H.A. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 2004, 364, 703–705. [Google Scholar] [CrossRef]
- Von Eiff, C.; Becker, K.; Machka, K.; Stammer, H.; Peters, G. Nasal Carriage as a Source of Staphylococcus aureus Bacteremia. N. Engl. J. Med. 2001, 344, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Wertheim, H.F.; Vos, M.C.; Ott, A.; Voss, A.; Kluytmans, J.A.; Vandenbroucke-Grauls, C.M.; Meester, M.H.; van Keulen, P.H.; Verbrugh, H.A. Mupirocin prophylaxis against nosocomial Staphylococcus aureus infections in nonsurgical patients: A randomized study. Ann. Intern. Med. 2004, 140, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Bode, L.G.; Kluytmans, J.A.; Wertheim, H.F.; Bogaers, D.; Vandenbroucke-Grauls, C.M.; Roosendaal, R.; Troelstra, A.; Box, A.T.; Voss, A.; van der Tweel, I.; et al. Preventing surgical-site infections in nasal carriers of Staphylococcus aureus. N. Engl. J. Med. 2010, 362, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Löffler, B.; Tuchscherr, L.; Niemann, S.; Peters, G. Staphylococcus aureus persistence in non-professional phagocytes. Int. J. Med. Microbiol. 2014, 304, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Tuchscherr, L.; Medina, E.; Hussain, M.; Volker, W.; Heitmann, V.; Niemann, S.; Holzinger, D.; Roth, J.; Proctor, R.A.; Becker, K.; et al. Staphylococcus aureus phenotype switching: An effective bacterial strategy to escape host immune response and establish a chronic infection. EMBO Mol. Med. 2011, 3, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, J.A.; Holden, M.T. Staphylococcus aureus: Superbug, super genome? Trends Microbiol. 2004, 12, 378–385. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, J.A.; Moore, C.E.; Day, N.P.; Peacock, S.J.; Witney, A.A.; Stabler, R.A.; Husain, S.E.; Butcher, P.D.; Hinds, J. Microarrays Reveal that Each of the Ten Dominant Lineages of Staphylococcus aureus Has a Unique Combination of Surface-Associated and Regulatory Genes. J. Bacteriol. 2006, 188, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Stranger-Jones, Y.K.; Bae, T.; Schneewind, O. Vaccine assembly from surface proteins of Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2006, 103, 16942–16947. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Q.F.; Yang, L.Y.; Feng, Q.; Lu, D.S.; Dong, Y.D.; Cai, C.Z.; Wu, Y.; Guo, Y.; Gu, J.; Zeng, H.; et al. Evaluation of the protective immunity of a novel subunit fusion vaccine in a murine model of systemic MRSA infection. PLoS ONE 2013, 8, e81212. [Google Scholar] [CrossRef] [PubMed]
- Spaulding, A.R.; Salgado-Pabon, W.; Merriman, J.A.; Stach, C.S.; Ji, Y.; Gillman, A.N.; Peterson, M.L.; Schlievert, P.M. Vaccination against Staphylococcus aureus pneumonia. J. Infect. Dis. 2014, 209, 1955–1962. [Google Scholar] [CrossRef] [PubMed]
- Burian, M.; Rautenberg, M.; Kohler, T.; Fritz, M.; Krismer, B.; Unger, C.; Hoffmann, W.H.; Peschel, A.; Wolz, C.; Goerke, C. Temporal expression of adhesion factors and activity of global regulators during establishment of Staphylococcus aureus nasal colonization. J. Infect. Dis. 2010, 201, 1414–1421. [Google Scholar] [CrossRef] [PubMed]
- Yarwood, J.M.; McCormick, J.K.; Paustian, M.L.; Kapur, V.; Schlievert, P.M. Repression of the Staphylococcus aureus Accessory Gene Regulator in Serum and In Vivo. J. Bacteriol. 2002, 184, 1095–1101. [Google Scholar] [CrossRef] [PubMed]
- Pragman, A.A.; Schlievert, P.M. Virulence regulation in Staphylococcus aureus: The need for in vivo analysis of virulence factor regulation. FEMS Immunol. Med. Microbiol. 2004, 42, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Hanses, F.; Roux, C.; Dunman, P.M.; Salzberger, B.; Lee, J.C. Staphylococcus aureus gene expression in a rat model of infective endocarditis. Genome Med. 2014, 6, 93. [Google Scholar] [CrossRef] [PubMed]
- Loffler, B.; Hussain, M.; Grundmeier, M.; Bruck, M.; Holzinger, D.; Varga, G.; Roth, J.; Kahl, B.C.; Proctor, R.A.; Peters, G. Staphylococcus aureus panton-valentine leukocidin is a very potent cytotoxic factor for human neutrophils. PLoS Pathog. 2010, 6, e1000715. [Google Scholar] [CrossRef] [PubMed]
- Rooijakkers, S.H.; Ruyken, M.; Roos, A.; Daha, M.R.; Presanis, J.S.; Sim, R.B.; van Wamel, W.J.; van Kessel, K.P.; van Strijp, J.A. Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat. Immunol. 2005, 6, 920–927. [Google Scholar] [CrossRef] [PubMed]
- Rooijakkers, S.H.; van Kessel, K.P.; van Strijp, J.A. Staphylococcal innate immune evasion. Trends Microbiol. 2005, 13, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Langley, R.; Wines, B.; Willoughby, N.; Basu, I.; Proft, T.; Fraser, J.D. The staphylococcal superantigen-like protein 7 binds IgA and complement C5 and inhibits IgA-Fc alpha RI binding and serum killing of bacteria. J. Immunol. 2005, 174, 2926–2933. [Google Scholar] [CrossRef] [PubMed]
- Salgado-Pabon, W.; Schlievert, P.M. Models matter: The search for an effective Staphylococcus aureus vaccine. Nat. Rev. Microbiol. 2014, 12, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Broker, B.M.; Holtfreter, S.; Bekeredjian-Ding, I. Immune control of Staphylococcus aureus—Regulation and counter-regulation of the adaptive immune response. Int. J. Med. Microbiol. 2014, 304, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Holtfreter, S.; Kolata, J.; Broker, B.M. Towards the immune proteome of Staphylococcus aureus—The anti-S. aureus antibody response. Int. J. Med. Microbiol. 2010, 300, 176–192. [Google Scholar] [CrossRef] [PubMed]
- Holtfreter, S.; Nguyen, T.T.; Wertheim, H.; Steil, L.; Kusch, H.; Truong, Q.P.; Engelmann, S.; Hecker, M.; Volker, U.; van Belkum, A.; et al. Human immune proteome in experimental colonization with Staphylococcus aureus. Clin. Vaccine Immunol. 2009, 16, 1607–1614. [Google Scholar] [CrossRef] [PubMed]
- Verkaik, N.J.; de Vogel, C.P.; Boelens, H.A.; Grumann, D.; Hoogenboezem, T.; Vink, C.; Hooijkaas, H.; Foster, T.J.; Verbrugh, H.A.; van Belkum, A.; et al. Anti-staphylococcal humoral immune response in persistent nasal carriers and noncarriers of Staphylococcus aureus. J. Infect. Dis. 2009, 199, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Oie, S.; Kamiya, A. Survival of methicillin-resistant Staphylococcus aureus (MRSA) on naturally contaminated dry mops. J. Hosp. Infect. 1996, 34, 145–149. [Google Scholar] [CrossRef]
- O’Connell, N.H.; Humphreys, H. Intensive care unit design and environmental factors in the acquisition of infection. J. Hosp. Infect. 2000, 45, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Koser, C.U.; Holden, M.T.; Ellington, M.J.; Cartwright, E.J.; Brown, N.M.; Ogilvy-Stuart, A.L.; Hsu, L.Y.; Chewapreecha, C.; Croucher, N.J.; Harris, S.R.; et al. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N. Engl. J. Med. 2012, 366, 2267–2275. [Google Scholar] [CrossRef] [PubMed]
