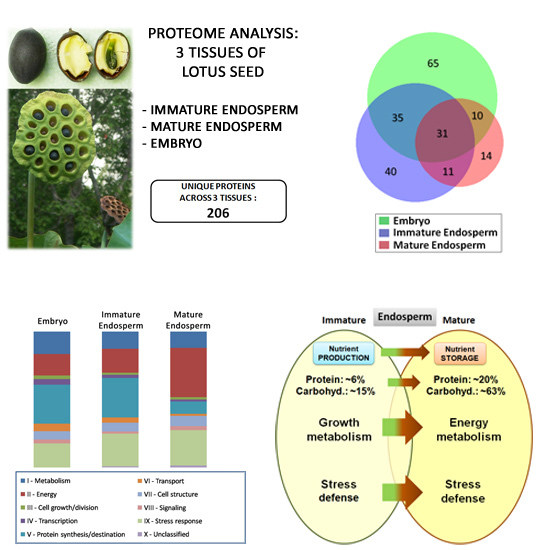
Immature Seed Endosperm and Embryo Proteomics of the Lotus (Nelumbo Nucifera Gaertn.) by One-Dimensional Gel-Based Tandem Mass Spectrometry and a Comparison with the Mature Endosperm Proteome
Abstract
:1. Introduction

2. Experimental Section
2.1. Plant Material and Tissues (Immature Endosperm and Embryo of Lotus Seed) Preparation
2.2. Extraction of the Lotus Seed Immature Endosperm and Embryo Proteins
2.3. Extraction of the Lotus Seed Immature Endosperm and Embryo Proteins
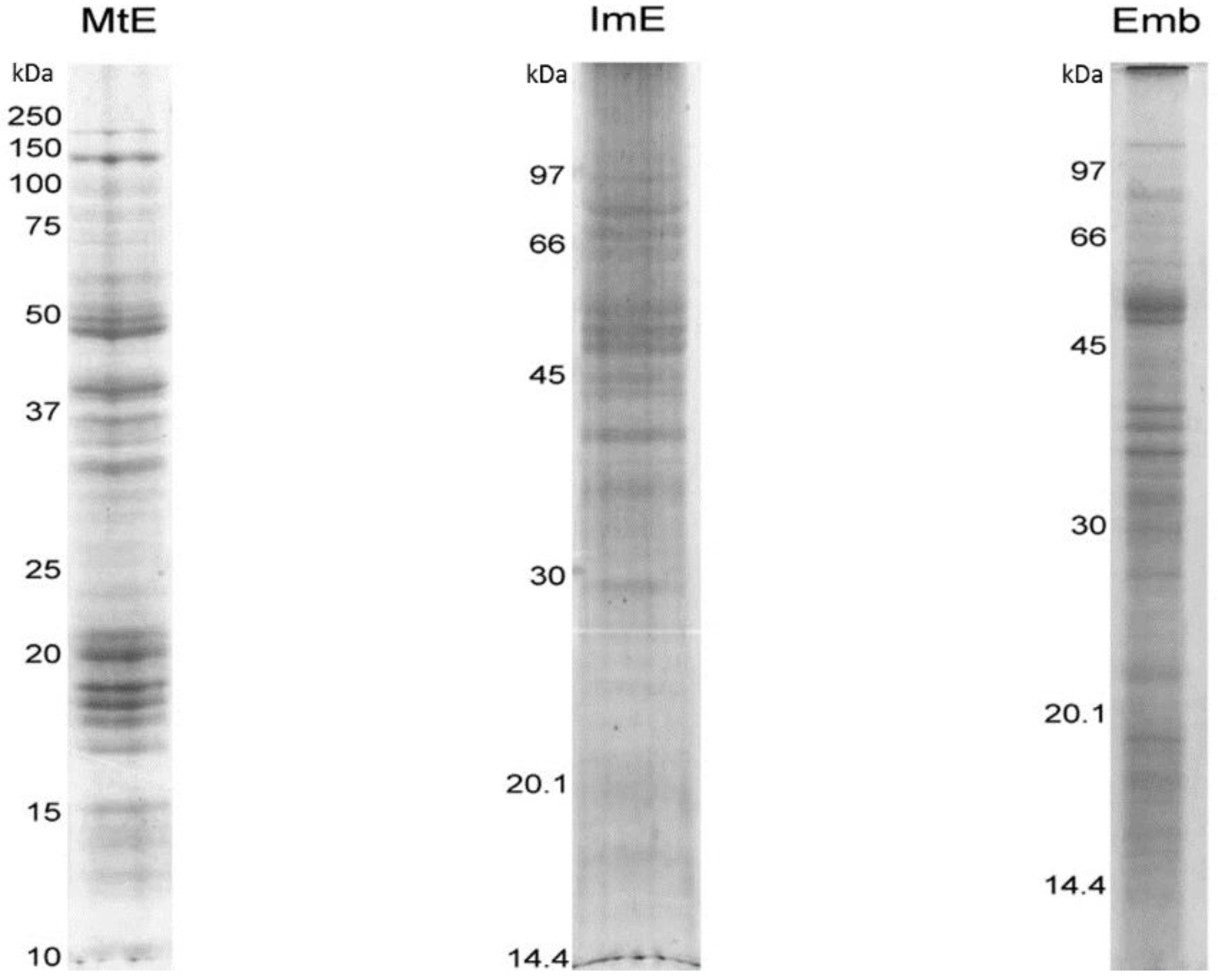
3. Results and Discussion
3.1. Protein Content of the Immature Endosperm and Embryo Tissues
3.2. Lotus Immature Endosperm Proteins Identified by 1-DGE and MS/MS Analyses
3.3. Lotus Embryo Proteins Identified by 1-DGE and MS/MS Analyses
| Fractions 1 | Protein Accession | Protein Description | Similar 2 | Score 3 | Cover (%) | PEPTIDE Sequences | Sig. Peptide Number | Func. Cat. 4 |
|---|---|---|---|---|---|---|---|---|
| 6,5,8,(7,4,3,1) | ENO1_HEVBR | Enolase 1 OS = Hevea brasiliensis | 11 | 1471 | 43.4 | TAIAK, YNQLLR, LTSEIGEK, ACNALLLK, DGGSDYLGK, AGWGVMASHR, EKACNALLLK, MGAEVYHHLK, RAGWGVMASHR, LGANAILAVSLAVCK, VQIVGDDLLVTNPK, AAVPSGASTGIYEALELR, LAMQEFMILPVGASSFK, SGETEDTFIADLSVGLATGQIK, YGQDATNVGDEGGFAPNIQENK, KYGQDATNVGDEGGFAPNIQENK, YGQDATNVGDEGGFAPNIQENKEGLELLK | 17 | II |
| 7,8,6,5,1,4,2 | G3PC_ANTMA | Glyceraldehyde-3-phosphate dehydrogenase, cytosolic OS = Antirrhinum majus | 23 | 1242 | 43.6 | AAAHLK, KATYEQIK, AAIKEESEGK, AGIALNDNFVK, DAPMFVVGVNEK, AASFNIIPSSTGAAK, VPTVDVSVVDLTVR, DAPMFVVGVNEKEYK, VPTVDVSVVDLTVRLEK, FGIVEGLMTTVHSITATQK, GILGYTEDDVVSTDFVGDSR, LTGMSFRVPTVDVSVVDLTVR, LKGILGYTEDDVVSTDFVGDSR, VINDRFGIVEGLMTTVHSITATQK | 14 | II |
| 4,8,6,7,5 | HSP7D_ARATH | Heat shock 70 kDa protein 4 OS = Arabidopsis thaliana | 10 | 625 | 23.8 | IEEVD, LSKEEIEK, ITITNDKGR, DAGVISGLNVMR, NALENYAYNMR, MVNHFVQEFKR, TTPSYVAFTDSER, IINEPTAAAIAYGLDK, ATAGDTHLGGEDFDNR, NAVVTVPAYFNDSQR, IINEPTAAAIAYGLDKK, EQIFSTYSDNQPGVLIQVYEGER | 12 | IX |
| 4 | HSP7E_SPIOL | Chloroplast envelope membrane 70 kDa heat shock-related protein OS = Spinacia oleracea | 1 | 580 | 21.7 | LSKEEIEK, DAGVISGLNVMR, EIAEAYLGSTVK, NALENYAYNMR, TTPSYVAFTDSER, IINEPTAAAIAYGLDK, ATAGDTHLGGEDFDNR, NAVVTVPAYFNDSQR, IINEPTAAAIAYGLDKK, EQVFSTYSDNQPGVLIQVYEGER | 10 | IX |
| 4 | BIP4_TOBAC | Luminal-binding protein 4 OS = Nicotiana tabacum | 5 | 542 | 21.6 | VQQLLK, NTVIPTKK, IMEYFIK, LSQEEIER, ITITNDKGR, DYFDGKEPNK, FEELNNDLFR, EAEEFAEEDKK, IVNKDGKPYIQVK, ARFEELNNDLFR, NGHVEIIANDQGNR, IINEPTAAAIAYGLDK, IINEPTAAAIAYGLDKK, IKDAVVTVPAYFNDAQR | 14 | IX |
| 4,5,6,7,8 | METE_ARATH | 5-methyltetrahydropteroyltriglutamate—homocysteine methyltransferase OS = Arabidopsis thaliana | 4 | 516 | 19.2 | AAAALK, VVEVNALAK, SWLAFAAQK, AVNEYKEAK, YLFAGVVDGR, SDEKLLSVFR, FALESFWDGK, GNASVPAMEMTK, YGAGIGPGVYDIHSPR, GMLTGPVTILNWSFVR | 10 | I |
| 6,1,5,(7,8,3,2,4) | EF1A_TOBAC | Elongation factor 1-alpha OS = Nicotiana tabacum | 8 | 484 | 34.7 | YDEIVK, GFVASNSK, QTVAVGVIK, EVSSYLKK, LPLQDVYK, ARYDEIVK, IGGIGTVPVGR, STNLDWYK, STTTGHLIYK, EHALLAFTLGVK, GFVASNSKDDPAK, YYCTVIDAPGHR, MIPTKPMVVETFSEYPPLGR, NMITGTSQADCAVLIIDSTTGGFEAGISK | 14 | V |
| 4 | HSP7L_ARATH | Heat shock 70 kDa protein 12 OS = Arabidopsis thaliana | 1 | 479 | 16.5 | VQQLLK, NTVIPTKK, IMEYFIK, FDLTGVPPAPR, FEELNNDLFR, EAEEFAEEDKK, ARFEELNNDLFR, NGHVEIIANDQGNR, IINEPTAAAIAYGLDK, IINEPTAAAIAYGLDKK, IKDAVVTVPAYFNDAQR | 11 | IX |
| 4,8,6 | HSP7N_ARATH | Heat shock 70 kDa protein 18 OS = Arabidopsis thaliana | 1 | 474 | 18.5 | ITITNDKGR, EIAEAYLGSSIK, MVNHFVQEFKR, TTPSYVAFTDSER, IINEPTAAAIAYGLDK, ATAGDTHLGGEDFDNR, NAVVTVPAYFNDSQR, IINEPTAAAIAYGLDKK | 8 | IX |
| 7 | MDHM_CITLA | Malate dehydrogenase, mitochondrial OS = Citrullus lanatus | 5 | 459 | 18.2 | TFYAGK, LFGVTTLDVVR, TQDGGTEVVEAK, DDLFNINAGIVK, KLFGVTTLDVVR, RTQDGGTEVVEAK, VAVLGAAGGIGQPLALLMK, KVAVLGAAGGIGQPLALLMK | 8 | II |
| 4,5 | HSP80_SOLLC | Heat shock cognate protein 80 OS = Solanum lycopersicum | 1 | 427 | 20.9 | AVENSPFLEK, LGIHEDSQNR, ADLVNNLGTIAR, KAVENSPFLEK, HFSVEGQLEFK, GIVDSEDLPLNISR, SLTNDWEEHLAVK, SGDEMTSLKDYVTR, KPEEITKEEYAAFYK, MKEGQNDIYYITGESK | 10 | IX |
| 5 | CH62_MAIZE | Chaperonin CPN60-2, mitochondrial OS = Zea mays | 6 | 422 | 23.3 | VTDALNATK, GVEELADAVK, IGGASEAEVGEK, SVAAGMNAMDLR, IGGASEAEVGEKK, NVVIEQSFGAPK, AAVEEGIVPGGGVALLYASK, TPVHTIASNAGVEGAVVVGK, QRPLLIVAEDVESEALGTLIINK | 9 | IX |
| 7,8,6,1 | ACT_GOSHI | Actin OS = Gossypium hirsutum | 19 | 414 | 32.6 | AGFAGDDAPR, GYSFTTTAER, EITALAPSSMK, DAYVGDEAQSK, AVFPSIVGRPR, DAYVGDEAQSKR, SYELPDGQVITIGAER, VAPEEHPVLLTEAPLNPK, TTGIVLDSGDGVSHTVPIYEGYALPHAILR | 9 | VII |
| 5,8,7 | ENO2_ARATH | Bifunctional enolase 2/transcriptional activator OS = Arabidopsis thaliana | 1 | 411 | 34.5 | YNQLLR, DGGSDYLGK, ISGDALKDLYK, LGANAILAVSLAVCK, VNQIGSVTESIEAVK, TYDLNFKEENNNGSQK, SGETEDTFIADLAVGLSTGQIK, YGQDATNVGDEGGFAPNIQENK, YGQDATNVGDEGGFAPNIQENKEGLELLK | 9 | IV |
| 4,5,6 | HSP83_IPONI | Heat shock protein 83 OS = Ipomoea nil | 1 | 405 | 23 | VIVTTK, VVVSDR, AILFVPK, DVDGEQLGR, APFDLFDTR, AVENSPFLER, LGIHEDSQNR, LDAQPELFIR, RAPFDLFDTR, ADLVNNLGTIAR, ELISNASDALDK, HFSVEGQLEFK, GVVDSDDLPLNISR, ELISNASDALDKIR, ITLFLKEDQLEYLEER | 14 | IX |
| 7,6,1,2 | ADH1_SOLTU | Alcohol dehydrogenase 1 OS = Solanum tuberosum | 7 | 390 | 23.7 | ELELEK, SDIPSVVEK, FGVTEFVNPK, GTFFGNYKPR, THPMNLLNER, KFGVTEFVNPK, YMNKELELEK, TLKGTFFGNYKPR, GSSVAIFGLGAVGLAAAEGAR | 9 | II |
| 4,5,7,8 | ENPL_CATRO | Endoplasmin homolog OS = Catharanthus roseus | 3 | 387 | 10.5 | FWNEFGK, ESFKELTK, YGWSSNMER, ELISNASDALDK, IMQSQTLSDASK, GLVDSDTLPLNVSR, ELISNASDALDKIR, VFISDEFDELLPK, RVFISDEFDELLPK | 9 | IX |
| 7,(8,6) | RBL_MAIZE | Ribulose bisphosphate carboxylase large chain OS = Zea mays | 52 | 382 | 27.5 | AMHAVIDR, AQAETGEIK, DTDILAAFR, DDFIEKDR, VALEACVQAR, EITLGFVDLLR, LTYYTPEYETK, MSGGDHIHSGTVVGK, YGRPLLGCTIKPK, GGLDFTKDDENVNSQPFMR | 10 | I |
| 6 | SAHH_MEDSA | Adenosylhomocysteinase OS = Medicago sativa | 3 | 369 | 15.1 | ATDVMIAGK, HSLPDGLMR, ITIKPQTDR, TEFGPSQPFK, VAVVCGYGDVGK, SKFDNLYGCR, IVGVSEETTTGVK, IVGVSEETTTGVKR | 8 | I |
| 4,(5,6) | HSP82_ORYSJ | Heat shock protein 81-2 OS = Oryza sativa subsp. Japonica | 2 | 368 | 21.5 | VVVSDR, IAELLR, AILFVPK, APFDLFDTR, AVENSPFLEK, RAPFDLFDTR, KAVENSPFLEK, SDLVNNLGTIAR, HFSVEGQLEFK, GIVDSEDLPLNISR, SLTNDWEEHLAVK, HSEFISYPISLWTEK, KPEEITKEEYAAFYK | 12 | IX |
| 4,6,7 | HSP70_DAUCA | Heat shock 70 kDa protein OS = Daucus carota | 1 | 335 | 15.7 | IEEVD, NALENYAYNMR, NQVAMNPSNTVFDAK, NQVAMNPSNTVFDAKR, SINPDEAVAYGAAVQAAILSGEGNER, EQIFSTYSDNQPGVLIQVYEGER | 6 | IX |
| 5 | CPNA1_ARATH | Chaperonin 60 subunit alpha 1, chloroplastic OS = Arabidopsis thaliana | 1 | 331 | 16.4 | KVTISK, VVNDGVTIAR, NVVLDEFGSPK, VGAATETELEDR, GYISPQFVTNPEK, TNDSAGDGTTTASILAR | 6 | IX |
| 4,5 | HSP82_MAIZE | Heat shock protein 82 OS = Zea mays | 2 | 297 | 14.4 | APFDLFDTR, AVENSPFLER, LGIHEDSQNR, RAPFDLFDTR, SDLVNNLGTIAR, ELISNASDALDK, HFSVEGQLEFK, GVVDSDDLPLNISR, ELISNASDALDKIR | 9 | IX |
| 8,7 | ALF_CICAR | Fructose-bisphosphate aldolase, cytoplasmic isozyme OS = Cicer arietinum | 2 | 289 | 14.2 | ANSEATLGTYK, GILAADESTGTIGK, GILAADESTGTIGKR, YHDELIANAAYIGTPGK | 4 | II |
| 4,(3,5) | CD48A_ARATH | Cell division control protein 48 homolog A OS = Arabidopsis thaliana | 3 | 285 | 14 | TLLAK, KGDLFLVR, ELVELPLR, LAEDVDLER, LAGESESNLR, GILLYGPPGSGK, IVSQLLTLMDGLK, ELVELPLRHPQLFK, NAPSIIFIDEIDSIAPK | 9 | III/IV |
| 7,8,6,1 | PGKH_TOBAC | Phosphoglycerate kinase, chloroplastic OS = Nicotiana tabacum | 6 | 284 | 15.4 | AAVPTIK, AHASTEGVTK, FAVGTEAIAK, VILSSHLGRPK, GVTTIIGGGDSVAAVEK, LASLADLYVNDAFGTAHR, KLASLADLYVNDAFGTAHR | 7 | II |
| 5,(4) | PGMC_POPTN | Phosphoglucomutase, cytoplasmic OS = Populus tremula | 1 | 281 | 12.2 | YLFEDGSR, FFEVPTGWK, LSGTGSEGATIR, SMPTSAALDVVAK, YDYENVDAGAAK, VETTPFGDQKPGTSGLR | 6 | II |