- Paterson, G.K.; Harrison, E.M.; Murray, G.G.; Welch, J.J.; Warland, J.H.; Holden, M.T.; Morgan, F.J.; Ba, X.; Koop, G.; Harris, S.R.; et al. Capturing the cloud of diversity reveals complexity and heterogeneity of MRSA carriage, infection and transmission. Nat. Commun. 2015, 6, 6560. [Google Scholar] [CrossRef] [PubMed]
- Laabei, M.; Recker, M.; Rudkin, J.K.; Aldeljawi, M.; Gulay, Z.; Sloan, T.J.; Williams, P.; Endres, J.L.; Bayles, K.W.; Fey, P.D.; et al. Predicting the virulence of MRSA from its genome sequence. Genome Res. 2014, 24, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Holden, M.T.; Hsu, L.Y.; Kurt, K.; Weinert, L.A.; Mather, A.E.; Harris, S.R.; Strommenger, B.; Layer, F.; Witte, W.; de Lencastre, H.; et al. A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res. 2013, 23, 653–664. [Google Scholar] [CrossRef] [PubMed]
- Harris, S.R.; Feil, E.J.; Holden, M.T.; Quail, M.A.; Nickerson, E.K.; Chantratita, N.; Gardete, S.; Tavares, A.; Day, N.; Lindsay, J.A.; et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science 2010, 327, 469–474. [Google Scholar] [CrossRef] [PubMed]
- DeLeo, F.R.; Kennedy, A.D.; Chen, L.; Wardenburg, J.B.; Kobayashi, S.D.; Mathema, B.; Braughton, K.R.; Whitney, A.R.; Villaruz, A.E.; Martens, C.A.; et al. Molecular differentiation of historic phage-type 80/81 and contemporary epidemic Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2011, 108, 18091–18096. [Google Scholar] [CrossRef] [PubMed]
- Kurt, K.; Rasigade, J.P.; Laurent, F.; Goering, R.V.; Zemlickova, H.; Machova, I.; Struelens, M.J.; Zautner, A.E.; Holtfreter, S.; Bröker, B.; et al. Subpopulations of Staphylococcus aureus clonal complex 121 are associated with distinct clinical entities. PLoS ONE 2013, 8, e58155. [Google Scholar] [CrossRef] [PubMed]
- McAdam, P.R.; Templeton, K.E.; Edwards, G.F.; Holden, M.T.; Feil, E.J.; Aanensen, D.M.; Bargawi, H.J.; Spratt, B.G.; Bentley, S.D.; Parkhill, J.; et al. Molecular tracing of the emergence, adaptation, and transmission of hospital-associated methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2012, 109, 9107–9112. [Google Scholar] [CrossRef] [PubMed]
- Luedicke, C.; Slickers, P.; Ehricht, R.; Monecke, S. Molecular fingerprinting of Staphylococcus aureus from bone and joint infections. Eur. J. Clin. Microbiol. Infect. Dis. 2010, 29, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Monecke, S.; Slickers, P.; Hotzel, H.; Richter-Huhn, G.; Pohle, M.; Weber, S.; Witte, W.; Ehricht, R. Microarray-based characterisation of a Panton-Valentine leukocidin-positive community-acquired strain of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 2006, 12, 718–728. [Google Scholar] [CrossRef] [PubMed]
- Monecke, S.; Luedicke, C.; Slickers, P.; Ehricht, R. Molecular epidemiology of Staphylococcus aureus in asymptomatic carriers. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 1159–1165. [Google Scholar] [CrossRef] [PubMed]
- Masoud-Landgraf, L.; Johler, S.; Badura, A.; Feierl, G.; Luxner, J.; Wagner-Eibel, U.; Eber, E.; Zarfel, G.; Grisold, A.J. Genetic and Phenotypic Characteristics of Staphylococcus aureus Isolates from Cystic Fibrosis Patients in Austria. Respir. Int. Rev. Thorac. Dis. 2015, 89, 390–395. [Google Scholar] [CrossRef] [PubMed]
- Monecke, S.; Ehricht, R. Rapid genotyping of methicillin-resistant Staphylococcus aureus (MRSA) isolates using miniaturised oligonucleotide arrays. Clin. Microbiol. Infect. 2005, 11, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Dunman, P.M.; Murphy, E.; Haney, S.; Palacios, D.; Tucker-Kellogg, G.; Wu, S.; Brown, E.L.; Zagursky, R.J.; Shlaes, D.; Projan, S.J. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 2001, 183, 7341–7353. [Google Scholar] [CrossRef] [PubMed]
- Witney, A.A.; Marsden, G.L.; Holden, M.T.; Stabler, R.A.; Husain, S.E.; Vass, J.K.; Butcher, P.D.; Hinds, J.; Lindsay, J.A. Design, validation, and application of a seven-strain Staphylococcus aureus PCR product microarray for comparative genomics. Appl. Environ. Microbiol. 2005, 71, 7504–7514. [Google Scholar] [CrossRef] [PubMed]
- McManus, B.A.; Coleman, D.C.; Deasy, E.C.; Brennan, G.I.; O’ Connell, B.; Monecke, S.; Ehricht, R.; Leggett, B.; Leonard, N.; Shore, A.C. Comparative Genotypes, Staphylococcal Cassette Chromosome mec (SCCmec) Genes and Antimicrobial Resistance amongst Staphylococcus epidermidis and Staphylococcus haemolyticus Isolates from Infections in Humans and Companion Animals. PLoS ONE 2015, 10, e0138079. [Google Scholar] [CrossRef] [PubMed]
- Thunberg, U.; Hugosson, S.; Monecke, S.; Ehricht, R.; Soderquist, B. Molecular characteristics of Staphylococcus aureus associated with chronic rhinosinusitis. APMIS 2015, 123, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, G.; Monecke, S.; Brus, O.; Ehricht, R.; Soderquist, B. Long term molecular epidemiology of methicillin-susceptible Staphylococcus aureus bacteremia isolates in Sweden. PLoS ONE 2014, 9, e114276. [Google Scholar] [CrossRef] [PubMed]
- Shore, A.C.; Brennan, O.M.; Deasy, E.C.; Rossney, A.S.; Kinnevey, P.M.; Ehricht, R.; Monecke, S.; Coleman, D.C. DNA microarray profiling of a diverse collection of nosocomial methicillin-resistant staphylococcus aureus isolates assigns the majority to the correct sequence type and staphylococcal cassette chromosome mec (SCCmec) type and results in the subsequent identification and characterization of novel SCCmec-SCCM1 composite islands. Antimicrob. Agents Chemother. 2012, 56, 5340–5355. [Google Scholar] [PubMed]
- Monecke, S.; Engelmann, I.; Archambault, M.; Coleman, D.C.; Coombs, G.W.; Cortez de Jackel, S.; Pelletier-Jacques, G.; Schwarz, S.; Shore, A.C.; Slickers, P.; et al. Distribution of SCCmec-associated phenol-soluble modulin in staphylococci. Mol. Cell. Probes 2012, 26, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.K.; Rose, W.; Schrodi, S.J. Complex host genetic susceptibility to Staphylococcus aureus infections. Trends Microbiol. 2015, 23, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Emonts, M.; Uitterlinden, A.G.; Nouwen, J.L.; Kardys, I.; Maat, M.P.; Melles, D.C.; Witteman, J.; Jong, P.T.; Verbrugh, H.A.; Hofman, A.; et al. Host polymorphisms in interleukin 4, complement factor H, and C-reactive protein associated with nasal carriage of Staphylococcus aureus and occurrence of boils. J. Infect. Dis. 2008, 197, 1244–1253. [Google Scholar] [CrossRef] [PubMed]
- Nurjadi, D.; Herrmann, E.; Hinderberger, I.; Zanger, P. Impaired beta-defensin expression in human skin links DEFB1 promoter polymorphisms with persistent Staphylococcus aureus nasal carriage. J. Infect. Dis. 2013, 207, 666–674. [Google Scholar] [CrossRef] [PubMed]
- Ruimy, R.; Angebault, C.; Djossou, F.; Dupont, C.; Epelboin, L.; Jarraud, S.; Lefevre, L.A.; Bes, M.; Lixandru, B.E.; Bertine, M.; et al. Are host genetics the predominant determinant of persistent nasal Staphylococcus aureus carriage in humans? J. Infect. Dis. 2010, 202, 924–934. [Google Scholar] [CrossRef] [PubMed]
- van den Akker, E.L.; Nouwen, J.L.; Melles, D.C.; van Rossum, E.F.; Koper, J.W.; Uitterlinden, A.G.; Hofman, A.; Verbrugh, H.A.; Pols, H.A.; Lamberts, S.W.; et al. Staphylococcus aureus nasal carriage is associated with glucocorticoid receptor gene polymorphisms. J. Infect. Dis. 2006, 194, 814–818. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.L.; Pelak, K.; Podgoreanu, M.V.; Ahn, S.H.; Scott, W.K.; Allen, A.S.; Cowell, L.G.; Rude, T.H.; Zhang, Y.; Tong, A.; et al. A genome-wide association study of variants associated with acquisition of Staphylococcus aureus bacteremia in a healthcare setting. BMC Infect Dis 2014, 14, 83. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Vasco, D.A.; Carter, T.C.; Brilliant, M.H.; Schrodi, S.J.; Shukla, S.K. Genome wide association study of SNP-, gene-, and pathway-based approaches to identify genes influencing susceptibility to Staphylococcus aureus infections. Front. Genet. 2014, 5, 125. [Google Scholar] [CrossRef] [PubMed]
- Cormier, C.; Mfuna Endam, L.; Filali-Mouhim, A.; Boisvert, P.; Boulet, L.P.; Boulay, M.E.; Vallee-Smedja, S.; Bosse, Y.; Desrosiers, M. A pooling-based genomewide association study identifies genetic variants associated with Staphylococcus aureus colonization in chronic rhinosinusitis patients. Int. Forum Allergy Rhinol. 2014, 4, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Six, A.; Mariotti-Ferrandiz, M.E.; Chaara, W.; Magadan, S.; Pham, H.P.; Lefranc, M.P.; Mora, T.; Thomas-Vaslin, V.; Walczak, A.M.; Boudinot, P. The past, present, and future of immune repertoire biology—The rise of next-generation repertoire analysis. Front. Immunol. 2013, 4, 413. [Google Scholar] [CrossRef] [PubMed]
- Han, A.; Glanville, J.; Hansmann, L.; Davis, M.M. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat. Biotechnol. 2014, 32, 684–692. [Google Scholar] [CrossRef] [PubMed]
- Newell, E.W.; Davis, M.M. Beyond model antigens: High-dimensional methods for the analysis of antigen-specific T-cells. Nat. Biotechnol. 2014, 32, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Calis, J.J.; Rosenberg, B.R. Characterizing immune repertoires by high throughput sequencing: Strategies and applications. Trends Immunol. 2014, 35, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Gao, G.; Li, Z.; Sun, W.; Li, X.; Chen, N.; Sun, J.; Yang, Y. Profiling the T-cell receptor repertoire of patient with pleural tuberculosis by high-throughput sequencing. Immunol. Lett. 2014, 162, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Diluvio, L.; Vollmer, S.; Besgen, P.; Ellwart, J.W.; Chimenti, S.; Prinz, J.C. Identical TCR beta-chain rearrangements in streptococcal angina and skin lesions of patients with psoriasis vulgaris. J. Immunol. 2006, 176, 7104–7111. [Google Scholar] [CrossRef] [PubMed]
- Westermann, A.J.; Forstner, K.U.; Amman, F.; Barquist, L.; Chao, Y.; Schulte, L.N.; Muller, L.; Reinhardt, R.; Stadler, P.F.; Vogel, J. Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature 2016, 529, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Westermann, A.J.; Gorski, S.A.; Vogel, J. Dual RNA-seq of pathogen and host. Nat. Rev. Microbiol. 2012, 10, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Humphrys, M.S.; Creasy, T.; Sun, Y.; Shetty, A.C.; Chibucos, M.C.; Drabek, E.F.; Fraser, C.M.; Farooq, U.; Sengamalay, N.; Ott, S.; et al. Simultaneous transcriptional profiling of bacteria and their host cells. PLoS ONE 2013, 8, e80597. [Google Scholar] [CrossRef] [PubMed]
- Chaves-Moreno, D.; Wos-Oxley, M.L.; Jauregui, R.; Medina, E.; Oxley, A.P.A.; Pieper, D.H. Application of a Novel “Pan-Genome”-Based Strategy for Assigning RNAseq Transcript Reads to Staphylococcus aureus Strains. PLoS ONE 2015, 10, e0145861. [Google Scholar]
- Cui, L.; Lian, J.Q.; Neoh, H.M.; Reyes, E.; Hiramatsu, K. DNA microarray-based identification of genes associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2005, 49, 3404–3413. [Google Scholar] [CrossRef] [PubMed]
- Stevens, D.L.; Ma, Y.; Salmi, D.B.; McIndoo, E.; Wallace, R.J.; Bryant, A.E. Impact of antibiotics on expression of virulence-associated exotoxin genes in methicillin-sensitive and methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 2007, 195, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.; Yang, S.J.; Bayer, A.S.; Vaezzadeh, A.R.; Herzig, S.; Stenz, L.; Girard, M.; Sakoulas, G.; Scherl, A.; Yeaman, M.R.; et al. Daptomycin resistance mechanisms in clinically derived Staphylococcus aureus strains assessed by a combined transcriptomics and proteomics approach. J. Antimicrob. Chemother. 2011, 66, 1696–1711. [Google Scholar] [CrossRef] [PubMed]
- Reiss, S.; Pane-Farre, J.; Fuchs, S.; Francois, P.; Liebeke, M.; Schrenzel, J.; Lindequist, U.; Lalk, M.; Wolz, C.; Hecker, M.; et al. Global analysis of the Staphylococcus aureus response to mupirocin. Antimicrob. Agents Chemother. 2012, 56, 787–804. [Google Scholar] [CrossRef] [PubMed]
- Palazzolo-Ballance, A.M.; Reniere, M.L.; Braughton, K.R.; Sturdevant, D.E.; Otto, M.; Kreiswirth, B.N.; Skaar, E.P.; DeLeo, F.R. Neutrophil microbicides induce a pathogen survival response in community-associated methicillin-resistant Staphylococcus aureus. J. Immunol. 2008, 180, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.; Small, D.A.; Toghrol, F.; Bentley, W.E. Global transcriptome analysis of Staphylococcus aureus response to hydrogen peroxide. J. Bacteriol. 2006, 188, 1648–1659. [Google Scholar] [CrossRef] [PubMed]
- Ishii, K.; Adachi, T.; Yasukawa, J.; Suzuki, Y.; Hamamoto, H.; Sekimizu, K. Induction of virulence gene expression in Staphylococcus aureus by pulmonary surfactant. Infect. Immun. 2014, 82, 1500–1510. [Google Scholar] [CrossRef] [PubMed]
- Windmuller, N.; Witten, A.; Block, D.; Bunk, B.; Sproer, C.; Kahl, B.C.; Mellmann, A. Transcriptional adaptations during long-term persistence of Staphylococcus aureus in the airways of a cystic fibrosis patient. Int. J. Med. Microbiol. 2015, 305, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Santiago-Rodriguez, T.M.; Naidu, M.; Jones, M.B.; Ly, M.; Pride, D.T. Identification of staphylococcal phage with reduced transcription in human blood through transcriptome sequencing. Front. Microbiol. 2015, 6, 216. [Google Scholar] [CrossRef] [PubMed]