| 4,(5) | HS903_ARATH | Heat shock protein 90-3 OS = Arabidopsis thaliana | 3 | 269 | 20.2 | IAELLR, AILFVPK, AVENSPFLEK, LGIHEDSQNR, ADLVNNLGTIAR, KAVENSPFLEK, HFSVEGQLEFK, GIVDSEDLPLNISR, HSEFISYPISLWIEK | 9 | IX |
| 5,6 | PMG2_ARATH | Probable 2,3-bisphosphoglycerate-independent phosphoglycerate mutase 2 OS = Arabidopsis thaliana | 1 | 257 | 13.6 | VHILTDGR, ARDAILSGK, LVDLALASGK, TFACSETVK, MKALEIAEK, GWDAQVLGEAPHK, RGWDAQVLGEAPHK, AVGPIVDGDAVVTFNFR | 8 | II |
| 5,6 | PMG1_ARATH | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase 1 OS = Arabidopsis thaliana | 5 | 224 | 10.2 | VHILTDGR, ARDAILSGK, LDQLQLLIK, GWDAQVLGEAPHK, RGWDAQVLGEAPHK, AVGPIVDGDAVVTFNFR | 6 | II |
| 5 | SSG1_HORVU | Granule-bound starch synthase 1, chloroplastic/amyloplastic OS = Hordeum vulgare | 5 | 214 | 7.5 | FFHCYK, EALQAEVGLPVDR, FSLLCQAALEAPR, VAFCIHNISYQGR | 4 | I |
| 6 | RL4_PRUAR | 60S ribosomal protein L4 OS = Prunus armeniaca | 3 | 200 | 20.8 | AGQGAFGNMCR, AGHQTSAESWGTGR, YAVVSAIAASAVPSLVLAR, AWYQTMISDSDYTEFDNFTK | 4 | V |
| 8 | H2B_GOSHI | Histone H2B OS = Gossypium hirsutum | 5 | 200 | 49 | IYIFK, LVLPGELAK, AMGIMNSFINDIFEK | 3 | VII |
| 5 | RUBA_RICCO | RuBisCO large subunit-binding protein subunit alpha (Fragment) OS = Ricinus communis | 2 | 200 | 16 | NVVLDEFGSPK VGAATETELEDR, GYISPQFVTNPEK, LGLLSVTSGANPVSIK | 4 | I |
| 8 | H2B1_MEDTR | Probable histone H2B.1 OS = Medicago truncatula | 2 | 199 | 45.3 | IYIFK, LVLPGELAK, AMGIMNSFINDIFEK | 3 | VII |
| 8 | RL182_ARATH | 60S ribosomal protein L18-2 OS = Arabidopsis thaliana | 1 | 185 | 13.4 | APLGQNTVLLR, AGGECLTFDQLALR | 2 | V |
| 5 | CALR_BERST | Calreticulin OS = Berberis stolonifera | 3 | 175 | 9.1 | LAEETWGK, LLSGDVDQK, KLAEETWGK, TLVFQFSVK, LLSGDVDQKK, YVGIELWQVK | 6 | V |
| 8 | 1433E_TOBAC | 14-3-3-like protein E OS = Nicotiana tabacum | 5 | 172 | 26.8 | NVIGAR, NLLSVAYK, DSTLIMQLLR, TVDVEELTVEER, IISSIEQKEESR, SAQDIALAELAPTHPIR | 6 | VIII |
| 8 | H4_ARATH | Histone H4 OS = Arabidopsis thaliana | 1 | 160 | 45.6 | TLYGFGG, IFLENVIR, DAVTYTEHAR, ISGLIYEETR, DNIQGITKPAIR | 5 | VII |
| 6,5,8 | KPYC_SOYBN | Pyruvate kinase, cytosolic isozyme OS = Glycine max | 2 | 157 | 9.2 | KGSDLVNVR, GDLGMEIPVEK, VENQEGVLNFDEILR | 3 | II |
| 8 | RS6_ASPOF | 40S ribosomal protein S6 OS = Asparagus officinalis | 2 | 155 | 11.6 | LVTPLTLQR, ISQEVSGDALGEEFK, ISQEVSGDALGEEFKGYVFK | 3 | V |
| 5 | TCPA_ARATH | T-complex protein 1 subunit alpha OS = Arabidopsis thaliana | 1 | 153 | 6.4 | YFVEAGAIAVR, VLVELAELQDR, NKIHPTSIISGYR | 3 | V |
| 1 | ADT1_GOSHI | ADP, ATP carrier protein 1, mitochondrial OS = Gossypium hirsutum | 1 | 143 | 5.7 | SSLDAFSQILK, LLIQNQDEMIK | 2 | VI |
| 4 | HSP7S_SPIOL | Stromal 70 kDa heat shock-related protein, chloroplastic (Fragment) OS = Spinacia oleracea | 2 | 142 | 7.2 | QFAAEEISAQVLR, AVVTVPAYFNDSQR, IINEPTAASLAYGFEK | 3 | IX |
| 7,8 | GCST_PEA | Aminomethyltransferase, mitochondrial OS = Pisum sativum | 2 | 140 | 14.7 | LYFGEFR, GGAIDDSVITK, SLLALQGPLAAPVLQHLTK, TGYTGEDGFEISVPSEHGVELAK | 4 | I |
| 3,2 | HSP7O_ARATH | Heat shock 70 kDa protein 14 OS = Arabidopsis thaliana | 1 | 139 | 7.7 | ILSHAFDR, AVLDAATIAGLHPLR, AVEKEFEMALQDR, RAVLDAATIAGLHPLR | 4 | IX |
| 4 | HSP7F_ARATH | Heat shock 70 kDa protein 6, chloroplastic OS = Arabidopsis thaliana | 1 | 139 | 7.5 | TTPSVVAYTK, QFAAEEISAQVLR, QAVVNPENTFFSVK, LSFKDIDEVILVGGSTR | 4 | IX |
| 8 | RS4_GOSHI | 40S ribosomal protein S4 OS = Gossypium hirsutum | 2 | 135 | 20.6 | LSIIEEAR, LGNVFTIGK, FDVGNVVMVTGGR, LGGAFAPKPSSGPHK | 4 | V |
| 3,4 | CLPA_BRANA | ATP-dependent Clp protease ATP-binding subunit clpA homolog, chloroplastic (Fragment) OS = Brassica napus | 1 | 135 | 5.8 | VIGQDEAVK, TAIAEGLAQR, YRGEFEER, VLELSLEEAR | 4 | I |
| 4,5,8 | EF2_BETVU | Elongation factor 2 OS = Beta vulgaris | 1 | 131 | 5.5 | GGGQIIPTAR, EGALAEENMR, RVFYASQLTAKPR, LWGENFFDPATKK | 4 | V |
| 8 | RL12_PRUAR | 60S ribosomal protein L12 OS = Prunus armeniaca | 1 | 129 | 22.3 | VSVVPSAAALVIK, VTGGEVGAASSLAPK | 2 | V |
| 6,(7,8) | ATPBM_NICPL | ATP synthase subunit beta, mitochondrial OS = Nicotiana plumbaginifolia | 4 | 129 | 12.5 | VLNTGSPITVPVGR, TVLIMELINNVAK, IPSAVGYQPTLATDLGGLQER | 3 | I |
| 4,(7) | PHSH_SOLTU | Alpha-glucan phosphorylase, H isozyme OS = Solanum tuberosum | 6 | 125 | 5.6 | AFATYTNAK, QLLNILGVIYR, HMEIIEEIDKR, TIAYTNHTVLPEALEK | 4 | II |
| 6 | RL3_ORYSJ | 60S ribosomal protein L3 OS = Oryza sativa subsp. Japonica | 2 | 124 | 6.9 | VIAHTQIR, HGSLGFLPR, GKGYEGVVTR | 3 | V |
| 8 | TPIS_MAIZE | Triosephosphate isomerase, cytosolic OS = Zea mays | 2 | 124 | 11.9 | FFVGGNWK, VAYALSQGLK, VIACVGETLEQR | 3 | VII |
| 8 | LE194_HORVU | Late embryogenesis abundant protein B19.4 OS = Hordeum vulgare | 1 | 121 | 9.2 | GGLSTMNESGGER, KGGLSTMNESGGER | 2 | IX |
| 1,(2) | AVP_VIGRR | Pyrophosphate-energized vacuolar membrane proton pump OS = Vigna radiata var. Radiata | 2 | 117 | 3.3 | AADVGADLVGK, YIEAGASEHAR, AADVGADLVGKVER | 3 | VI |
| 8 | RL6_MESCR | 60S ribosomal protein L6 OS = Mesembryanthemum crystallinum | 1 | 115 | 10.7 | VDISGVNVEK, ASITPGTVLIILAGR | 2 | V |
| 5 | SSG1_ARATH | Probable granule-bound starch synthase 1, chloroplastic/amyloplastic OS = Arabidopsis thaliana | 1 | 115 | 3.9 | FFHCYK, YGTVPIVASTGGLVDTVK | 2 | I |
| 8 | RL10_VITRI | 60S ribosomal protein L10 OS = Vitis riparia | 1 | 114 | 10.5 | VSIGQVLLSVR, ENVSSEALEAAR | 2 | V |
| 8 | RS18_ARATH | 40S ribosomal protein S18 OS = Arabidopsis thaliana | 1 | 111 | 21.7 | LRDDLER, VLNTNVDGK, IMFALTSIK, IPDWFLNR | 4 | V |
| 7 | AATM_LUPAN | Aspartate aminotransferase P2, mitochondrial (Fragment) OS = Lupinus angustifolius | 1 | 111 | 6.8 | IADVIQEK, LNLGVGAYR, VATVQGLSGTGSLR | 3 | I |
| 8 | GBLPA_ORYSJ | Guanine nucleotide-binding protein subunit beta-like protein A OS = Oryza sativa subsp. Japonica | 1 | 110 | 9 | DGVTLLWDLAEGK, FSPNTFQPTIVSGSWDR | 2 | VIII |
| 8 | H2AX_CICAR | Histone H2AX OS = Cicer arietinum | 1 | 108 | 15.1 | AGLQFPVGR, GKGEIGSASQEF | 2 | VII |
| 4 | VATA_GOSHI | V-type proton ATPase catalytic subunit A OS = Gossypium hirsutum | 2 | 108 | 5 | LAADTPLLTGQR, LVSQKFEDPAEGEEALVAK | 2 | VI |
| 8 | PARP3_SOYBN | Poly [ADP-ribose] polymerase 3 OS = Glycine max | 1 | 107 | 4.2 | VLCSQEIYK, LEPLVANFMK, LFEEITGNEFEPWER | 3 | III |
| 3,4,(5) | CLPC1_ARATH | Chaperone protein ClpC1, chloroplastic OS = Arabidopsis thaliana | 5 | 106 | 5.4 | TAIAEGLAQR, YRGEFEER, VLELSLEEAR | 3 | IX |
| 8 | NDK1_ARATH | Nucleoside diphosphate kinase 1 OS = Arabidopsis thaliana | 2 | 103 | 9.4 | NVIHGSDSVESAR, NVIHGSDSVESARK | 2 | I |
| 8 | RL13_TOBAC | 60S ribosomal protein L13 OS = Nicotiana tabacum | 1 | 98 | 16.3 | SLEGLQTNVQR, KLAPTIGIAVDHR | 2 | V |
| 8 | RS5_CICAR | 40S ribosomal protein S5 (Fragment) OS = Cicer arietinum | 2 | 95 | 15.2 | GSSNSYAIK, AQCPIVER, VNQAIYLLTTGAR | 3 | V |
| 5,6 | PDC2_ORYSI | Pyruvate decarboxylase isozyme 2 OS = Oryza sativa subsp. Indica | 2 | 94 | 4.5 | AVKPVLVGGPK, ILHHTIGLPDFSQELR | 2 | II |
| 8 | HSP14_SOYBN | 17.5 kDa class I heat shock protein OS = Glycine max | 4 | 92 | 24.7 | AIEISG, ADIPGLK, VLQISGER, FRLPENAK | 4 | IX |
| 6 | AMPL1_ARATH | Leucine aminopeptidase 1 OS = Arabidopsis thaliana | 2 | 92 | 4.6 | GLTFDSGGYNIK, TIEVNNTDAEGR | 2 | I/IX |
| 6 | ACT5_ARATH | Putative actin-5 OS = Arabidopsis thaliana | 1 | 92 | 15.9 | AGFAGDDAPR, IWHHTFYNELR | 2 | VII |
| 8 | RS14_CHLRE | 40S ribosomal protein S14 OS = Chlamydomonas reinhardtii | 1 | 87 | 15.7 | TPGPGAQSALR, IEDVTPIPTDSTR | 2 | V |
| 8 | RS3A1_VITVI | 40S ribosomal protein S3a-1 OS = Vitis vinifera | 2 | 86 | 6.5 | TTDNYTLR, LRAEDVQGR | 2 | V |
| 7 | AAT3_ARATH | Aspartate aminotransferase, chloroplastic OS = Arabidopsis thaliana | 1 | 83 | 4.9 | LNLGVGAYR, TEEGKPLVLNVVR | 2 | I |
| 1 | COB21_ORYSJ | Coatomer subunit beta-1 OS = Oryza sativa subsp. Japonica | 1 | 83 | 4.5 | HNEIQTVNIK, DTNTFASASLDR | 2 | VI |
| 8 | GRDH1_ARATH | Glucose and ribitol dehydrogenase homolog 1 OS = Arabidopsis thaliana | 3 | 83 | 8 | GAIVAFTR, EGSSIINTTSVNAYK | 2 | II |