- Oogai, Y.; Matsuo, M.; Hashimoto, M.; Kato, F.; Sugai, M.; Komatsuzawa, H. Expression of virulence factors by Staphylococcus aureus grown in serum. Appl. Environ. Microbiol. 2011, 77, 8097–8105. [Google Scholar] [CrossRef] [PubMed]
- Malachowa, N.; Kobayashi, S.D.; Sturdevant, D.E.; Scott, D.P.; DeLeo, F.R. Insights into the Staphylococcus aureus-host interface: Global changes in host and pathogen gene expression in a rabbit skin infection model. PLoS ONE 2015, 10, e0117713. [Google Scholar] [CrossRef] [PubMed]
- Resch, A.; Rosenstein, R.; Nerz, C.; Gotz, F. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl. Environ. Microbiol. 2005, 71, 2663–2676. [Google Scholar] [CrossRef] [PubMed]
- Seggewiss, J.; Becker, K.; Kotte, O.; Eisenacher, M.; Yazdi, M.R.; Fischer, A.; McNamara, P.; Al Laham, N.; Proctor, R.; Peters, G.; et al. Reporter metabolite analysis of transcriptional profiles of a Staphylococcus aureus strain with normal phenotype and its isogenic hemB mutant displaying the small-colony-variant phenotype. J. Bacteriol. 2006, 188, 7765–7777. [Google Scholar] [CrossRef] [PubMed]
- Garzoni, C.; Francois, P.; Huyghe, A.; Couzinet, S.; Tapparel, C.; Charbonnier, Y.; Renzoni, A.; Lucchini, S.; Lew, D.P.; Vaudaux, P.; et al. A global view of Staphylococcus aureus whole genome expression upon internalization in human epithelial cells. BMC Genom. 2007, 8, 171. [Google Scholar] [CrossRef] [PubMed]
- Scherr, T.D.; Roux, C.M.; Hanke, M.L.; Angle, A.; Dunman, P.M.; Kielian, T. Global transcriptome analysis of Staphylococcus aureus biofilms in response to innate immune cells. Infect. Immun. 2013, 81, 4363–4376. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xiu, L.; Hu, Q.; Cui, X.; Liu, B.; Tao, L.; Wang, T.; Wu, J.; Chen, Y.; Chen, Y. Deep sequencing-based transcriptional analysis of bovine mammary epithelial cells gene expression in response to in vitro infection with Staphylococcus aureus stains. PLoS ONE 2013, 8, e82117. [Google Scholar] [CrossRef] [PubMed]
- Lewandowska-Sabat, A.M.; Boman, G.M.; Downing, A.; Talbot, R.; Storset, A.K.; Olsaker, I. The early phase transcriptome of bovine monocyte-derived macrophages infected with Staphylococcus aureus in vitro. BMC Genom. 2013, 14, 891. [Google Scholar] [CrossRef] [PubMed]
- Koziel, J.; Maciag-Gudowska, A.; Mikolajczyk, T.; Bzowska, M.; Sturdevant, D.E.; Whitney, A.R.; Shaw, L.N.; DeLeo, F.R.; Potempa, J. Phagocytosis of Staphylococcus aureus by macrophages exerts cytoprotective effects manifested by the upregulation of antiapoptotic factors. PLoS ONE 2009, 4, e5210. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.D.; Braughton, K.R.; Palazzolo-Ballance, A.M.; Kennedy, A.D.; Sampaio, E.; Kristosturyan, E.; Whitney, A.R.; Sturdevant, D.E.; Dorward, D.W.; Holland, S.M.; et al. Rapid neutrophil destruction following phagocytosis of Staphylococcus aureus. J. Innate Immun. 2010, 2, 560–575. [Google Scholar] [CrossRef] [PubMed]
- Toufeer, M.; Bonnefont, C.M.; Foulon, E.; Caubet, C.; Tasca, C.; Aurel, M.R.; Robert-Granie, C.; Rupp, R.; Foucras, G. Gene expression profiling of dendritic cells reveals important mechanisms associated with predisposition to Staphylococcus infections. PLoS ONE 2011, 6, e22147. [Google Scholar] [CrossRef] [PubMed]
- Brady, R.A.; Bruno, V.M.; Burns, D.L. RNA-Seq Analysis of the Host Response to Staphylococcus aureus Skin and Soft Tissue Infection in a Mouse Model. PLoS ONE 2015, 10, e0124877. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhang, C.L.; Liao, X.X.; Chen, D.; Wang, W.Q.; Zhu, Y.H.; Geng, X.H.; Ji, D.J.; Mao, Y.J.; Gong, Y.C.; et al. Transcriptome microRNA profiling of bovine mammary glands infected with Staphylococcus aureus. Int. J. Mol. Sci. 2015, 16, 4997–5013. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Feng, G.; Guo, Q.; Wardenburg, J.B.; Lin, S.; Inoshima, I.; Deaton, R.; Yuan, J.X.; Garcia, J.G.; Machado, R.F.; et al. Transcriptional events during the recovery from MRSA lung infection: A mouse pneumonia model. PLoS ONE 2013, 8, e70176. [Google Scholar] [CrossRef] [PubMed]
- Depke, M.; Burian, M.; Schafer, T.; Broker, B.M.; Ohlsen, K.; Volker, U. The alternative sigma factor B modulates virulence gene expression in a murine Staphylococcus aureus infection model but does not influence kidney gene expression pattern of the host. Int. J. Med. Microbiol. 2012, 302, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Perez Novo, C.A.; Jedrzejczak-Czechowicz, M.; Lewandowska-Polak, A.; Claeys, C.; Holtappels, G.; Van Cauwenberge, P.; Kowalski, M.L.; Bachert, C. T-cell inflammatory response, Foxp3 and TNFRS18-L regulation of peripheral blood mononuclear cells from patients with nasal polyps-asthma after staphylococcal superantigen stimulation. Clin. Exp. Allergy 2010, 40, 1323–1332. [Google Scholar] [CrossRef] [PubMed]
- Niebuhr, M.; Scharonow, H.; Gathmann, M.; Mamerow, D.; Werfel, T. Staphylococcal exotoxins are strong inducers of IL-22: A potential role in atopic dermatitis. J. Allergy Clin. Immunol. 2010, 126, 1176–1183.e4. [Google Scholar] [CrossRef] [PubMed]
- Matsui, K.; Nishikawa, A. Effects of the macrolide antibiotic, midecamycin, on Staphylococcus aureus product-induced Th2 cytokine response in patients with atopic dermatitis. J. Interferon Cytokine Res. 2004, 24, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Matsui, K.; Nishikawa, A. Percutaneous application of peptidoglycan from Staphylococcus aureus induces infiltration of CCR4+ cells into mouse skin. J. Investig. Allergol. Clin. Immunol. 2011, 21, 354–362. [Google Scholar] [PubMed]
- Breuer, K.; Wittmann, M.; Kempe, K.; Kapp, A.; Mai, U.; Dittrich-Breiholz, O.; Kracht, M.; Mrabet-Dahbi, S.; Werfel, T. Alpha-toxin is produced by skin colonizing Staphylococcus aureus and induces a T helper type 1 response in atopic dermatitis. Clin. Exp. Allergy 2005, 35, 1088–1095. [Google Scholar] [CrossRef] [PubMed]
- Burian, M.; Grumann, D.; Holtfreter, S.; Wolz, C.; Goerke, C.; Bröker, B.M. Expression of staphylococcal superantigens during nasal colonization is not sufficient to induce a systemic neutralizing antibody response in humans. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Krismer, B.; Liebeke, M.; Janek, D.; Nega, M.; Rautenberg, M.; Hornig, G.; Unger, C.; Weidenmaier, C.; Lalk, M.; Peschel, A. Nutrient limitation governs Staphylococcus aureus metabolism and niche adaptation in the human nose. PLoS Pathog. 2014, 10, e1003862. [Google Scholar] [CrossRef] [PubMed]
- Chaffin, D.O.; Taylor, D.; Skerrett, S.J.; Rubens, C.E. Changes in the Staphylococcus aureus transcriptome during early adaptation to the lung. PLoS ONE 2012, 7, e41329. [Google Scholar] [CrossRef] [PubMed]