| 8 | ANXD1_ARATH | Annexin D1 OS = Arabidopsis thaliana | 1 | 80 | 5 | AQINATFNR, SKAQINATFNR | 2 | IX |
| 7 | PDI21_ORYSJ | Protein disulfide isomerase-like 2-1 OS = Oryza sativa subsp. Japonica | 1 | 80 | 10.9 | KLAPEYEK, YGVSGFPTLK, YGVSGYPTIQWFPK | 3 | V |
| 8,(6) | ATPAM_NICPL | ATP synthase subunit alpha, mitochondrial OS = Nicotiana plumbaginifolia | 4 | 79 | 9.4 | VVSVGDGIAR, TAIAIDTILNQK | 2 | I |
| 8,(1) | CB2_PHYPA | Chlorophyll a-b binding protein, chloroplastic OS = Physcomitrella patens subsp. Patens | 1 | 75 | 3.7 | ELEVIHAR, NRELEVIHAR | 2 | II |
| 8 | HSP12_SOYBN | Class I heat shock protein (Fragment) OS = Glycine max | 1 | 75 | 18.9 | AIEISG, ILQISGER | 2 | IX |
| 8 | BAS1_ORYSJ | 2-Cys peroxiredoxin BAS1, chloroplastic OS = Oryza sativa subsp. Japonica | 1 | 69 | 9.6 | LSDYIGKK, SGGLGDLKYPLISDVTK | 2 | IX |
| 8 | RLA0_LUPLU | 60S acidic ribosomal protein P0 OS = Lupinus luteus | 1 | 69 | 7.5 | VGSSEAALLAK, GTVEIITPVELIK | 2 | V |
| 7 | EF1G2_ORYSJ | Elongation factor 1-gamma 2 OS = Oryza sativa subsp. Japonica | 1 | 68 | 5 | NPLDLLPPSK, SFTSEFPHVER | 2 | V |
| 1 | MDAR_SOLLC | Monodehydroascorbate reductase OS = Solanum lycopersicum | 1 | 68 | 9.7 | AYLFPEGAAR, IVGAFLESGSPEENKAIAK | 2 | IX |
| 7 | RSSA_BRANA | 40S ribosomal protein SA OS = Brassica napus | 1 | 65 | 10.3 | LLILTDPR, VIVAIENPQDIIVQSARPYGQR | 2 | V |
| 6,8 | IF4A1_ARATH | Eukaryotic initiation factor 4A-1 OS = Arabidopsis thaliana | 1 | 65 | 8.3 | ELAQQIEK, VLITTDLLAR | 2 | V |
| 7 | HSP11_PEA | 18.1 kDa class I heat shock protein OS = Pisum sativum | 1 | 64 | 14.6 | SIEISG, VLQISGER | 2 | IX |
| 8 | RS16_FRIAG | 40S ribosomal protein S16 OS = Fritillaria agrestis | 1 | 64 | 12.4 | ALVAYYQK, AFEPILLLGR | 2 | V |
| 8 | RL51_ARATH | 60S ribosomal protein L5-1 OS = Arabidopsis thaliana | 1 | 64 | 7.3 | KLTYEER, GALDGGLDIPHSDKR | 2 | V |
| 4 | HSP7M_PHAVU | Heat shock 70 kDa protein, mitochondrial OS = Phaseolus vulgaris | 1 | 64 | 6.1 | HLNITLTR, SSGGLSEDEIEK | 2 | IX |
| 8,7 | HSP12_MEDSA | 18.2 kDa class I heat shock protein OS = Medicago sativa | 1 | 63 | 22.8 | TIDISG, VLQISGER, FRLPENAK | 3 | IX |
| 8 | RS102_ARATH | 40S ribosomal protein S10-2 OS = Arabidopsis thaliana | 1 | 61 | 8.9 | TYLNLPSEIVPATLK, TYLNLPSEIVPATLKK | 2 | V |
| 8 | RS193_ARATH | 40S ribosomal protein S19-3 OS = Arabidopsis thaliana | 1 | 60 | 15.4 | DVSPHEFVK, ELAPYDPDWYYIR | 2 | V |
| 1 | CYF_AETCO | Apocytochrome f OS = Aethionema cordifolium | 2 | 60 | 8.7 | NILVIGPVPGQK, SNNTVYNATAGGIISK | 2 | II |
| 8 | UBIQP_ACECL | Polyubiquitin (Fragment) OS = Acetabularia cliftonii | 1 | 60 | 8.7 | IIFAGK, TLADYNIQK, ESTLHLVLR | 3 | V |
| 8 | RL40A_ARATH | Ubiquitin-60S ribosomal protein L40-1 OS = Arabidopsis thaliana | 1 | 60 | 37.5 | LIFAGK, TLADYNIQK, ESTLHLVLR | 3 | V |
| 3 | UREA_CANEN | Urease OS = Canavalia ensiformis | 1 | 59 | 3 | NYFLF, TIHTYHSEGAGGGHAPDIIK | 2 | I |
| 8 | RS13_PEA | 40S ribosomal protein S13 OS = Pisum sativum | 1 | 58 | 17.2 | DSHGIAQVK, AHGLAPEIPEDLYHLIK | 2 | V |
| 1,8 | RAN_VICFA | GTP-binding nuclear protein Ran/TC4 OS = Vicia faba | 2 | 58 | 13.1 | HLTGEFEK, NLQYYEISAK | 2 | III/VI |
| 7 | PYRB_ARATH | Aspartate carbamoyltransferase, chloroplastic OS = Arabidopsis thaliana | 1 | 57 | 5.4 | GETLEDTIR, LGGEVLTTENAR | 2 | I |
| 8 | HSP11_CHERU | 18.3 kDa class I heat shock protein OS = Chenopodium rubrum | 1 | 52 | 18.6 | FRLPENAK, IDWKETPEAHVFK | 2 | IX |
| 5 | CLAH1_ARATH | Clathrin heavy chain 1 OS = Arabidopsis thaliana | 1 | 51 | 0.9 | ILALK, SPEQVSAAVK | 2 | VI |
| 6 | PDI_RICCO | Protein disulfide-isomerase OS = Ricinus communis | 1 | 49 | 4.2 | FFNSPDAK, SEPIPEVNNEPVK | 2 | V |
| 7 | PDIA6_MEDSA | Probable protein disulfide-isomerase A6 OS = Medicago sativa | 1 | 48 | 7.4 | KLAPEYEK, YGVSGYPTIQWFPK | 2 | V |
| 4 | SUSY_MEDSA | Sucrose synthase OS = Medicago sativa | 1 | 46 | 2.9 | NITGLVEWYGK, SGFHIDPYHGDR | 2 | II |
| 7 | FKB62_ARATH | Peptidyl-prolyl cis-trans isomerase FKBP62 OS = Arabidopsis thaliana | 1 | 42 | 4.4 | SDGVEFTVK, FTLGQGQVIK | 2 | V |
| 5 | DLDH2_ARATH | Dihydrolipoyl dehydrogenase 2, mitochondrial OS = Arabidopsis thaliana | 1 | 40 | 3.4 | AAQLGLK, SLPGITIDEK | 2 | II |
| 7 | WIT2_ARATH | WPP domain-interacting tail-anchored protein 2 OS = Arabidopsis thaliana | 1 | 39 | 3.3 | ELELEK, AESGEAKIK | 2 | III |
| 8 | TBA_PRUDU | Tubulin alpha chain OS = Prunus dulcis | 2 | 37 | 7.8 | DVNAAVATIK, LVSQVISSLTASLR | 2 | VII |
| 7 | PER1B_ARMRU | Peroxidase C1B OS = Armoracia rusticana | 1 | 35 | 5.7 | VPLGR, MGNITPLTGTQGEIR | 2 | IX |
| 7,(8,6) | YCF1_IPOPU | Putative membrane protein ycf1 OS = Ipomoea purpurea | 3 | 35 | 1.3 | ALILK, IVIEK, VIQEKER | 3 | X |
| 6 | RFS_ORYSJ | Galactinol--sucrose galactosyltransferase OS = Oryza sativa subsp. Japonica | 1 | 27 | 2.8 | VELAK, LMEEK | 2 | II |
| 7 | Y1497_ARATH | Probable receptor-like protein kinase At1g49730 OS = Arabidopsis thaliana | 1 | 20 | 1.7 | FLLAK, NLVALK | 2 | V |
| Fractions 1 | Protein Accession | Protein Description | Similar 2 | Score 3 | Cover (%) | Peptide sequences | Sig. Peptide Number | Func. Cat. 4 |
|---|---|---|---|---|---|---|---|---|
| 6,(5,2,1,7,3) | ENO1_HEVBR | Enolase 1 OS = Hevea brasiliensis | 19 | 1125 | 42 | TAIAK, YNQLLR, LTSEIGEK, DGGSDYLGK, AGWGVMASHR, MGAEVYHHLK, DGGSDYLGKGVSK, VQIVGDDLLVTNPK, VNQIGSVTESIEAVK, EAMKMGAEVYHHLK, AAVPSGASTGIYEALELR, LAMQEFMILPVGASSFK, SGETEDTFIADLSVGLATGQIK, YGQDATNVGDEGGFAPNIQENK, KYGQDATNVGDEGGFAPNIQENK, YGQDATNVGDEGGFAPNIQENKEGLELLK | 16 | II |
| 4,(1,2,3,5) | HSP7C_PETHY | Heat shock cognate 70 kDa protein OS = Petunia hybrida | 33 | 922 | 33.6 | IEEVD, DISGNPR, NTTIPTKK, ITITNDKGR, DAGVIAGLNVMR, MVNHFVQEFK, NALENYAYNMR, MVNHFVQEFKR, TTPSYVGFTDTER, ARFEELNMDLFR, IINEPTAAAIAYGLDK, NQVAMNPINTVFDAK ATAGDTHLGGEDFDNR, NQVAMNPINTVFDAK, NAVVTVPAYFNDSQR, EQVFSTYSDNQPGVLIQVYEGER | 16 | IX |
| 1,2,3,4,5,6,7 | ACT_GOSHI | Actin OS = Gossypium hirsutum | 14 | 903 | 50.7 | DLTDALMK, AGFAGDDAPR, IKVVAPPER, GYSFTTTAER, HTGVMVGMGQK, EITALAPSSMK, DAYVGDEAQSK, AVFPSIVGRPR, IWHHTFYNELR, LDLAGRDLTDALMK, GYSFTTTAEREIVR, SYELPDGQVITIGAER, VAPEEHPVLLTEAPLNPK, VAPEEHPVLLTEAPLNPK, TTGIVLDSGDGVSHTVPIYEGYALPHAILR | 15 | VII |
| 6,(1,2,3,7) | ACT12_SOLTU | Actin-100 (Fragment) OS = Solanum tuberosum | 5 | 872 | 53.5 | AGFAGDDAPR, IKVVAPPER, HTGVMVGMGQK, EITALAPSSMK, DAYVGDEAQSK, AVFPSIVGRPR, DAYVGDEAQSKR, GEYDESGPSIVHR, IWHHTFYNELR, SYELPDGQVITIGAER, LAYVALDYEQELETAK, YPIEHGIVSNWDDMEK, TTGIVLDSGDGVSHTVPIYEGYALPHAILR | 13 | VII |
| 7,(8,2) | G3PC_ANTMA | Glyceraldehyde-3-phosphate dehydrogenase, cytosolic OS = Antirrhinum majus | 20 | 749 | 43.6 | VALQR, SSIFDAK, KATYEQIK, AAIKEESEGK, AGIALNDNFVK, DAPMFVVGVNEK, AASFNIIPSSTGAAK, VPTVDVSVVDLTVR, VPTVDVSVVDLTVRLEK, FGIVEGLMTTVHSITATQK, GILGYTEDDVVSTDFVGDSR, LTGMSFRVPTVDVSVVDLTVR, LKGILGYTEDDVVSTDFVGDSR, VINDRFGIVEGLMTTVHSITATQK | 14 | II |
| 4,(1,5,2) | HSP83_IPONI | Heat shock protein 83 OS = Ipomoea nil | 4 | 717 | 31 | VIVTTK, VVVSDR, KLVSATK, AILFVPK, EMLQQNK, DVDGEQLGR, FESLTDKSK, APFDLFDTR, AVENSPFLER, LGIHEDSQNR, DIYYITGESK, LDAQPELFIR, RAPFDLFDTR, ADLVNNLGTIAR, ELISNASDALDK, KAVENSPFLER, HFSVEGQLEFK, GVVDSDDLPLNISR, ELISNASDALDKIR, SGDELTSLKDYVTR, KPEEITKEEYASFYK, HSEFISYPIYLWTEK, ITLFLKEDQLEYLEER | 23 | IX |
| 6 | ATPBM_MAIZE | ATP synthase subunit beta, mitochondrial OS = Zea mays | 3 | 712 | 31.8 | IGLFGGAGVGK, VVDLLAPYQR, TIAMDGTEGLVR, AHGGFSVFAGVGER, VGLTGLTVAEHFR, VLNTGSPITVPVGR, TVLIMELINNVAK, FTQANSEVSALLGR, QISELGIYPAVDPLDSTSR, EAPAFVEQATEQQILVTGIK, IPSAVGYQPTLATDLGGLQER | 11 | I |
| 4,(5,2,1,3) | HSP7E_SPIOL | Chloroplast envelope membrane 70 kDa heat shock-related protein OS = Spinacia oleracea | 5 | 631 | 31.2 | NTTIPTKK, LSKEEIEK, TRDNNLLGK, DAGVISGLNVMR, EIAEAYLGSTVK, NALENYAYNMR, TTPSYVAFTDSER, IINEPTAAAIAYGLDK, ATAGDTHLGGEDFDNR, NQVAMNPINTVFDAK, NAVVTVPAYFNDSQR, EQVFSTYSDNQPGVLIQVYEGER | 12 | IX |
| 4,(2,1,5) | HSP81_ORYSI | Heat shock protein 81-1 OS = Oryza sativa subsp. Indica | 9 | 610 | 35.1 | NLVKK, VVVTTK, IAELLR, KLVSATK, EMLQQNK, FESLTDKSK, APFDLFDTR, DSSMAGYMSSK, RAPFDLFDTR, KAVENSPFLEK, SDLVNNLGTIAR, HFSVEGQLEFK, EVSHEWSLVNK, GIVDSEDLPLNISR, SLTNDWEEHLAVK, SGDELTSLKDYVTR, LDAQPELFIHIVPDK, HSEFISYPISLWTEK, KPEEITKEEYAAFYK, MKEGQNDIYYITGESK, KHSEFISYPISLWTEK | 21 | IX |
| 6,(3,1,2) | TBB_HORVU | Tubulin beta chain OS = Hordeum vulgare | 21 | 583 | 39.1 | YLTASAMFR, IREEYPDR, LAVNLIPFPR, VSEQFTAMFR, YTGTSDLQLER, MMLTFSVFPSPK, EVDEQMINVQNK, LHFFMVGFAPLTSR, AVLMDLEPGTMDSVR, LHFFMVGFAPLTSR, NSSYFVEWIPNNVK, ALTVPELTQQMWDAK, GHYTEGAELIDSVLDVVRK, TGPYGQIFRPDNFVFGQSGAGNNWAK | 14 | VII |
| 6,(1,7,3,4,2) | EF1A_TOBAC | Elongation factor 1-alpha OS = Nicotiana tabacum | 17 | 574 | 38.5 | YDEIVK, GFVASNSK, EVSSYLK, QTVAVGVIK, EVSSYLKK, RGFVASNSK, LPLQDVYK, ARYDEIVK, IGGIGTVPVGR, STNLDWYK, STTTGHLIYK, EHALLAFTLGVK, GFVASNSKDDPAK, YYCTVIDAPGHR, YDEIVKEVSSYLK, YYCTVIDAPGHRDFIK, MIPTKPMVVETFSEYPPLGR, NMITGTSQADCAVLIIDSTTGGFEAGISK | 18 | V |
| 4,(1,2,5,4) | HS901_ARATH | Heat shock protein 90-1 OS = Arabidopsis thaliana | 6 | 539 | 26.4 | VVVTTKVVVTTK, VVVSDR, KLVSATK, AILFVPK, FESLTDKSK, APFDLFDTR, AVENSPFLER, LGIHEDSQNR, DSSMSGYMSSK, RAPFDLFDTR, ADLVNNLGTIAR, KAVENSPFLER, HFSVEGQLEFK, TLSIIDSGIGMTK, GVVDSDDLPLNISR, KPEEITKEEYAAFYK, HSEFISYPIYLWTEK | 17 | IX |
| 4,(2,1,3) | METE_ARATH | 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase OS = Arabidopsis thaliana | 12 | 536 | 20.9 | AAAALK, VVEVNALAK, SWLAFAAQK, AVNEYKEAK, YLFAGVVDGR, SDEKLLSVFR, FALESFWDGK, GNASVPAMEMTK, YGAGIGPGVYDIHSPR, GMLTGPVTILNWSFVR | 10 | I |
| 4 | ENPL_CATRO | Endoplasmin homolog OS = Catharanthus roseus | 3 | 472 | 12.6 | NLGTIAK, FWNEFGK, YGWSSNMER, ELISNASDALDK, IMQSQTLSDASK, GLVDSDTLPLNVSR, ELISNASDALDKIR, VFISDEFDELLPK, RVFISDEFDELLPK, LMDIIINSLYSNKDIFLR | 10 | IX |
| 4,(5,2) | BIP4_TOBAC | Luminal-binding protein 4 OS = Nicotiana tabacum | 6 | 454 | 21 | LIGEAAK, NTVIPTKK, IMEYFIK, LSQEEIER, ITITNDKGR, ALSSQHQVR, EAEEFAEEDKK, IVNKDGKPYIQVK, ARFEELNNDLFR, IINEPTAAAIAYGLDK, IKDAVVTVPAYFNDAQR | 11 | IX |
| 4,(1,3,2,7) | EF2_BETVU | Elongation factor 2 OS = Beta vulgaris | 6 | 454 | 16.6 | DLYVK, VASDLPK, GGGQIIPTAR, MIPASDKGR, IRPVLTVNK, EGALAEENMR, NMSVIAHVDHGK, FGVDESKMMER, VFYASQLTAKPR, LWGENFFDPATK, IRPVLTVNKMDR, RVFYASQLTAKPR, GHVFEEMQRPGTPLYNIK, RGHVFEEMQRPGTPLYNIK | 15 | V |
| 4,(1,2,3) | HSP82_MAIZE | Heat shock protein 82 OS = Zea mays | 5 | 429 | 16.1 | VVVSDR, KLVSATK, APFDLFDTR, AVENSPFLER, LGIHEDSQNR, RAPFDLFDTR, SDLVNNLGTIAR, ELISNASDALDK, KAVENSPFLER, HFSVEGQLEFK, GVVDSDDLPLNISR, ELISNASDALDKIR, HSEFISYPIYLWTEK | 13 | IX |
| 6,(2,3) | IF4A1_ORYSJ | Eukaryotic initiation factor 4A-1 OS = Oryza sativa subsp. Japonica | 4 | 418 | 31.4 | ALGDYLGVK, ELAQQIEK, KGVAINFVTR, VLITTDLLAR, QSLRPDYIK, RDELTLEGIK, GLDVIQQAQSGTGK, GIYAYGFEKPSAIQQR, GFKDQIYDIFQLLPSK | 9 | V |
| 3,(2,1) | CAPPC_FLATR | Phosphoenolpyruvate carboxylase 2 OS = Flaveria trinervia | 32 | 417 | 18 | GIAAGMQNTG, MNIGSRPSK, VILGDVRDK, KPSGGIESLR, LSAAWQLYK, SPEEVFDALK, RPLFGPDLPK, TPPTPQDEMR, QVSTFGLSLVR, VTIDLVEMVFAK, AGMSYFHETIWK, AIPWIFAWTQTR, VPYNAPLIQFSSWMGGDRDGNPR | 13 | I |
| 6,(2) | ENO2_ARATH | Bifunctional enolase 2/transcriptional activator OS = Arabidopsis thaliana | 2 | 391 | 33.3 | YNQLLR, DGGSDYLGK, ISGDALKDLYK, DGGSDYLGKGVSK, VNQIGSVTESIEAVK, IVLPVPAFNVINGGSHAGNK, SGETEDTFIADLAVGLSTGQIK, YGQDATNVGDEGGFAPNIQENK, KYGQDATNVGDEGGFAPNIQENK, YGQDATNVGDEGGFAPNIQENKEGLELLK | 10 | IV |
| 5,(2) | CH61_CUCMA | Chaperonin CPN60-1, mitochondrial OS = Cucurbita maxima | 6 | 387 | 33.33 | ISSINAVVK, VTDALNATK, VTKDGVTVAK, KISSINAVVK, IGGASEAEVGEK, IGVQIIQNALK, IGGASEAEVGEKK, GYISPYFITNQK, AAVEEGIVPGGGVALLYASK, TPVHTIASNAGVEGAVVVGK | 10 | IX |
| 4 | HSP7L_ARATH | Heat shock 70 kDa protein 12 OS = Arabidopsis thaliana | 1 | 382 | 16.8 | NTVIPTKK, IMEYFIK, ALSSQHQVR, EAEEFAEEDKK, ARFEELNNDLFR, ARFEELNNDLFR, IINEPTAAAIAYGLDK, IKDAVVTVPAYFNDAQR | 8 | IX |
| 2,(1) | CLAH1_ARATH | Clathrin heavy chain 1 OS = Arabidopsis thaliana | 4 | 374 | 10.3 | TVDNDLALK, SPEQVSAAVK, VANVELYYK, DPTLAVVAYR, FQELFAQTK, VEEDAVWSQVAK, GNLPGAENLVVQR, EGLVSDAIESFIR, GNMQLFSVDQQR, KNLLENWLAEDK, RGNLPGAENLVVQR, QLIDQVVSTALPESK, YKEAAELAAESPQGILR | 13 | VI |
| 6,(2,5,1) | ATPAM_PEA | ATP synthase subunit alpha, mitochondrial OS = Pisum sativum | 1 | 364 | 29.4 | VVSVGDGIAR, TGSIVDVPAGK, AAELTTLLESR, VVDALGVPIDGR, TAIAIDTILNQK, KSVHEPMQTGLK, GIRPAINVGLSVSR, EAFPGDVFYLHSR, ITNFYTNFQVDEIGR, LTEVLKQPQYAPLPIEK, EVAAFAQFGSDLDAATQALLNR | 11 | I |
| 5 | RUBB_PEA | RuBisCO large subunit-binding protein subunit beta, chloroplastic OS = Pisum sativum | 2 | 357 | 23.98 | IAALK, VVLTK, NVVLESK, VEDALNATK, IVNDGVTVAK, KGVVTLEEGK, LADLVGVTLGPK, GYISPYFVTDSEK, EVELEDPVENIGAK, TNDLAGDGTTTSVVLAQGLIAEGVK, IVNDGVTVAKEVELEDPVENIGAK | 11 | I |
| 5,(2) | PGMC_PEA | Phosphoglucomutase, cytoplasmic OS = Pisum sativum | 5 | 333 | 41.16 | YLFEDGSR, FFEVPTGWK, LSGTGSEGATIR, SMPTSAALDVVAK, YDYENVDAGAAK | 5 | II |
| 6,(2) | SAHH_MESCR | Adenosylhomocysteinase OS = Mesembryanthemum crystallinum | 6 | 330 | 11.1 | ATDVMIAGK, HSLPDGLMR, ITIKPQTDR, TEFGPSQPFK, LVGVSEETTTGVK, TEFGPSQPFKGAK, LVGVSEETTTGVKR | 7 | I |
| 7 | PGKY_TOBAC | Phosphoglycerate kinase, cytosolic OS = Nicotiana tabacum | 7 | 302 | 21.7 | LAELSGK, YSLKPLVPR, YLKPAVAGFLMQK, GVSLLLPTDVVIADK, GVTTIIGGGDSVAAVEK, LASLADLYVNDAFGTAHR, KLASLADLYVNDAFGTAHR | 7 | II |
| 4 | SUSY_SOYBN | Sucrose synthase OS = Glycine max | 12 | 295 | 13.4 | YLEMFYALK, VVHGIDVFDPK, NITGLVEWYGK, ELVNLVVVAGDR, LLPDAVGTTCGQR, SGFHIDPYHGDR, LGVTQCTIAHALEK | 7 | II |
| 5 | CPNB3_ARATH | Chaperonin 60 subunit beta 3, chloroplastic OS = Arabidopsis thaliana | 1 | 292 | 25.35 | VVLTK, NVVLESK, VEDALNATK, KGVVTLEEGK, LADLVGVTLGPK, GYISPYFVTDSEK, EVELEDPVENIGAK, TNDLAGDGTTTSVVLAQGLIAEGVK | 8 | IX |
| 7,(8) | MDHC2_ARATH | Malate dehydrogenase, cytoplasmic 2 OS = Arabidopsis thaliana | 2 | 288 | 23.2 | GAAIIK, NVSIYK, SQASALEK, EFAPSIPEK, MELVDAAFPLLK, VLVVANPANTNALILK, VLVTGAAGQIGYALVPMIAR | 7 | II |
| 8,(1) | 1433E_TOBAC | 14-3-3-like protein E OS = Nicotiana tabacum | 19 | 284 | 29.8 | NVIGAR, VFYLK, YLAEFK, MKGDYHR, NLLSVAYK, IISSIEQK, TVDVEELTVEER, IISSIEQKEESR, SAQDIALAELAPTHPIR | 9 | VIII |
| 5 | VATA_GOSHI | V-type proton ATPase catalytic subunit A OS = Gossypium hirsutum | 2 | 272 | 49.1 | SGDVYIPR, TVISQALSK, LAADTPLLTGQR, LAEMPADSGYPAYLAAR, LTTFEDSEKESEYGYVR, LVSQKFEDPAEGEEALVAK | 6 | VI |
| 4,(3,2,1) | CD48A_ARATH | Cell division control protein 48 homolog A OS = Arabidopsis thaliana | 3 | 265 | 18.5 | TLLAK, KGDLFLVR, RSVSDADIR, DFSTAILER, LAEDVDLER, GILLYGPPGSGK, LAGESESNLRK, IVSQLLTLMDGLK, ELVELPLRHPQLFK, NAPSIIFIDEIDSIAPK | 10 | III/IV |
| 7 | ALF_CICAR | Fructose-bisphosphate aldolase, cytoplasmic isozyme OS = Cicer arietinum | 1 | 245 | 11.1 | GILAADESTGTIGK, GILAADESTGTIGKR, YHDELIANAAYIGTPGK | 3 | II |
| 7 | MDHM_CITLA | Malate dehydrogenase, mitochondrial OS = Citrullus lanatus | 2 | 235 | 17.6 | LFGVTTLDVVR, TQDGGTEVVEAK, DDLFNINAGIVK, RTQDGGTEVVEAK, VAVLGAAGGIGQPLALLMK | 5 | II |
| 6,7 | ACT5_ARATH | Putative actin-5 OS = Arabidopsis thaliana | 1 | 213 | 20.4 | AGFAGDDAPR, IKVVAPPER, IWHHTFYNELR, TTGIVLDSGDGVSHTVPIYEGYALPHAILR | 4 | VII |
| 6 | UGPA_MUSAC | UTP--glucose-1-phosphate uridylyltransferase OS = Musa acuminata | 3 | 199 | 18 | VANFLSR, GGTLISYEGR, VLQLETAAGAAIR, FFDHAIGINVPR, LQSAVAELNQISENEK | 5 | II |
| 8 | ADT1_GOSHI | ADP, ATP carrier protein 1, mitochondrial OS = Gossypium hirsutum | 2 | 194 | 8.8 | SSLDAFSQILK, LLIQNQDEMIK, YFPTQALNFAFK | 3 | VI |
| 8 | RAN_VICFA | GTP-binding nuclear protein Ran/TC4 OS = Vicia faba | 1 | 193 | 26.2 | NVPTWHR, HLTGEFEK, AKQVTFHR, LVIVGDGGTGK, NLQYYEISAK, SNYNFEKPFLYLAR | 6 | VIII |
| 3 | CLPB1_ARATH | Chaperone protein ClpB1 OS = Arabidopsis thaliana | 2 | 192 | 11.1 | TAVVEGLAQR, YRGEFEER, TKNNPVLIGEPGVGK, KVESASGDTNFQALK, VQLDSQPEEIDNLER, LIGAPPGYVGHEEGGQLTEAVR | 6 | IX |