- Szafranska, A.K.; Oxley, A.P.; Chaves-Moreno, D.; Horst, S.A.; Rosslenbroich, S.; Peters, G.; Goldmann, O.; Rohde, M.; Sinha, B.; Pieper, D.H.; et al. High-resolution transcriptomic analysis of the adaptive response of staphylococcus aureus during acute and chronic phases of osteomyelitis. mBio 2014, 5, e01775-14. [Google Scholar] [CrossRef] [PubMed]
- Date, S.V.; Modrusan, Z.; Lawrence, M.; Morisaki, J.H.; Toy, K.; Shah, I.M.; Kim, J.; Park, S.; Xu, M.; Basuino, L.; et al. Global gene expression of methicillin-resistant Staphylococcus aureus USA300 during human and mouse infection. J. Infect. Dis. 2014, 209, 1542–1550. [Google Scholar] [CrossRef] [PubMed]
- Weinrick, B.; Dunman, P.M.; McAleese, F.; Murphy, E.; Projan, S.J.; Fang, Y.; Novick, R.P. Effect of mild acid on gene expression in Staphylococcus aureus. J. Bacteriol. 2004, 186, 8407–8423. [Google Scholar] [CrossRef] [PubMed]
- Maass, S.; Sievers, S.; Zuhlke, D.; Kuzinski, J.; Sappa, P.K.; Muntel, J.; Hessling, B.; Bernhardt, J.; Sietmann, R.; Volker, U.; et al. Efficient, global-scale quantification of absolute protein amounts by integration of targeted mass spectrometry and two-dimensional gel-based proteomics. Anal. Chem. 2011, 83, 2677–2684. [Google Scholar] [CrossRef] [PubMed]
- Becher, D.; Hempel, K.; Sievers, S.; Zuhlke, D.; Pane-Farre, J.; Otto, A.; Fuchs, S.; Albrecht, D.; Bernhardt, J.; Engelmann, S.; et al. A proteomic view of an important human pathogen--towards the quantification of the entire Staphylococcus aureus proteome. PLoS ONE 2009, 4, e8176. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, F.; Scharf, S.S.; Hildebrandt, P.; Burian, M.; Bernhardt, J.; Dhople, V.; Kalinka, J.; Gutjahr, M.; Hammer, E.; Volker, U. Time-resolved quantitative proteome profiling of host-pathogen interactions: The response of Staphylococcus aureus RN1HG to internalisation by human airway epithelial cells. Proteomics 2010, 10, 2801–2811. [Google Scholar] [CrossRef] [PubMed]
- Otto, A.; Becher, D.; Schmidt, F. Quantitative proteomics in the field of microbiology. Proteomics 2014, 14, 547–565. [Google Scholar] [CrossRef] [PubMed]
- Otto, A.; Bernhardt, J.; Hecker, M.; Becher, D. Global relative and absolute quantitation in microbial proteomics. Curr. Opin. Microbiol. 2012, 15, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Dreisbach, A.; van Dijl, J.M.; Buist, G. The cell surface proteome of Staphylococcus aureus. Proteomics 2011, 11, 3154–3168. [Google Scholar] [CrossRef] [PubMed]
- Kusch, H.; Engelmann, S. Secrets of the secretome in Staphylococcus aureus. Int. J. Med. Microbiol. 2014, 304, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Depke, M.; Michalik, S.; Rabe, A.; Surmann, K.; Brinkmann, L.; Jehmlich, N.; Bernhardt, J.; Hecker, M.; Wollscheid, B.; Sun, Z.; et al. A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies. Proteomics 2015, 15, 3648–3661. [Google Scholar] [CrossRef] [PubMed]
- Hecker, M.; Becher, D.; Fuchs, S.; Engelmann, S. A proteomic view of cell physiology and virulence of Staphylococcus aureus. Int. J. Med. Microbiol. 2010, 300, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Jayaswal, R.K.; Wilkinson, B.J. Cell wall-active antibiotic induced proteins of Staphylococcus aureus identified using a proteomic approach. FEMS Microbiol. Lett. 2001, 199, 79–84. [Google Scholar] [CrossRef]
- Scherl, A.; Francois, P.; Charbonnier, Y.; Deshusses, J.M.; Koessler, T.; Huyghe, A.; Bento, M.; Stahl-Zeng, J.; Fischer, A.; Masselot, A.; et al. Exploring glycopeptide-resistance in Staphylococcus aureus: A combined proteomics and transcriptomics approach for the identification of resistance-related markers. BMC Genom. 2006, 7, 296. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.; Pane-Farre, J.; Kohler, C.; Hecker, M.; Engelmann, S. Anaerobic gene expression in Staphylococcus aureus. J. Bacteriol. 2007, 189, 4275–4289. [Google Scholar] [CrossRef] [PubMed]
- Wolf, C.; Hochgrafe, F.; Kusch, H.; Albrecht, D.; Hecker, M.; Engelmann, S. Proteomic analysis of antioxidant strategies of Staphylococcus aureus: Diverse responses to different oxidants. Proteomics 2008, 8, 3139–3153. [Google Scholar] [CrossRef] [PubMed]
- Hochgrafe, F.; Wolf, C.; Fuchs, S.; Liebeke, M.; Lalk, M.; Engelmann, S.; Hecker, M. Nitric oxide stress induces different responses but mediates comparable protein thiol protection in Bacillus subtilis and Staphylococcus aureus. J. Bacteriol. 2008, 190, 4997–5008. [Google Scholar] [CrossRef] [PubMed]
- Resch, A.; Leicht, S.; Saric, M.; Pasztor, L.; Jakob, A.; Gotz, F.; Nordheim, A. Comparative proteome analysis of Staphylococcus aureus biofilm and planktonic cells and correlation with transcriptome profiling. Proteomics 2006, 6, 1867–1877. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.; Zühlke, D.; Pane-Farre, J.; Kusch, H.; Wolf, C.; Reiss, S.; Binh le, T.N.; Albrecht, D.; Riedel, K.; Hecker, M.; et al. Aureolib—A proteome signature library: Towards an understanding of Staphylococcus aureus pathophysiology. PLoS ONE 2013, 8, e70669. [Google Scholar] [CrossRef] [PubMed]
- Stentzel, S.; Sundaramoorthy, N.; Michalik, S.; Nordengrun, M.; Schulz, S.; Kolata, J.; Kloppot, P.; Engelmann, S.; Steil, L.; Hecker, M.; et al. Specific serum IgG at diagnosis of Staphylococcus aureus bloodstream invasion is correlated with disease progression. J. Proteom. 2015, 128, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ziebandt, A.K.; Kusch, H.; Degner, M.; Jaglitz, S.; Sibbald, M.J.; Arends, J.P.; Chlebowicz, M.A.; Albrecht, D.; Pantucek, R.; Doskar, J.; et al. Proteomics uncovers extreme heterogeneity in the Staphylococcus aureus exoproteome due to genomic plasticity and variant gene regulation. Proteomics 2010, 10, 1634–1644. [Google Scholar] [CrossRef] [PubMed]
- Liew, Y.K.; Awang Hamat, R.; van Belkum, A.; Chong, P.P.; Neela, V. Comparative Exoproteomics and Host Inflammatory Response in Staphylococcus aureus Skin and Soft Tissue Infections, Bacteremia, and Subclinical Colonization. Clin. Vaccine Immunol. 2015, 22, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Bumann, D. Pathogen proteomes during infection: A basis for infection research and novel control strategies. J. Proteom. 2010, 73, 2267–2276. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, F.; Volker, U. Proteome analysis of host-pathogen interactions: Investigation of pathogen responses to the host cell environment. Proteomics 2011, 11, 3203–3211. [Google Scholar] [CrossRef] [PubMed]