| 5 | CPNA1_ARATH | Chaperonin 60 subunit alpha 1, chloroplastic OS = Arabidopsis thaliana | 1 | 189 | 82.82 | VVNDGVTIAR, NVVLDEFGSPK, VGAATETELEDR | 3 | IX |
| 3 | ACOC_CUCMA | Aconitate hydratase, cytoplasmic OS = Cucurbita maxima | 6 | 187 | 9.2 | NFEGR, ILLESAIR, STYESITK, DFNSYGSR, RGNDEVMAR, TSLAPGSGVVTK, ATIANMSPEYGATMGFFPVDHVTLQYLK | 7 | II |
| 3 | SYA_ARATH | Alanine--tRNA ligase OS = Arabidopsis thaliana | 1 | 181 | 5.4 | LTSVLQNK, HVDTGMGFER, ESDGSLKPLPAK, AFALLSEEGIAK, AVFGEVYPDPVR | 5 | IV |
| 6,(7) | PRS6A_SOLLC | 26S protease regulatory subunit 6A homolog OS = Solanum lycopersicum | 4 | 170 | 8.5 | IIKEELQR, GVLLYGPPGTGK, LAGPQLVQMFIGDGAK | 3 | I |
| 7 | AATM_LUPAN | Aspartate aminotransferase P2, mitochondrial (Fragment) OS = Lupinus angustifolius | 1 | 170 | 10.8 | IADVIQEK, NLGLYAER, LNLGVGAYR, ISLAGLSLAK, VATVQGLSGTGSLR | 5 | I |
| 6 | UGDH_SOYBN | UDP-glucose 6-dehydrogenase OS = Glycine max | 1 | 168 | 6.5 | IAILGFAFK, LAANAFLAQR, AADLTYWESAAR | 3 | II |
| 2,8 | ANX4_FRAAN | Annexin-like protein RJ4 OS = Fragaria ananassa | 1 | 163 | 9.6 | VGTDEDALTR, LLVALVTAYR | 2 | IX |
| 8 | RS4_GOSHI | 40S ribosomal protein S4 OS = Gossypium hirsutum | 4 | 163 | 22.9 | LSIIEEAR, LGNVFTIGK, GIPYLNTYDGR, LGGAFAPKPSSGPHK, TDKTYPAGFMDVVSIPK | 5 | V |
| 7,(8) | RSSA_SOYBN | 40S ribosomal protein SA OS = Glycine max | 2 | 161 | 19 | LLILTDPR, YVDIGIPANNK, HTPGTFTNQLQTSFSEPR, VIVAIENPQDIIVQSARPYGQR | 4 | V |
| 8 | RAA1D_ARATH | Ras-related protein RABA1d OS = Arabidopsis thaliana | 9 | 161 | 26.6 | AITSAYYR, VVLIGDSGVGK, STIGVEFATR, HSTFENVER, AQIWDTAGQER | 5 | VIII |
| 8 | RS18_ARATH | 40S ribosomal protein S18 OS = Arabidopsis thaliana | 1 | 155 | 23.7 | LRDDLER, VLNTNVDGK, IPDWFLNR, YSQVVSNALDMK | 4 | V |
| 1 | AVP_VIGRR | Pyrophosphate-energized vacuolar membrane proton pump OS = Vigna radiata var. Radiata | 1 | 151 | 9.5 | TDALDAAGNTTAAIGK, AAVIGDTIGDPLKDTSGPSLNILIK | 2 | VI |
| 5 | ILV5_ARATH | Ketol-acid reductoisomerase, chloroplastic OS = Arabidopsis thaliana | 1 | 150 | 27.45 | SDIVVK, SVVLAGR, QIGVIGWGSQGPAQAQNLR | 3 | I |
| 7 | AATC_DAUCA | Aspartate aminotransferase, cytoplasmic OS = Daucus carota | 2 | 149 | 10.4 | ISMAGLSSR, LNLGVGAYR, LIFGADSPAIQENR | 3 | I |
| 8 | RL13_TOBAC | 60S ribosomal protein L13 OS = Nicotiana tabacum | 1 | 149 | 23.3 | GFSLEELK, TWFNQPAR, SLEGLQTNVQR, KLAPTIGIAVDHR | 4 | V |
| 5 | ACLB1_ORYSJ | ATP-citrate synthase beta chain protein 1 OS = Oryza sativa subsp. Japonica | 1 | 148 | 35.6 | FNNIPQVK, FGGAIDDAAR, SEVQFGHAGAK, SIGLIGHTFDQKR, VVAIIAEGVPESDTK | 5 | I |
| 2,(3) | COPA1_ARATH | Coatomer subunit alpha-1 OS = Arabidopsis thaliana | 3 | 147 | 4.4 | VWDIGALR, YVLEGHDR, AWEVDTLR, VVIFDLQQR, TLDVPIYITK, QDIIVSNSEDK | 6 | VI |
| 6 | CATA2_RICCO | Catalase isozyme 2 OS = Ricinus communis | 2 | 146 | 9.3 | FSTVIHER, APGVQTPVIVR, EGNFDIVGNNFPVFFIR | 3 | IX |
| 8 | RAN3_ORYSI | GTP-binding nuclear protein Ran-3 OS = Oryza sativa subsp. Indica | 1 | 145 | 14.2 | HITGEFEK, NLQYYEISAK, SNYNFEKPFLYLAR | 3 | III/VI |
| 6 | MDAR_SOLLC | Monodehydroascorbate reductase OS = Solanum lycopersicum | 1 | 139 | 12.5 | AYLFPEGAAR, LSDFGVQGADSK, IVGAFLESGSPEENKAIAK | 3 | IX |
| 6,(1) | RL3_ORYSJ | 60S ribosomal protein L3 OS = Oryza sativa subsp. Japonica | 4 | 138 | 9.3 | VIAHTQIR, HGSLGFLPR, LALEEIKLK, GKGYEGVVTR | 4 | V |
| 5 | PMGI_RICCO | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase OS = Ricinus communis | 4 | 136 | 43.5 | ARDAILSGK, LVDLALASGK, LDQLQLLLK, AHGTAVGLPTEDDMGNSEVGHNALGAGR | 4 | II |
| 5 | RUBA_RICCO | RuBisCO large subunit-binding protein subunit alpha (Fragment) OS = Ricinus communis | 2 | 134 | 71.19 | NVVLDEFGSPK, VGAATETELEDR, LGLLSVTSGANPVSIK | 3 | I |
| 7 | RL4_PRUAR | 60S ribosomal protein L4 OS = Prunus armeniaca | 1 | 134 | 14.5 | AGHQTSAESWGTGR, YAVVSAIAASAVPSLVLAR | 2 | V |
| 5 | G6PI_SPIOL | Glucose-6-phosphate isomerase, cytosolic OS = Spinacia oleracea | 2 | 131 | 27 | SQQPVYLK, FLANVDPIDVAK, TFTTAETMLNAR | 3 | II |
| 8 | RS8_MAIZE | 40S ribosomal protein S8 OS = Zea mays | 1 | 130 | 21.7 | LDTGNYSWGSEAVTR, ILDVVYNASNNELVR | 2 | V |
| 8 | RL11_MEDSA | 60S ribosomal protein L11 OS = Medicago sativa | 1 | 129 | 17.7 | YEGVILNK, AMQLLESGLK, VLEQLSGQTPVFSK | 3 | V |
| 8,(7) | H4_ARATH | Histone H4 OS = Arabidopsis thaliana | 2 | 128 | 46.6 | TLYGFGG, IFLENVIR, DAVTYTEHAR, ISGLIYEETR | 4 | VII |
| 5 | TCPE_ARATH | T-complex protein 1 subunit epsilon OS = Arabidopsis thaliana | 1 | 123 | 73.41 | IAEGYEMASR, QQQILLATQVVK | 2 | V |
| 5 | TCPA_ARATH | T-complex protein 1 subunit alpha OS = Arabidopsis thaliana | 1 | 119 | 71.23 | YFVEAGAIAVR, NKIHPTSIISGYR | 2 | V |
| 4 | TKTC_SPIOL | Transketolase, chloroplastic OS = Spinacia oleracea | 4 | 118 | 6.1 | FLAIDAVEK, ALPTYTPETPGDATR, VIPGLLGGSADLASSNMTLLK | 3 | II |
| 8 | TPIS_MAIZE | Triosephosphate isomerase, cytosolic OS = Zea mays | 1 | 117 | 11.9 | FFVGGNWK, VAYALSQGLK, VIACVGETLEQR | 3 | VII |
| 8 | PROF3_ARATH | Profilin-3 OS = Arabidopsis thaliana | 2 | 117 | 17.2 | LGDYLLEQGL, YMVIQGEPGAVIR | 2 | VII |
| 8 | RS92_ARATH | 40S ribosomal protein S9-2 OS = Arabidopsis thaliana | 1 | 115 | 19.3 | LVGEYGLR, ERLDAELK, RPYEKER, RLQTIVFK, IFEGEALLR | 5 | V |
| 6 | VATB1_ARATH | V-type proton ATPase subunit B1 OS = Arabidopsis thaliana | 1 | 115 | 10.5 | YQEIVNIR, TVSGVAGPLVILDK, QIYPPINVLPSLSR | 3 | VI |
| 6 | ERF1X_ARATH | Eukaryotic peptide chain release factor subunit 1-1 OS = Arabidopsis thaliana | 1 | 113 | 9.9 | GFGGIGGILR, QSVLGAITSAQQR | 2 | V |
| 5 | HSP7M_PHAVU | Heat shock 70 kDa protein, mitochondrial OS = Phaseolus vulgaris | 3 | 111 | 24.87 | HLNITLTR, VIENSEGAR, TTPSVVAFNQK, SSGGLSEDEIEK | 4 | IX |
| 8 | ANXD1_ARATH | Annexin D1 OS = Arabidopsis thaliana | 1 | 110 | 5 | AQINATFNR, SKAQINATFNR | 2 | IX |
| 8 | RS16_FRIAG | 40S ribosomal protein S16 OS = Fritillaria agrestis | 3 | 109 | 13.8 | ALVAYYQK, AFEPILLLGR, YKAFEPILLLGR | 3 | V |
| 8 | RS5_CICAR | 40S ribosomal protein S5 (Fragment) OS = Cicer arietinum | 1 | 108 | 15.7 | IGSAGVVRR, GSSNSYAIK, VNQAIYLLTTGAR | 3 | V |
| 8 | ARF_VIGUN | ADP-ribosylation factor OS = Vigna unguiculata | 1 | 104 | 28.7 | ILMVGLDAAGK, NISFTVWDVGGQDK | 2 | VIII |
| 4 | SYGM1_ARATH | Glycine--tRNA ligase 1, mitochondrial OS = Arabidopsis thaliana | 1 | 103 | 5.1 | LFYIPSFK, VFTPSVIEPSFGIGR | 2 | IV/VI |
| 7 | RGP1_ORYSJ | UDP-arabinopyranose mutase 1 OS = Oryza sativa subsp. Japonica | 1 | 101 | 6.9 | ILGPK, ASNPFVNLK, ASNPFVNLKK, YVDAVMTVPK | 4 | I |
| 3 | HSP7O_ARATH | Heat shock 70 kDa protein 14 OS = Arabidopsis thaliana | 1 | 101 | 5.1 | ILSHAFDR, NAVESYVYDMR, AVLDAATIAGLHPLR | 3 | IX |
| 8 | RS15A_DAUCA | 40S ribosomal protein S15a OS = Daucus carota | 1 | 100 | 29.2 | VSVLNDALK, HGYIGEFEYVDDHR | 2 | V |
| 8 | RS61_ARATH | 40S ribosomal protein S6-1 OS = Arabidopsis thaliana | 2 | 100 | 18 | LVTPLTLQR, KGENDLPGLTDTEKPR, ISQEVSGDALGEEFKGYVFK | 3 | V |
| 6 | ACCC2_POPTR | Biotin carboxylase 2, chloroplastic OS = Populus trichocarpa | 1 | 97 | 7.8 | LLEEAPSPALTPELR, ALDDTVITGVPTTIDYHK | 2 | I |
| 8 | RLA2_PARAR | 60S acidic ribosomal protein P2 OS = Parthenium argentatum | 1 | 97 | 10.5 | DITELIASGR, GKDITELIASGR | 2 | V |
| 8 | RS33_ARATH | 40S ribosomal protein S3-3 OS = Arabidopsis thaliana | 3 | 96 | 10.9 | ELAEDGYSGVEVR, FKFPQDSVELYAEK | 2 | V |
| 5 | KPYC_SOYBN | Pyruvate kinase, cytosolic isozyme OS = Glycine max | 1 | 94 | 55.66 | KGSDLVNVR, STPLPMSPLESLASSAVR | 2 | II |
| 7 | GMD1_ARATH | GDP-mannose 4,6 dehydratase 1 OS = Arabidopsis thaliana | 2 | 93 | 5 | RGENFVTR, LFLGNIQASR | 2 | II |
| 4 | HSP7S_SPIOL | Stromal 70 kDa heat shock-related protein, chloroplastic (Fragment) OS = Spinacia oleracea | 2 | 93 | 5.3 | HIETTLTR, IINEPTAASLAYGFEK | 2 | IX |
| 6 | EF1G2_ORYSJ | Elongation factor 1-gamma 2 OS = Oryza sativa subsp. Japonica | 3 | 93 | 16.7 | EVAIK, LYSNTK, NPLDLLPPSK, MILDEWKR, SFTSEFPHVER | 5 | V |
| 8 | RLA0_LUPLU | 60S acidic ribosomal protein P0 OS = Lupinus luteus | 1 | 90 | 9.9 | EYLKDPSK, VGSSEAALLAK | 2 | V |
| 7 | GCST_PEA | Aminomethyltransferase, mitochondrial OS = Pisum sativum | 1 | 89 | 8.3 | GGAIDDSVITK, TGYTGEDGFEISVPSEHGVELAK | 2 | I |
| 8 | APX1_ORYSJ | L-ascorbate peroxidase 1, cytosolic OS = Oryza sativa subsp. Japonica | 1 | 89 | 15.2 | TGGPFGTMK, LSELGFADA, ALLSDPAFRPLVEK | 3 | IX |
| 8 | RS193_ARATH | 40S ribosomal protein S19-3 OS = Arabidopsis thaliana | 1 | 88 | 23.1 | AYAAHLKR, TVKDVSPHEFVK, ELAPYDPDWYYIR | 3 | V |
| 1,(2) | RPN1A_ARATH | 26S proteasome non-ATPase regulatory subunit 2 1A OS = Arabidopsis thaliana | 3 | 87 | 5.5 | VGQAVDVVGQAGRPK, NLAGEIAQEYTKR | 2 | I |
| 4,(3) | PPDK_FLABR | Pyruvate, phosphate dikinase, chloroplastic OS = Flaveria brownii | 8 | 87 | 2.9 | SDFEGIFR, AALIADEIAK, AMDGLPVTIR | 3 | II |
| 8 | RS13_PEA | 40S ribosomal protein S13 OS = Pisum sativum | 1 | 87 | 25.8 | DSHGIAQVK, GLTPSQIGVILR, KGLTPSQIGVILR, AHGLAPEIPEDLYHLIK | 4 | V |
| 8 | RS14_CHLRE | 40S ribosomal protein S14 OS = Chlamydomonas reinhardtii | 1 | 85 | 18.3 | TPGPGAQSALR, IEDVTPIPTDSTRR | 2 | V |
| 6 | VATB2_GOSHI | V-type proton ATPase subunit B 2 (Fragment) OS = Gossypium hirsutum | 1 | 84 | 10.1 | FVTQGAYDTR, QIYPPINVLPSLSR | 2 | VI |
| 8,(5) | CYPH_MAIZE | Peptidyl-prolyl cis-trans isomerase OS = Zea mays | 3 | 83 | 16.3 | SGKPLHYK, VFFDMTVGGAPAGR | 2 | V |
| 8 | NDK1_ARATH | Nucleoside diphosphate kinase 1 OS = Arabidopsis thaliana | 1 | 82 | 9.4 | NVIHGSDSVESAR, NVIHGSDSVESARK | 2 | I |
| 7 | GLN11_ORYSJ | Glutamine synthetase cytosolic isozyme 1-1 OS = Oryza sativa subsp. Japonica | 1 | 79 | 7.6 | DIVDSHYK, HKEHISAYGEGNER | 2 | I |
| 7 | SERC_SPIOL | Phosphoserine aminotransferase, chloroplastic OS = Spinacia oleracea | 1 | 79 | 5.3 | FGLIYAGAQK, NVGPSGVTIVIVR | 2 | I |
| 7 | PDI21_ARATH | Protein disulfide-isomerase like 2-1 OS = Arabidopsis thaliana | 1 | 78 | 8.9 | KLAPEYEK, YGVSGFPTLK, YGVSGYPTIQWFPK | 3 | V |
| 8 | RS254_ARATH | 40S ribosomal protein S25-4 OS = Arabidopsis thaliana | 2 | 76 | 29.6 | LITPSILSDR, MVAAHSSQQIYTR | 2 | V |
| 6 | IDHC_TOBAC | Isocitrate dehydrogenase [NADP] OS = Nicotiana tabacum | 2 | 75 | 7.5 | HAFGDQYR, DLALIIHGSK, TIEAEAAHGTVTR | 3 | II |
| 6 | OPD22_ARATH | Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial OS = Arabidopsis thaliana | 1 | 73 | 3.9 | ISVNDLVIK, VIDGAIGAEWLK | 2 | II |
| 8 | RL40A_ARATH | Ubiquitin-60S ribosomal protein L40-1 OS = Arabidopsis thaliana | 1 | 70 | 45.3 | ESTLHLVLR, TITLEVESSDTIDNVK | 2 | V |
| 8 | RL24_PRUAV | 60S ribosomal protein L24 OS = Prunus avium | 1 | 69 | 7 | SIVGATLEVIQK, SIVGATLEVIQKR | 2 | V |
| 7,(5) | SAPK6_ORYSJ | Serine/threonine-protein kinase SAPK6 OS = Oryza sativa subsp. Japonica | 2 | 68 | 7.4 | DIGSGNFGVAR, STVGTPAYIAPEVLSR | 2 | III |
| 6 | GME2_ORYSJ | GDP-mannose 3,5-epimerase 2 OS = Oryza sativa subsp. Japonica | 1 | 67 | 7 | NSDNTLIKEK, ISITGAGGFIASHIAR | 2 | II |
| 8 | EF1D1_ORYSJ | Elongation factor 1-delta 1 OS = Oryza sativa subsp. Japonica | 2 | 66 | 7.9 | LVPVGYGIK, KLDEYLLTR | 2 | V |
| 8 | IF5A1_ARATH | Eukaryotic translation initiation factor 5A-1 OS = Arabidopsis thaliana | 1 | 64 | 12 | VVEVSTSK, TYPQQAGTIR, TYPQQAGTIRK | 3 | V |
| 8 | H2B11_ARATH | Histone H2B.11 OS = Arabidopsis thaliana | 1 | 62 | 30 | LVLPGELAK, QVHPDIGISSK, YNKKPTITSR | 3 | VII |
| 8 | PSA3_ARATH | Proteasome subunit alpha type-3 OS = Arabidopsis thaliana | 1 | 60 | 7.6 | VFQIEYAAK, VPDDLLEEAK | 2 | I |
| 7 | AAT3_ARATH | Aspartate aminotransferase, chloroplastic OS = Arabidopsis thaliana | 1 | 60 | 4.9 | LNLGVGAYR, TEEGKPLVLNVVR | 2 | I |
| 1 | PDR4_ORYSJ | Pleiotropic drug resistance protein 4 OS = Oryza sativa subsp. Japonica | 1 | 60 | 1.5 | TTLLLALAGK, VTTGEMLVGPAR | 2 | IX |
| 2,(8) | UBQ12_ARATH | Polyubiquitin 12 OS = Arabidopsis thaliana | 3 | 60 | 23.9 | MQIFLKTLTGK, IQDKEGIPPDQQR, TITLEVESSDTIDNVK | 3 | V |
| 2,(1) | UBIQ_AVESA | Ubiquitin OS = Avena sativa | 1 | 60 | 57.9 | TLADYNIQK, IQDKEGIPPDQQR, TITLEVESSDTIDNVK | 3 | V |
| 5,(1) | DIM_PEA | Delta(24)-sterol reductase OS = Pisum sativum | 2 | 59 | 45.72 | NILDIDKER, SDLEAPLRPK | 2 | I |
| 8 | HSP11_PEA | 18.1 kDa class I heat shock protein OS = Pisum sativum | 2 | 56 | 8.9 | SIEISG, VLQISGER | 2 | IX |
| 6 | MPPA_SOLTU | Mitochondrial-processing peptidase subunit alpha OS = Solanum tuberosum | 1 | 52 | 3.4 | QLLTYGER, MVASEDIGR | 2 | I |
| 8 | RL17_MAIZE | 60S ribosomal protein L17 OS = Zea mays | 1 | 52 | 10.5 | NAESNADVK, YLEDVIAHK | 2 | V |
| 8 | RL51_ARATH | 60S ribosomal protein L5-1 OS = Arabidopsis thaliana | 2 | 52 | 9.3 | VFGALK, KLTYEER, GALDGGLDIPHSDKR | 3 | V |
| 3 | PHSL1_SOLTU | Alpha-1,4 glucan phosphorylase L-1 isozyme, chloroplastic/amyloplastic OS = Solanum tuberosum | 1 | 50 | 1.8 | NDVSYPIK, AFATYVQAK | 2 | II |
| 6 | PRS4A_ARATH | 26S proteasome regulatory subunit 4 homolog A OS = Arabidopsis thaliana | 1 | 49 | 9.5 | VVGSELIQK, GVILYGEPGTGK | 2 | II |
| 8 | YPTC1_CHLRE | GTP-binding protein YPTC1 OS = Chlamydomonas reinhardtii | 2 | 49 | 17.2 | TITSSYYR, LLLIGDSGVGK | 2 | III/VI |
| 4 | HSP7G_ARATH | Heat shock 70 kDa protein 7, chloroplastic OS = Arabidopsis thaliana | 1 | 47 | 9.5 | HIETTLTR, TTPSVVAYTK, QAVVNPENTFFSVKR | 3 | IX |
| 8 | SODM_HEVBR | Superoxide dismutase [Mn], mitochondrial OS = Hevea brasiliensis | 1 | 46 | 11.2 | HHQTYITNYNK, LVVETTANQDPLVTK | 2 | IX |
| 5 | CALX2_ARATH | Calnexin homolog 2 OS = Arabidopsis thaliana | 2 | 45 | 30.3 | NPAYK, SEGHDDYGLLVSEK | 2 | V |
| 8 | RS30_ARATH | 40S ribosomal protein S30 OS = Arabidopsis thaliana | 1 | 44 | 30.6 | GKVHGSLAR, FVTAVVGFGK | 2 | V |
| 8 | RL7A1_ARATH | 60S ribosomal protein L7a-1 OS = Arabidopsis thaliana | 1 | 44 | 9.3 | TLDKNLATSLFK, LKVPPALNQFTK | 2 | V |
| 7 | METK4_POPTR | S-adenosylmethionine synthase 4 OS = Populus trichocarpa | 1 | 42 | 12.3 | FVIGGPHGDAGLTGR, VLVNIEQQSPDIAQGVHGHLTK | 2 | I |
| 8 | RL18A_CASSA | 60S ribosomal protein L18a OS = Castanea sativa | 1 | 41 | 9.6 | ASRPNLFM, FHQYQVVGR | 2 | V |
| 7 | EFTM_ARATH | Elongation factor Tu, mitochondrial OS = Arabidopsis thaliana | 1 | 41 | 9.7 | QAILK, VLAEEGKAK, GITIATAHVEYETAKR | 3 | V |
| 1 | CALSB_ARATH | Callose synthase 11 OS = Arabidopsis thaliana | 1 | 38 | 1.5 | ILFNEAFSR, LGEGKPENQNHALIFTR | 2 | IX |
| 3 | APBLB_ARATH | Beta-adaptin-like protein B OS = Arabidopsis thaliana | 1 | 27 | 2 | EAENIVER, DSQDPNPLIR | 2 | VII |
3.4. Comparative Analysis of Lotus Seed (Immature Endosperm, Mature Endosperm, and Embryo) Proteins
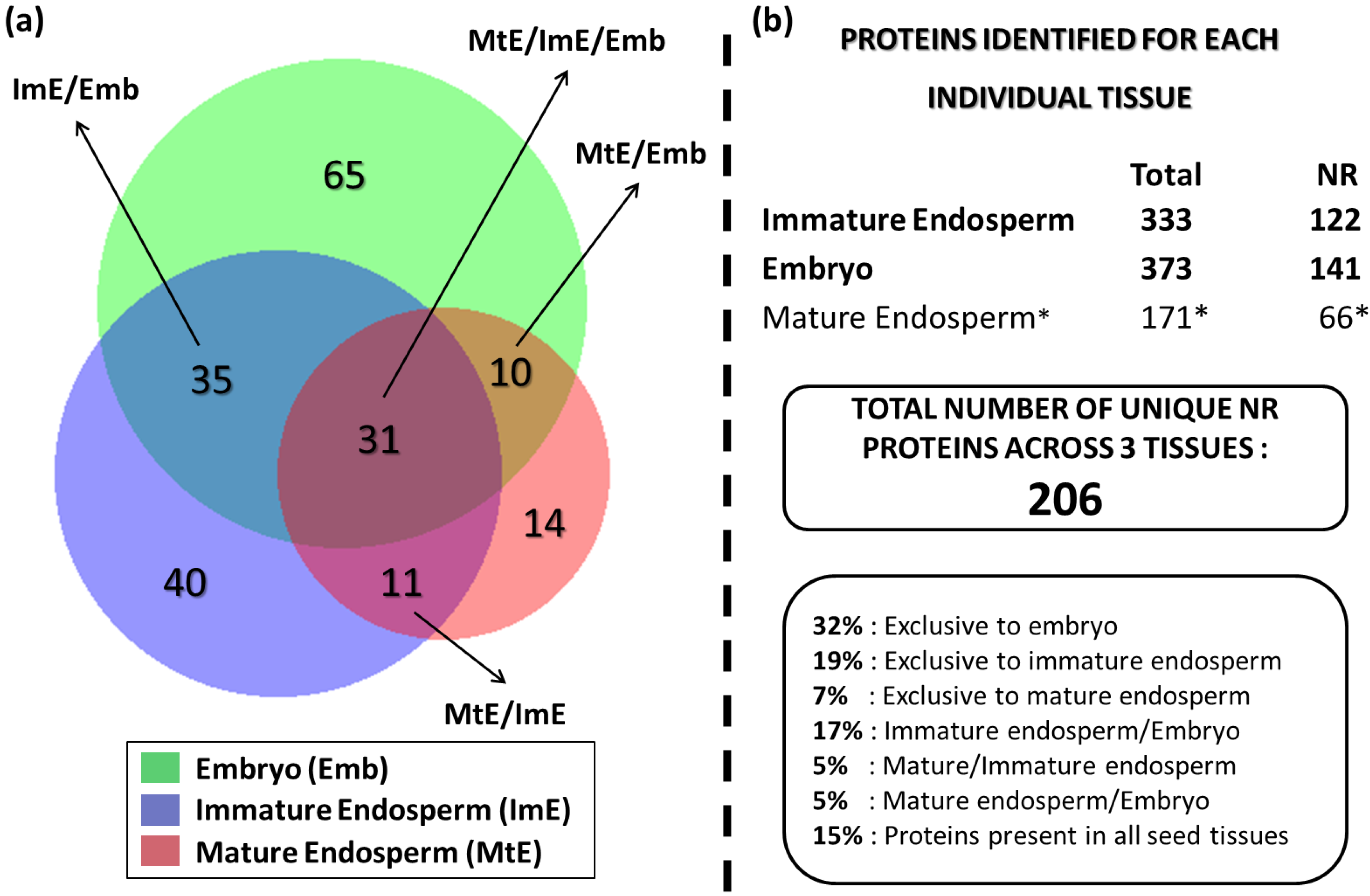
3.5. Functional Significance of the Identified Seed Proteins
| Protein Accession | Protein Description | Tissues 1 |
|---|---|---|
| 1433E_TOBAC | 14-3-3-like protein E OS = Nicotiana tabacum | M/I/E |
| HSP14_SOYBN | 17.5 kDa class I heat shock protein OS = Glycine max | I |
| HSP11_SOLLC | 17.8 kDa class I heat shock protein OS = Solanum lycopersicum | M |
| HSP11_PEA | 18.1 kDa class I heat shock protein OS = Pisum sativum | M/I/E |
| HSP12_MEDSA | 18.2 kDa class I heat shock protein OS = Medicago sativa | M/I |
| HSP11_CHERU | 18.3 kDa class I heat shock protein OS = Chenopodium rubrum | I |
| PMG1_ARATH | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase 1 OS = Arabidopsis thaliana | M/I/E |
| PRS6A_SOLLC | 26S protease regulatory subunit 6A homolog OS = Solanum lycopersicum | E |
| RPN1A_ARATH | 26S proteasome non-ATPase regulatory subunit 2 1A OS = Arabidopsis thaliana | E |
| PRS4A_ARATH | 26S proteasome regulatory subunit 4 homolog A OS = Arabidopsis thaliana | E |
| BAS1_ORYSJ | 2-Cys peroxiredoxin BAS1, chloroplastic OS = Oryza sativa subsp. Japonica | I |
| RS102_ARATH | 40S ribosomal protein S10-2 OS = Arabidopsis thaliana | I |
| RS13_PEA | 40S ribosomal protein S13 OS = Pisum sativum | I/E |
| RS14_CHLRE | 40S ribosomal protein S14 OS = Chlamydomonas reinhardtii | I/E |
| RS15A_DAUCA | 40S ribosomal protein S15a OS = Daucus carota | E |