- Pförtner, H.; Burian, M.S.; Michalik, S.; Depke, M.; Hildebrandt, P.; Dhople, V.M.; Pane-Farre, J.; Hecker, M.; Schmidt, F.; Völker, U. Activation of the alternative sigma factor SigB of Staphylococcus aureus following internalization by epithelial cells—An in vivo proteomics perspective. Int. J. Med. Microbiol. 2014, 304, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Surmann, K.; Michalik, S.; Hildebrandt, P.; Gierok, P.; Depke, M.; Brinkmann, L.; Bernhardt, J.; Salazar, M.G.; Sun, Z.; Shteynberg, D.; et al. Comparative proteome analysis reveals conserved and specific adaptation patterns of Staphylococcus aureus after internalization by different types of human non-professional phagocytic host cells. Front. Microbiol. 2014, 5, 392. [Google Scholar] [CrossRef] [PubMed]
- Steinhauser, C.; Heigl, U.; Tchikov, V.; Schwarz, J.; Gutsmann, T.; Seeger, K.; Brandenburg, J.; Fritsch, J.; Schroeder, J.; Wiesmuller, K.H.; et al. Lipid-labeling facilitates a novel magnetic isolation procedure to characterize pathogen-containing phagosomes. Traffic 2013, 14, 321–336. [Google Scholar] [CrossRef] [PubMed]
- Mattow, J.; Siejak, F.; Hagens, K.; Becher, D.; Albrecht, D.; Krah, A.; Schmidt, F.; Jungblut, P.R.; Kaufmann, S.H.; Schaible, U.E. Proteins unique to intraphagosomally grown Mycobacterium tuberculosis. Proteomics 2006, 6, 2485–2494. [Google Scholar] [CrossRef] [PubMed]
- Sturgill-Koszycki, S.; Schlesinger, P.H.; Chakraborty, P.; Haddix, P.L.; Collins, H.L.; Fok, A.K.; Allen, R.D.; Gluck, S.L.; Heuser, J.; Russell, D.G. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science 1994, 263, 678–681. [Google Scholar] [CrossRef] [PubMed]
- Luhrmann, A.; Haas, A. A method to purify bacteria-containing phagosomes from infected macrophages. Methods Cell Sci. Off. J. Soc. Vitr. Biol. 2000, 22, 329–341. [Google Scholar] [CrossRef]
- Surmann, K.; Simon, M.; Hildebrandt, P.; Pförtner, H.; Michalik, S.; Stentzel, S.; Steil, L.; Dhople, V.M.; Bernhardt, J.; Schlüter, R.; et al. A proteomic perspective of the interplay of Staphylococcus aureus and human alveolar epithelial cells during infection. J. Proteom. 2015, 128, 203–217. [Google Scholar] [CrossRef] [PubMed]
- Ventura, C.L.; Higdon, R.; Hohmann, L.; Martin, D.; Kolker, E.; Liggitt, H.D.; Skerrett, S.J.; Rubens, C.E. Staphylococcus aureus elicits marked alterations in the airway proteome during early pneumonia. Infect. Immun. 2008, 76, 5862–5872. [Google Scholar] [CrossRef] [PubMed]
- van Schaik, B.; Klarenbeek, P.; Doorenspleet, M.; van Kampen, A.; Moody, D.B.; de Vries, N.; Van Rhijn, I. Discovery of invariant T-cells by next-generation sequencing of the human TCR alpha-chain repertoire. J. Immunol. 2014, 193, 5338–5344. [Google Scholar] [CrossRef] [PubMed]
- Wine, Y.; Horton, A.P.; Ippolito, G.C.; Georgiou, G. Serology in the 21st century: The molecular-level analysis of the serum antibody repertoire. Curr. Opin. Immunol. 2015, 35, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Lavinder, J.J.; Horton, A.P.; Georgiou, G.; Ippolito, G.C. Next-generation sequencing and protein mass spectrometry for the comprehensive analysis of human cellular and serum antibody repertoires. Curr. Opin. Chem. Biol. 2015, 24, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Al-Yahya, S.; Mahmoud, L.; Al-Zoghaibi, F.; Al-Tuhami, A.; Amer, H.; Almajhdi, F.N.; Polyak, S.J.; Khabar, K.S. Human Cytokinome Analysis for Interferon Response. J. Virol. 2015, 89, 7108–7119. [Google Scholar] [CrossRef] [PubMed]
- Capone, F.; Guerriero, E.; Colonna, G.; Maio, P.; Mangia, A.; Marfella, R.; Paolisso, G.; Izzo, F.; Potenza, N.; Tomeo, L.; et al. The Cytokinome Profile in Patients with Hepatocellular Carcinoma and Type 2 Diabetes. PLoS ONE 2015, 10, e0134594. [Google Scholar] [CrossRef] [PubMed]
- Derycke, L.; Eyerich, S.; Van Crombruggen, K.; Perez-Novo, C.; Holtappels, G.; Deruyck, N.; Gevaert, P.; Bachert, C. Mixed T helper cell signatures in chronic rhinosinusitis with and without polyps. PLoS ONE 2014, 9, e97581. [Google Scholar] [CrossRef] [PubMed]
- Kolata, J.; Kühbandner, I.; Link, C.; Normann, N.; Weidenmaier, C.; Bröker, B. The fall of a dogma? Unexpectedly high T-cell memory response to Staphylococcus aureus in humans. J. Infect. Dis. 2015, 212, 830–838. [Google Scholar] [CrossRef] [PubMed]
- Ricciardi-Castagnoli, P.; Granucci, F. Opinion: Interpretation of the complexity of innate immune responses by functional genomics. Nat. Rev. Immunol. 2002, 2, 881–889. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, G.; Melamed, R.; Park, R.; Seguritan, R.; Laplace, C.; Poirot, L.; Zucchelli, S.; Obst, R.; Matos, M.; Venanzi, E.; et al. Gene expression microarrays: Glimpses of the immunological genome. Nat. Immunol. 2006, 7, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, G.; Ippolito, G.C.; Beausang, J.; Busse, C.E.; Wardemann, H.; Quake, S.R. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat. Biotechnol. 2014, 32, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Doolan, D.L. Plasmodium immunomics. Int. J. Parasitol. 2011, 41, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Bröker, B.M.; van Belkum, A. Immune proteomics of Staphylococcus aureus. Proteomics 2011, 11, 3221–3231. [Google Scholar] [CrossRef] [PubMed]
- Kolata, J.; Bode, L.G.; Holtfreter, S.; Steil, L.; Kusch, H.; Holtfreter, B.; Albrecht, D.; Hecker, M.; Engelmann, S.; van Belkum, A.; et al. Distinctive patterns in the human antibody response to Staphylococcus aureus bacteremia in carriers and non-carriers. Proteomics 2011, 11, 3914–3927. [Google Scholar] [CrossRef] [PubMed]
- Tjalsma, H.; Schaeps, R.M.; Swinkels, D.W. Immunoproteomics: From biomarker discovery to diagnostic applications. Proteom. Clin. Appl. 2008, 2, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Etz, H.; Minh, D.; Henics, T.; Dryla, A.; Winkler, B.; Triska, C.; Boyd, A.; Söllner, J.; Schmidt, W.; von Ahsen, U.; et al. Identification of in vivo expressed vaccine candidate antigens from Staphylococcus aureus. Proc. Natl. Acad. Sci. USA. 2002, 99, 6573–6578. [Google Scholar] [CrossRef] [PubMed]
- Verkaik, N.; Brouwer, E.; Hooijkaas, H.; van Belkum, A.; van Wamel, W. Comparison of carboxylated and Penta-His microspheres for semi-quantitative measurement of antibody responses to His-tagged proteins. J. Immunol. Methods 2008, 335, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Verkaik, N.J.; Boelens, H.A.; de Vogel, C.P.; Tavakol, M.; Bode, L.G.; Verbrugh, H.A.; van Belkum, A.; van Wamel, W.J. Heterogeneity of the humoral immune response following Staphylococcus aureus bacteremia. Eur. J. Clin. Microbiol. Infect. Dis. 2010, 29, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Kloppot, P.; Selle, M.; Kohler, C.; Stentzel, S.; Fuchs, S.; Liebscher, V.; Muller, E.; Kale, D.; Ohlsen, K.; Broker, B.M.; et al. Microarray-based identification of human antibodies against Staphylococcus aureus antigens. Proteomics Clin. Appl. 2015, 9, 1003–1011. [Google Scholar] [CrossRef] [PubMed]