| RS16_FRIAG | 40S ribosomal protein S16 OS = Fritillaria agrestis | I/E |
| RS18_ARATH | 40S ribosomal protein S18 OS = Arabidopsis thaliana | I/E |
| RS193_ARATH | 40S ribosomal protein S19-3 OS = Arabidopsis thaliana | I/E |
| RS254_ARATH | 40S ribosomal protein S25-4 OS = Arabidopsis thaliana | E |
| RS30_ARATH | 40S ribosomal protein S30 OS = Arabidopsis thaliana | E |
| RS33_ARATH | 40S ribosomal protein S3-3 OS = Arabidopsis thaliana | E |
| RS3A1_VITVI | 40S ribosomal protein S3a-1 OS = Vitis vinifera | I |
| RS4_GOSHI | 40S ribosomal protein S4 OS = Gossypium hirsutum | I/E |
| RS5_CICAR | 40S ribosomal protein S5 (fragment) OS = Cicer arietinum | I/E |
| RS6_ASPOF | 40S ribosomal protein S6 OS = Asparagus officinalis | I |
| RS61_ARATH | 40S ribosomal protein S6-1 OS = Arabidopsis thaliana | E |
| RS8_MAIZE | 40S ribosomal protein S8 OS = Zea mays | E |
| RS91_ARATH | 40S ribosomal protein S9-1 OS = Arabidopsis thaliana | M |
| RS92_ARATH | 40S ribosomal protein S9-2 OS = Arabidopsis thaliana | E |
| RSSA_SOYBN | 40S ribosomal protein SA OS = Glycine max | I/E |
| METE_ARATH | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase OS = Arabidopsis thaliana | M/I/E |
| RLA0_LUPLU | 60S acidic ribosomal protein P0 OS = Lupinus luteus | I/E |
| RLA2_PARAR | 60S acidic ribosomal protein P2 OS = Parthenium argentatum | E |
| RL10_VITRI | 60S ribosomal protein L10 OS = Vitis riparia | I |
| RL11_MEDSA | 60S ribosomal protein L11 OS = Medicago sativa | E |
| RL12_PRUAR | 60S ribosomal protein L12 OS = Prunus armeniaca | I |
| RL13_TOBAC | 60S ribosomal protein L13 OS = Nicotiana tabacum | I/E |
| RL17_MAIZE | 60S ribosomal protein L17 OS = Zea mays | E |
| RL182_ARATH | 60S ribosomal protein L18-2 OS = Arabidopsis thaliana | I |
| RL18A_CASSA | 60S ribosomal protein L18a OS = Castanea sativa | E |
| RL24_PRUAV | 60S ribosomal protein L24 OS = Prunus avium | E |
| RL3_ORYSJ | 60S ribosomal protein L3 OS = Oryza sativa subsp. Japonica | I/E |
| RL4_PRUAR | 60S ribosomal protein L4 OS = Prunus armeniaca | I/E |
| RL51_ARATH | 60S ribosomal protein L5-1 OS = Arabidopsis thaliana | I/E |
| RL6_MESCR | 60S ribosomal protein L6 OS = Mesembryanthemum crystallinum | I |
| RL7A1_ARATH | 60S ribosomal protein L7a-1 OS = Arabidopsis thaliana | E |
| ACOC_CUCMA | Aconitate hydratase, cytoplasmic OS = Cucurbita maxima | M/E |
| ACT_GOSHI | Actin OS = Gossypium hirsutum | I/E |
| ACT1_ORYSI | Actin-1 OS = Oryza sativa subsp. Indica | M |
| ACT12_SOLTU | Actin-100 (fragment) OS = Solanum tuberosum | M/E |
| ACT1_SOLLC | Actin-41 (fragment) OS = Solanum lycopersicum | M |
| ACT7_ARATH | Actin-7 OS = Arabidopsis thaliana | M |
| SAHH_MEDSA | Adenosylhomocysteinase OS = Medicago sativa | M/I/E |
| ADT1_GOSHI | ADP, ATP carrier protein 1, mitochondrial OS = Gossypium hirsutum | M/I/E |
| ARF_VIGUN | ADP-ribosylation factor OS = Vigna unguiculata | E |
| SYA_ARATH | Alanine--tRNA ligase OS = Arabidopsis thaliana | E |
| ADH1_SOLTU | Alcohol dehydrogenase 1 OS = Solanum tuberosum | M/I |
| PHSL_IPOBA | Alpha-1,4 glucan phosphorylase L isozyme, chloroplastic/amyloplastic OS = Ipomoea batatas | M/E |
| PHSH_ARATH | Alpha-glucan phosphorylase, H isozyme OS = Arabidopsis thaliana | M/I |
| GCST_PEA | Aminomethyltransferase, mitochondrial OS = Pisum sativum | I/E |
| ANXD1_ARATH | Annexin D1 OS = Arabidopsis thaliana | M/I/E |
| ANX4_FRAAN | Annexin-like protein RJ4 OS = Fragaria ananassa | E |
| CYF_AETCO | Apocytochrome f OS = Aethionema cordifolium | I |
| AATM_LUPAN | Aspartate aminotransferase P2, mitochondrial (fragment) OS = Lupinus angustifolius | E |
| AATM_LUPAN | Aspartate aminotransferase P2, mitochondrial (fragment) OS = Lupinus angustifolius | I |
| AAT3_ARATH | Aspartate aminotransferase, chloroplastic OS = Arabidopsis thaliana | I/E |
| AATC_DAUCA | Aspartate aminotransferase, cytoplasmic OS = Daucus carota | E |
| PYRB_ARATH | Aspartate carbamoyltransferase, chloroplastic OS = Arabidopsis thaliana | I |
| ATPAM_HELAN | ATP synthase subunit alpha, mitochondrial OS = Helianthus annuus | M/I/E |
| ATPBM_NICPL | ATP synthase subunit beta, mitochondrial OS = Nicotiana plumbaginifolia | M/I/E |
| ACLB1_ORYSJ | ATP-citrate synthase beta chain protein 1 OS = Oryza sativa subsp. Japonica | E |
| CLPA_BRANA | ATP-dependent Clp protease ATP-binding subunit clpA homolog, chloroplastic (fragment) OS = Brassica napus | I |
| APBLB_ARATH | Beta-adaptin-like protein B OS = Arabidopsis thaliana | E |
| ENO2_ARATH | Bifunctional enolase 2/transcriptional activator OS = Arabidopsis thaliana | I/E |
| ACCC2_POPTR | Biotin carboxylase 2, chloroplastic OS = Populus trichocarpa | E |
| CALSB_ARATH | Callose synthase 11 OS = Arabidopsis thaliana | E |
| CALX2_ARATH | Calnexin homolog 2 OS = Arabidopsis thaliana | E |
| CALR_BERST | Calreticulin OS = Berberis stolonifera | I |
| CATA2_RICCO | Catalase isozyme 2 OS = Ricinus communis | E |
| CD48A_ARATH | Cell division control protein 48 homolog A OS = Arabidopsis thaliana | M/I/E |
| CLPB1_ARATH | Chaperone protein ClpB1 OS = Arabidopsis thaliana | E |
| CLPC1_ARATH | Chaperone protein ClpC1, chloroplastic OS = Arabidopsis thaliana | I |
| CPNA1_ARATH | Chaperonin 60 subunit alpha 1, chloroplastic OS = Arabidopsis thaliana | I/E |
| CPNB3_ARATH | Chaperonin 60 subunit beta 3, chloroplastic OS = Arabidopsis thaliana | E |
| CH60A_ARATH | Chaperonin CPN60, mitochondrial OS = Arabidopsis thaliana | M/I/E |
| CB2_PHYPA | Chlorophyll a-b binding protein, chloroplastic OS = Physcomitrella patens subsp. patens | I |
| HSP7E_SPIOL | Chloroplast envelope membrane 70 kDa heat shock-related protein OS = Spinacia oleracea | M/I/E |
| HSP12_SOYBN | Class I heat shock protein (fragment) OS = Glycine max | I |
| CLAH1_ARATH | Clathrin heavy chain 1 OS = Arabidopsis thaliana | I/E |
| COPA1_ARATH | Coatomer subunit alpha-1 OS = Arabidopsis thaliana | E |
| COB21_ORYSJ | Coatomer subunit beta-1 OS = Oryza sativa subsp. Japonica | I |
| RH2_ORYSJ | DEAD-box ATP-dependent RNA helicase 2 OS = Oryza sativa subsp. Japonica | M |
| DIM_PEA | Delta(24)-sterol reductase OS = Pisum sativum | E |
| DLDH2_ARATH | Dihydrolipoyl dehydrogenase 2, mitochondrial OS = Arabidopsis thaliana | I |
| OPD22_ARATH | Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial OS = Arabidopsis thaliana | E |
| EF1A_TOBAC | Elongation factor 1-alpha OS = Nicotiana tabacum | M/I/E |
| EF1D1_ORYSJ | Elongation factor 1-delta 1 OS = Oryza sativa subsp. Japonica | E |
| EF1G2_ORYSJ | Elongation factor 1-gamma 2 OS = Oryza sativa subsp. Japonica | M/I/E |
| EF2_BETVU | Elongation factor 2 OS = Beta vulgaris | M/I/E |
| EFTM_ARATH | Elongation factor Tu, mitochondrial OS = Arabidopsis thaliana | E |
| ENPL_CATRO | Endoplasmin homolog OS = Catharanthus roseus | M/I/E |
| ENO1_HEVBR | Enolase 1 OS = Hevea brasiliensis | M/I/E |
| IF4A1_ARATH | Eukaryotic initiation factor 4A-1 OS = Arabidopsis thaliana | M/I/E |
| ERF1X_ARATH | Eukaryotic peptide chain release factor subunit 1-1 OS = Arabidopsis thaliana | E |
| IF5A1_ARATH | Eukaryotic translation initiation factor 5A-1 OS = Arabidopsis thaliana | E |
| ALF_CICAR | Fructose-bisphosphate aldolase, cytoplasmic isozyme OS = Cicer arietinum | M/I/E |
| RFS_ORYSJ | Galactinol--sucrose galactosyltransferase OS = Oryza sativa subsp. Japonica | I |
| GME2_ORYSJ | GDP-mannose 3,5-epimerase 2 OS = Oryza sativa subsp. Japonica | E |
| GMD1_ARATH | GDP-mannose 4,6 dehydratase 1 OS = Arabidopsis thaliana | E |
| GRDH1_ARATH | Glucose and ribitol dehydrogenase homolog 1 OS = Arabidopsis thaliana | M/I |
| GLGS_BETVU | Glucose-1-phosphate adenylyltransferase small subunit, chloroplastic/amyloplastic (fragment) OS = Beta vulgaris | M |
| G6PI2_CLACO | Glucose-6-phosphate isomerase, cytosolic 2 OS = Clarkia concinna | M/E |
| GPT2_ARATH | Glucose-6-phosphate/phosphate translocator 2, chloroplastic OS = Arabidopsis thaliana | M |