- Colque-Navarro, P.; Palma, M.; Soderquist, B.; Flock, J.I.; Mollby, R. Antibody responses in patients with staphylococcal septicemia against two Staphylococcus aureus fibrinogen binding proteins: Clumping factor and an extracellular fibrinogen binding protein. Clin. Diagn. Lab. Immunol. 2000, 7, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Verkaik, N.J.; Lebon, A.; de Vogel, C.P.; Hooijkaas, H.; Verbrugh, H.A.; Jaddoe, V.W.; Hofman, A.; Moll, H.A.; van Belkum, A.; van Wamel, W.J. Induction of antibodies by Staphylococcus aureus nasal colonization in young children. Clin. Microbiol. Infect. 2009, 16, 1312–1317. [Google Scholar] [CrossRef] [PubMed]
- Colque-Navarro, P.; Jacobsson, G.; Andersson, R.; Flock, J.I.; Mollby, R. Levels of antibody against 11 Staphylococcus aureus antigens in a healthy population. Clin. Vaccine Immunol. 2010, 17, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Holtfreter, S.; Roschack, K.; Eichler, P.; Eske, K.; Holtfreter, B.; Kohler, C.; Engelmann, S.; Hecker, M.; Greinacher, A.; Bröker, B.M. Staphylococcus aureus carriers neutralize superantigens by antibodies specific for their colonizing strain: A potential explanation for their improved prognosis in severe sepsis. J. Infect. Dis. 2006, 193, 1275–1278. [Google Scholar] [CrossRef] [PubMed]
- Ghasemzadeh-Moghaddam, H.; Neela, V.; van Wamel, W.; Hamat, R.A.; Shamsudin, M.N.; Hussin, N.S.; Aziz, M.N.; Haspani, M.S.; Johar, A.; Thevarajah, S.; et al. Nasal carriers are more likely to acquire exogenous Staphylococcus aureus strains than non-carriers. Clin. Microbiol. Infect. 2015, 21, 998.e1–998.e7. [Google Scholar] [CrossRef] [PubMed]
- Van der Kooi-Pol, M.M.; de Vogel, C.P.; Westerhout-Pluister, G.N.; Veenstra-Kyuchukova, Y.K.; Duipmans, J.C.; Glasner, C.; Buist, G.; Elsinga, G.S.; Westra, H.; Bonarius, H.P.; et al. High anti-staphylococcal antibody titers in patients with epidermolysis bullosa relate to long-term colonization with alternating types of Staphylococcus aureus. J. Investig. Dermatol. 2013, 133, 847–850. [Google Scholar] [CrossRef] [PubMed]
- Van der Kooi-Pol, M.M.; Duipmans, J.C.; Jonkman, M.F.; van Dijl, J.M. Host-pathogen interactions in epidermolysis bullosa patients colonized with Staphylococcus aureus. Int. J. Med. Microbiol. 2014, 304, 195–203. [Google Scholar] [CrossRef] [PubMed]
- O’hea, J. Helper T-cell differentiation and plasticity. In Fundamental Immunology, 7th ed.; Paul, W.E., Ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 708–717. [Google Scholar]
- McHeyzer-Williams, M.; Okitsu, S.; Wang, N.; McHeyzer-Williams, L. Molecular programming of B-cell memory. Nat. Rev. Immunol. 2012, 12, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, C.P.; David, M.Z.; Daum, R.S. Host factors that contribute to recurrent staphylococcal skin infection. Curr. Opin. Infect. Dis. 2015, 28, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Murphy, A.G.; O’Keeffe, K.M.; Lalor, S.J.; Maher, B.M.; Mills, K.H.; McLoughlin, R.M. Staphylococcus aureus infection of mice expands a population of memory gammadelta T-cells that are protective against subsequent infection. J. Immunol. 2014, 192, 3697–3708. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.S.; Pietras, E.M.; Garcia, N.C.; Ramos, R.I.; Farzam, D.M.; Monroe, H.R.; Magorien, J.E.; Blauvelt, A.; Kolls, J.K.; Cheung, A.L.; et al. IL-17 is essential for host defense against cutaneous Staphylococcus aureus infection in mice. J. Clin. Investig. 2010, 120, 1762–1773. [Google Scholar] [CrossRef] [PubMed]
- Ishigame, H.; Kakuta, S.; Nagai, T.; Kadoki, M.; Nambu, A.; Komiyama, Y.; Fujikado, N.; Tanahashi, Y.; Akitsu, A.; Kotaki, H.; et al. Differential roles of interleukin-17A and -17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity 2009, 30, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Kudva, A.; Scheller, E.V.; Robinson, K.M.; Crowe, C.R.; Choi, S.M.; Slight, S.R.; Khader, S.A.; Dubin, P.J.; Enelow, R.I.; Kolls, J.K.; et al. Influenza A inhibits Th17-mediated host defense against bacterial pneumonia in mice. J. Immunol. 2011, 186, 1666–1674. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Ibrahim, A.S.; Xu, X.; Farber, J.M.; Avanesian, V.; Baquir, B.; Fu, Y.; French, S.W.; Edwards, J.E., Jr.; Spellberg, B. Th1-Th17 cells mediate protective adaptive immunity against Staphylococcus aureus and Candida albicans infection in mice. PLoS Pathog. 2009, 5, e1000703. [Google Scholar] [CrossRef] [PubMed]
- Archer, N.K.; Harro, J.M.; Shirtliff, M.E. Clearance of Staphylococcus aureus nasal carriage is T-cell dependent and mediated through interleukin-17A expression and neutrophil influx. Infect. Immun. 2013, 81, 2070–2075. [Google Scholar] [CrossRef] [PubMed]
- Fraunholz, M.; Sinha, B. Intracellular Staphylococcus aureus: Live-in and let die. Front. Cell. Infect. Microbiol. 2012, 2, 43. [Google Scholar] [CrossRef] [PubMed]
- Grumann, D.; Nubel, U.; Broker, B.M. Staphylococcus aureus toxins--their functions and genetics. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2014, 21, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.X.; McCormick, J.K. Staphylococcal superantigens in colonization and disease. Front. Cell. Infect. Microbiol. 2012, 2, 52. [Google Scholar] [CrossRef] [PubMed]
- Alonzo, F., III; Torres, V.J. The bicomponent pore-forming leucocidins of Staphylococcus aureus. Microbiol. Mol. Biol. Rev. 2014, 78, 199–230. [Google Scholar] [CrossRef] [PubMed]
- Berube, B.J.; Bubeck Wardenburg, J. Staphylococcus aureus alpha-toxin: Nearly a century of intrigue. Toxins 2013, 5, 1140–1166. [Google Scholar] [CrossRef] [PubMed]
- Braga-Neto, U.M.; Marques, E.T., Jr. From functional genomics to functional immunomics: New challenges, old problems, big rewards. PLoS Comput. Biol. 2006, 2, e81. [Google Scholar] [CrossRef] [PubMed]
- Liebeke, M.; Lalk, M. Staphylococcus aureus metabolic response to changing environmental conditions—A metabolomics perspective. Int. J. Med. Microbiol. 2014, 304, 222–229. [Google Scholar] [CrossRef] [PubMed]
- Lochner, M.; Berod, L.; Sparwasser, T. Fatty acid metabolism in the regulation of T-cell function. Trends Immunol. 2015, 36, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Ammons, M.C.; Tripet, B.P.; Carlson, R.P.; Kirker, K.R.; Gross, M.A.; Stanisich, J.J.; Copie, V. Quantitative NMR metabolite profiling of methicillin-resistant and methicillin-susceptible Staphylococcus aureus discriminates between biofilm and planktonic phenotypes. J. Proteome Res. 2014, 13, 2973–2985. [Google Scholar] [CrossRef] [PubMed]
- Dorries, K.; Schlueter, R.; Lalk, M. Impact of antibiotics with various target sites on the metabolome of Staphylococcus aureus. Antimicrob. Agents Chemother. 2014, 58, 7151–7163. [Google Scholar] [CrossRef] [PubMed]
- Keaton, M.A.; Rosato, R.R.; Plata, K.B.; Singh, C.R.; Rosato, A.E. Exposure of clinical MRSA heterogeneous strains to beta-lactams redirects metabolism to optimize energy production through the TCA cycle. PLoS ONE 2013, 8, e71025. [Google Scholar] [CrossRef] [PubMed]
- Ledala, N.; Zhang, B.; Seravalli, J.; Powers, R.; Somerville, G.A. Influence of iron and aeration on Staphylococcus aureus growth, metabolism, and transcription. J. Bacteriol. 2014, 196, 2178–2189. [Google Scholar] [CrossRef] [PubMed]
- Hoerr, V.; Zbytnuik, L.; Leger, C.; Tam, P.P.; Kubes, P.; Vogel, H.J. Gram-negative and Gram-positive bacterial infections give rise to a different metabolic response in a mouse model. J. Proteome Res. 2012, 11, 3231–3245. [Google Scholar] [CrossRef] [PubMed]
- Slupsky, C.M.; Cheypesh, A.; Chao, D.V.; Fu, H.; Rankin, K.N.; Marrie, T.J.; Lacy, P. Streptococcus pneumoniae and Staphylococcus aureus pneumonia induce distinct metabolic responses. J. Proteome Res. 2009, 8, 3029–3036. [Google Scholar] [CrossRef] [PubMed]
- Bruno, L.; Cortese, M.; Rappuoli, R.; Merola, M. Lessons from Reverse Vaccinology for viral vaccine design. Curr. Opin. Virol. 2015, 11, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Flower, D.R.; Macdonald, I.K.; Ramakrishnan, K.; Davies, M.N.; Doytchinova, I.A. Computer aided selection of candidate vaccine antigens. Immunome Res. 2010, 6 (Suppl. 2), S1. [Google Scholar] [CrossRef] [PubMed]
- Rappuoli, R. Reverse vaccinology, a genome-based approach to vaccine development. Vaccine 2001, 19, 2688–2691. [Google Scholar] [CrossRef]
- Bagnoli, F.; Baudner, B.; Mishra, R.P.; Bartolini, E.; Fiaschi, L.; Mariotti, P.; Nardi-Dei, V.; Boucher, P.; Rappuoli, R. Designing the next generation of vaccines for global public health. Omics J. Integr. Biol. 2011, 15, 545–566. [Google Scholar] [CrossRef] [PubMed]
- Patronov, A.; Doytchinova, I. T-cell epitope vaccine design by immunoinformatics. Open Biol. 2013, 3, 120139. [Google Scholar] [CrossRef] [PubMed]
- Ponomarenko, J.V.; Bourne, P.E. Antibody-protein interactions: Benchmark datasets and prediction tools evaluation. BMC Struct. Biol. 2007, 7, 64. [Google Scholar] [CrossRef] [PubMed]
- Sela-Culang, I.; Ofran, Y.; Peters, B. Antibody specific epitope prediction-emergence of a new paradigm. Curr. Opin. Virol. 2015, 11, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Vita, R.; Overton, J.A.; Greenbaum, J.A.; Ponomarenko, J.; Clark, J.D.; Cantrell, J.R.; Wheeler, D.K.; Gabbard, J.L.; Hix, D.; Sette, A.; et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015, 43, D405–D412. [Google Scholar] [CrossRef] [PubMed]
- Yao, B.; Zheng, D.; Liang, S.; Zhang, C. Conformational B-cell epitope prediction on antigen protein structures: A review of current algorithms and comparison with common binding site prediction methods. PLoS ONE 2013, 8, e62249. [Google Scholar] [CrossRef] [PubMed]
- Doytchinova, I.A.; Flower, D.R. Identifying candidate subunit vaccines using an alignment-independent method based on principal amino acid properties. Vaccine 2007, 25, 856–866. [Google Scholar] [CrossRef] [PubMed]
- Altindis, E.; Cozzi, R.; di Palo, B.; Necchi, F.; Mishra, R.P.; Fontana, M.R.; Soriani, M.; Bagnoli, F.; Maione, D.; Grandi, G.; et al. Protectome analysis: A new selective bioinformatics tool for bacterial vaccine candidate discovery. Mol. Cell. Proteom. 2015, 14, 418–429. [Google Scholar] [CrossRef] [PubMed]
- Oprea, M.; Antohe, F. Reverse-vaccinology strategy for designing T-cell epitope candidates for Staphylococcus aureus endocarditis vaccine. Biol. J. Int. Assoc. Biol. Stand. 2013, 41, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Glowalla, E.; Tosetti, B.; Krönke, M.; Krut, O. Proteomics-based identification of anchorless cell wall proteins as vaccine candidates against Staphylococcus aureus. Infect. Immun. 2009, 77, 2719–2729. [Google Scholar] [CrossRef] [PubMed]
- Vytvytska, O.; Nagy, E.; Bluggel, M.; Meyer, H.; Kurzbauer, R.; Huber, L.; Klade, C. Identification of vaccine candidate antigens of Staphylococcus aureus by serological proteome analysis. Proteomics 2002, 2, 580–590. [Google Scholar] [CrossRef]
- Glasner, C.; van Timmeren, M.M.; Stobernack, T.; Omansen, T.F.; Raangs, E.C.; Rossen, J.W.; de Goffau, M.C.; Arends, J.P.; Kampinga, G.A.; Koedijk, D.G.; et al. Low anti-staphylococcal IgG responses in granulomatosis with polyangiitis patients despite long-term Staphylococcus aureus exposure. Sci. Rep. 2015, 5, 8188. [Google Scholar] [CrossRef] [PubMed]
- Swierstra, J.; Debets, S.; de Vogel, C.; Lemmens-den Toom, N.; Verkaik, N.; Ramdani-Bouguessa, N.; Jonkman, M.F.; van Dijl, J.M.; Fahal, A.; van Belkum, A.; et al. IgG4 subclass-specific responses to Staphylococcus aureus antigens shed new light on host-pathogen interaction. Infect. Immun. 2015, 83, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Den Reijer, P.M.; Lemmens-den Toom, N.; Kant, S.; Snijders, S.V.; Boelens, H.; Tavakol, M.; Verkaik, N.J.; van Belkum, A.; Verbrugh, H.A.; van Wamel, W.J. Characterization of the humoral immune response during Staphylococcus aureus bacteremia and global gene expression by Staphylococcus aureus in human blood. PLoS ONE 2013, 8, e53391. [Google Scholar]
| Genomics | Transcriptomics | Proteomics | Immunomics | Metabolomics | |
|---|---|---|---|---|---|
| Omics technologies |
|
|
|
|
|
| Approaches for deciphering the behavior of S. aureus |
|
|
|
|
|
| Approaches for deciphering the host response |
|
|
|
|
|
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Holtfreter, S.; Kolata, J.; Stentzel, S.; Bauerfeind, S.; Schmidt, F.; Sundaramoorthy, N.; Bröker, B.M. Omics Approaches for the Study of Adaptive Immunity to Staphylococcus aureus and the Selection of Vaccine Candidates. Proteomes 2016, 4, 11. https://doi.org/10.3390/proteomes4010011
Holtfreter S, Kolata J, Stentzel S, Bauerfeind S, Schmidt F, Sundaramoorthy N, Bröker BM. Omics Approaches for the Study of Adaptive Immunity to Staphylococcus aureus and the Selection of Vaccine Candidates. Proteomes. 2016; 4(1):11. https://doi.org/10.3390/proteomes4010011
Chicago/Turabian StyleHoltfreter, Silva, Julia Kolata, Sebastian Stentzel, Stephanie Bauerfeind, Frank Schmidt, Nandakumar Sundaramoorthy, and Barbara M. Bröker. 2016. "Omics Approaches for the Study of Adaptive Immunity to Staphylococcus aureus and the Selection of Vaccine Candidates" Proteomes 4, no. 1: 11. https://doi.org/10.3390/proteomes4010011
APA StyleHoltfreter, S., Kolata, J., Stentzel, S., Bauerfeind, S., Schmidt, F., Sundaramoorthy, N., & Bröker, B. M. (2016). Omics Approaches for the Study of Adaptive Immunity to Staphylococcus aureus and the Selection of Vaccine Candidates. Proteomes, 4(1), 11. https://doi.org/10.3390/proteomes4010011