| GLN11_ORYSJ | Glutamine synthetase cytosolic isozyme 1-1 OS = Oryza sativa subsp. Japonica | E |
| G3PC_ANTMA | Glyceraldehyde-3-phosphate dehydrogenase, cytosolic OS = Antirrhinum majus | M/I/E |
| SYGM1_ARATH | Glycine--tRNA ligase 1, mitochondrial OS = Arabidopsis thaliana | E |
| SSG1_HORVU | Granule-bound starch synthase 1, chloroplastic/amyloplastic OS = Hordeum vulgare | M/I |
| RAN_VICFA | GTP-binding nuclear protein Ran/TC4 OS = Vicia faba | M/I/E |
| RAN3_ORYSI | GTP-binding nuclear protein Ran-3 OS = Oryza sativa subsp. Indica | E |
| YPTC1_CHLRE | GTP-binding protein YPTC1 OS = Chlamydomonas reinhardtii | E |
| GBLPA_ORYSJ | Guanine nucleotide-binding protein subunit beta-like protein A OS = Oryza sativa subsp. Japonica | I |
| HSP7L_ARATH | Heat shock 70 kDa protein 12 OS = Arabidopsis thaliana | I/E |
| HSP7O_ARATH | Heat shock 70 kDa protein 14 OS = Arabidopsis thaliana | I/E |
| HSP7N_ARATH | Heat shock 70 kDa protein 18 OS = Arabidopsis thaliana | I |
| HSP7D_ARATH | Heat shock 70 kDa protein 4 OS = Arabidopsis thaliana | I |
| HSP7F_ARATH | Heat shock 70 kDa protein 6, chloroplastic OS = Arabidopsis thaliana | I |
| HSP7G_ARATH | Heat shock 70 kDa protein 7, chloroplastic OS = Arabidopsis thaliana | E |
| HSP70_DAUCA | Heat shock 70 kDa protein OS = Daucus carota | M/I/E |
| HSP7M_PHAVU | Heat shock 70 kDa protein, mitochondrial OS = Phaseolus vulgaris | I/E |
| HSP80_SOLLC | Heat shock cognate protein 80 OS = Solanum lycopersicum | M/I |
| HS101_ARATH | Heat shock protein 101 OS = Arabidopsis thaliana | M |
| HS101_ORYSJ | Heat shock protein 101 OS = Oryza sativa subsp. Japonica | M |
| HSP81_ORYSI | Heat shock protein 81-1 OS = Oryza sativa subsp. Indica | M/I/E |
| HSP82_TOBAC | Heat shock protein 82 (fragment) OS = Nicotiana tabacum | M |
| HSP82_MAIZE | Heat shock protein 82 OS = Zea mays | M/I/E |
| HSP83_IPONI | Heat shock protein 83 OS = Ipomoea nil | M/I/E |
| HS901_ARATH | Heat shock protein 90-1 OS = Arabidopsis thaliana | E |
| HS903_ARATH | Heat shock protein 90-3 OS = Arabidopsis thaliana | I |
| H2AX_CICAR | Histone H2AX OS = Cicer arietinum | I |
| H2B_GOSHI | Histone H2B OS = Gossypium hirsutum | I/E |
| H4_ARATH | Histone H4 OS = Arabidopsis thaliana | I/E |
| IDHC_TOBAC | Isocitrate dehydrogenase [NADP] OS = Nicotiana tabacum | E |
| ILV5_ARATH | Ketol-acid reductoisomerase, chloroplastic OS = Arabidopsis thaliana | E |
| APX1_ORYSJ | L-ascorbate peroxidase 1, cytosolic OS = Oryza sativa subsp. Japonica | E |
| LE194_HORVU | Late embryogenesis abundant protein B19.4 OS = Hordeum vulgare | I |
| AMPL1_ARATH | Leucine aminopeptidase 1 OS = Arabidopsis thaliana | M/I |
| BIP4_TOBAC | Luminal-binding protein OS = Nicotiana tabacum | M/I/E |
| MDHC2_ARATH | Malate dehydrogenase, cytoplasmic 2 OS = Arabidopsis thaliana | M/E |
| MDHM_CITLA | Malate dehydrogenase, mitochondrial OS = Citrullus lanatus | M/I/E |
| MPPA_SOLTU | Mitochondrial-processing peptidase subunit alpha OS = Solanum tuberosum | E |
| MDAR_SOLLC | Monodehydroascorbate reductase OS = Solanum lycopersicum | I/E |
| MAOX_POPTR | NADP-dependent malic enzyme OS = Populus trichocarpa | M |
| NDK1_ARATH | Nucleoside diphosphate kinase 1 OS = Arabidopsis thaliana | M/I/E |
| FKB62_ARATH | Peptidyl-prolyl cis-trans isomerase FKBP62 OS = Arabidopsis thaliana | I/E |
| PER1B_ARMRU | Peroxidase C1B OS = Armoracia rusticana | I |
| CAPPC_FLATR | Phosphoenolpyruvate carboxylase 2 OS = Flaveria trinervia | E |
| PGMC_PEA | Phosphoglucomutase, cytoplasmic OS = Pisum sativum | M/I/E |
| PGKH_TOBAC | Phosphoglycerate kinase, chloroplastic OS = Nicotiana tabacum | M/I |
| PGKY_TOBAC | Phosphoglycerate kinase, cytosolic OS = Nicotiana tabacum | M/E |
| SERC_SPIOL | Phosphoserine aminotransferase, chloroplastic OS = Spinacia oleracea | E |
| PDR4_ORYSJ | Pleiotropic drug resistance protein 4 OS = Oryza sativa subsp. Japonica | E |
| PARP3_SOYBN | Poly [ADP-ribose] polymerase 3 OS = Glycine max | I |
| UBIQP_ACECL | Polyubiquitin (fragment) OS = Acetabularia cliftonii | I |
| UBQ12_ARATH | Polyubiquitin 12 OS = Arabidopsis thaliana | E |
| PMG2_ARATH | Probable 2,3-bisphosphoglycerate-independent phosphoglycerate mutase 2 OS = Arabidopsis thaliana | M/I |
| SSG1_ARATH | Probable granule-bound starch synthase 1, chloroplastic/amyloplastic OS = Arabidopsis thaliana | I |
| H2B1_MEDTR | Probable histone H2B.1 OS = Medicago truncatula | I |
| PDIA6_MEDSA | Probable protein disulfide-isomerase A6 OS = Medicago sativa | I |
| Y1497_ARATH | Probable receptor-like protein kinase At1g49730 OS = Arabidopsis thaliana | I |
| PROF3_ARATH | Profilin-3 OS = Arabidopsis thaliana | E |
| PSA3_ARATH | Proteasome subunit alpha type-3 OS = Arabidopsis thaliana | E |
| PDI21_ORYSJ | Protein disulfide isomerase-like 2-1 OS = Oryza sativa subsp. Japonica | I |
| PDI21_ARATH | Protein disulfide-isomerase like 2-1 OS = Arabidopsis thaliana | M/I/E |
| ACT5_ARATH | Putative actin-5 OS = Arabidopsis thaliana | I/E |
| YCF1_IPOPU | Putative membrane protein ycf1 OS = Ipomoea purpurea | M/I |
| AVP_VIGRR | Pyrophosphate-energized vacuolar membrane proton pump OS = Vigna radiata var. radiata | I/E |
| PDC1_TOBAC | Pyruvate decarboxylase isozyme 1 (fragment) OS = Nicotiana tabacum | M/I |
| KPYC_SOYBN | Pyruvate kinase, cytosolic isozyme OS = Glycine max | M/I/E |
| PPDK2_ORYSJ | Pyruvate, phosphate dikinase 2 OS = Oryza sativa subsp. Japonica | M |
| PPDK_FLABR | Pyruvate, phosphate dikinase, chloroplastic OS = Flaveria brownii | M/E |
| RAA1D_ARATH | Ras-related protein RABA1d OS = Arabidopsis thaliana | E |
| RBL_MAIZE | Ribulose bisphosphate carboxylase large chain OS = Zea mays | I |
| RUBA_RICCO | RuBisCO large subunit-binding protein subunit alpha (fragment) OS = Ricinus communis | I/E |
| RUBB_PEA | RuBisCO large subunit-binding protein subunit beta, chloroplastic OS = Pisum sativum | E |
| METK4_POPTR | S-adenosylmethionine synthase 4 OS = Populus trichocarpa | E |
| SAPK6_ORYSJ | Serine/threonine-protein kinase SAPK6 OS = Oryza sativa subsp. Japonica | E |
| HSP7S_SPIOL | Stromal 70 kDa heat shock-related protein, chloroplastic (fragment) OS = Spinacia oleracea | I/E |
| SUSY_SOYBN | Sucrose synthase OS = Glycine max | I/E |
| SODM_HEVBR | Superoxide dismutase [Mn], mitochondrial OS = Hevea brasiliensis | E |
| TCPA_ARATH | T-complex protein 1 subunit alpha OS = Arabidopsis thaliana | I/E |
| TCPE_ARATH | T-complex protein 1 subunit epsilon OS = Arabidopsis thaliana | M/E |
| TKTC_SPIOL | Transketolase, chloroplastic OS = Spinacia oleracea | E |
| TCTP_TOBAC | Translationally-controlled tumor protein homolog OS = Nicotiana tabacum | M |
| TPIS_MAIZE | Triosephosphate isomerase, cytosolic OS = Zea mays | I/E |
| TBA_PRUDU | Tubulin alpha chain OS = Prunus dulcis | I |
| TBB_HORVU | Tubulin beta chain OS = Hordeum vulgare | E |
| UBIQ_ARATH | Ubiquitin OS = Arabidopsis thaliana | M/E |
| RL40A_ARATH | Ubiquitin-60S ribosomal protein L40-1 OS = Arabidopsis thaliana | I/E |
| RGP1_ORYSJ | UDP-arabinopyranose mutase 1 OS = Oryza sativa subsp. Japonica | E |
| UGDH_SOYBN | UDP-glucose 6-dehydrogenase OS = Glycine max | E |
| UREA_CANEN | Urease OS = Canavalia ensiformis | I |
| UGPA1_ARATH | UTP--glucose-1-phosphate uridylyltransferase 1 OS = Arabidopsis thaliana | M/E |
| VATA_GOSHI | V-type proton ATPase catalytic subunit A OS = Gossypium hirsutum | I/E |
| VATB2_GOSHI | V-type proton ATPase subunit B 2 (fragment) OS = Gossypium hirsutum | E |
| VATB1_ARATH | V-type proton ATPase subunit B1 OS = Arabidopsis thaliana | E |
| WIT2_ARATH | WPP domain-interacting tail-anchored protein 2 OS = Arabidopsis thaliana | I |

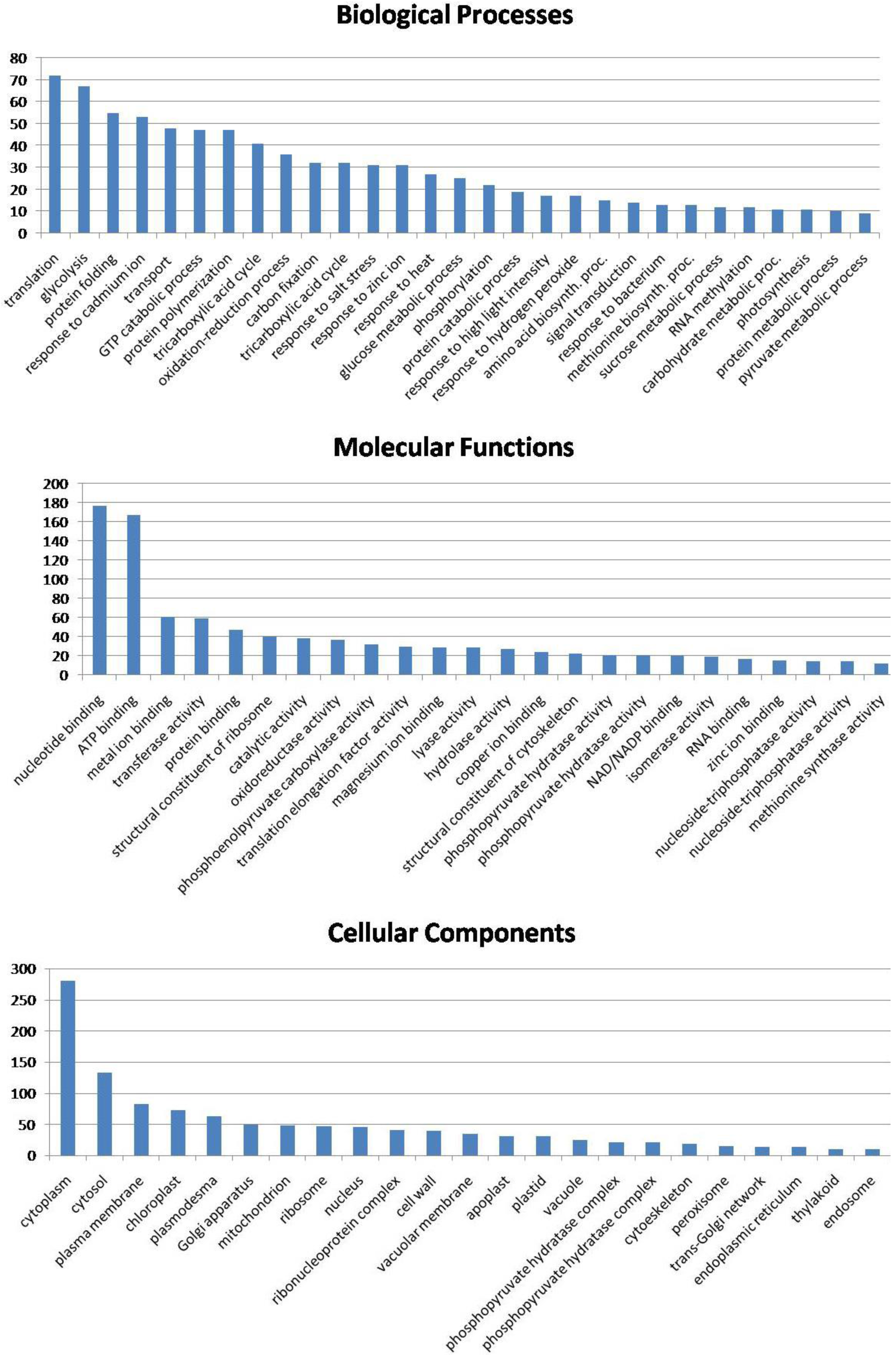
3.6. Biological Function of the Identified Seed Proteins
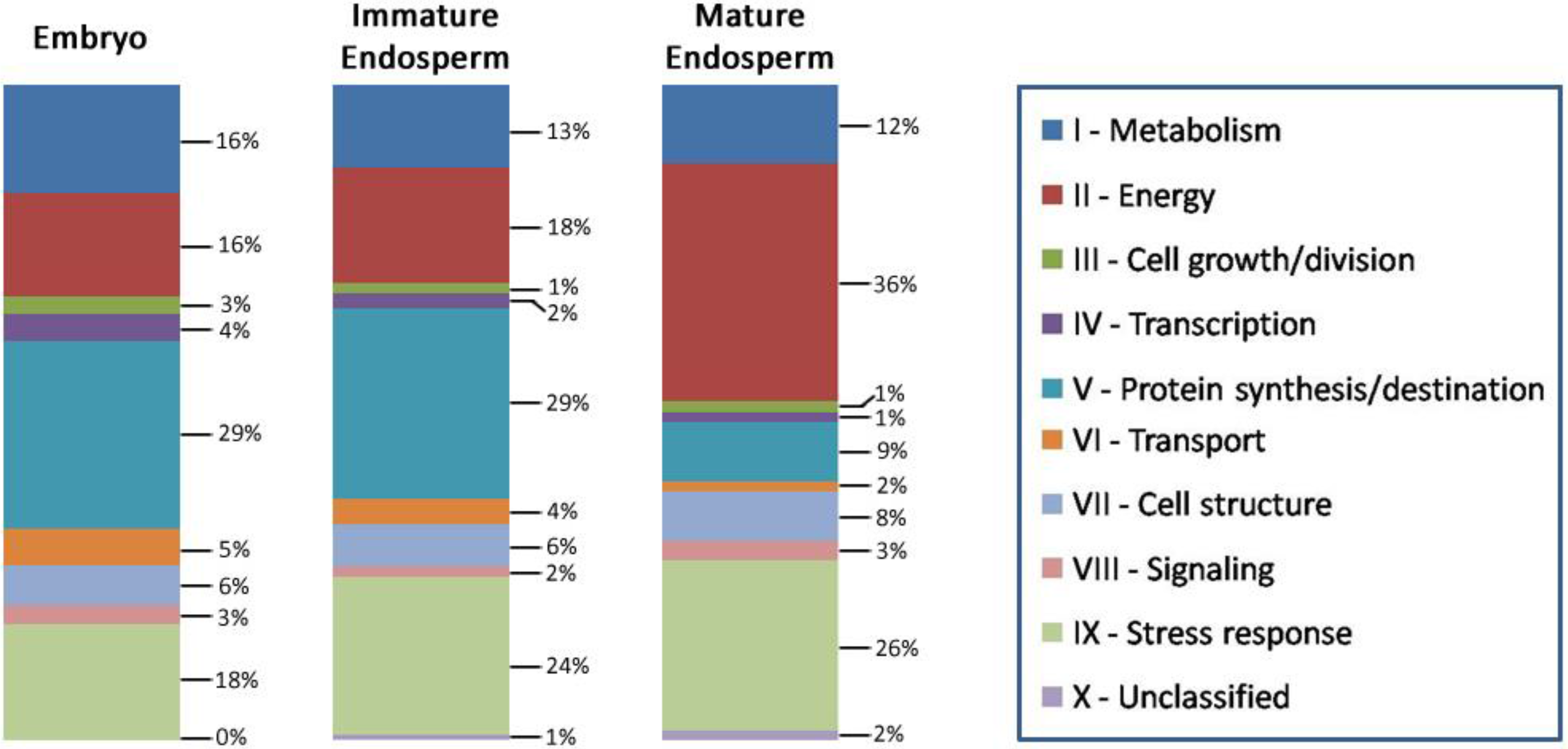
3.7. Lotus Seed Proteome Compared with Other Seed Proteomes
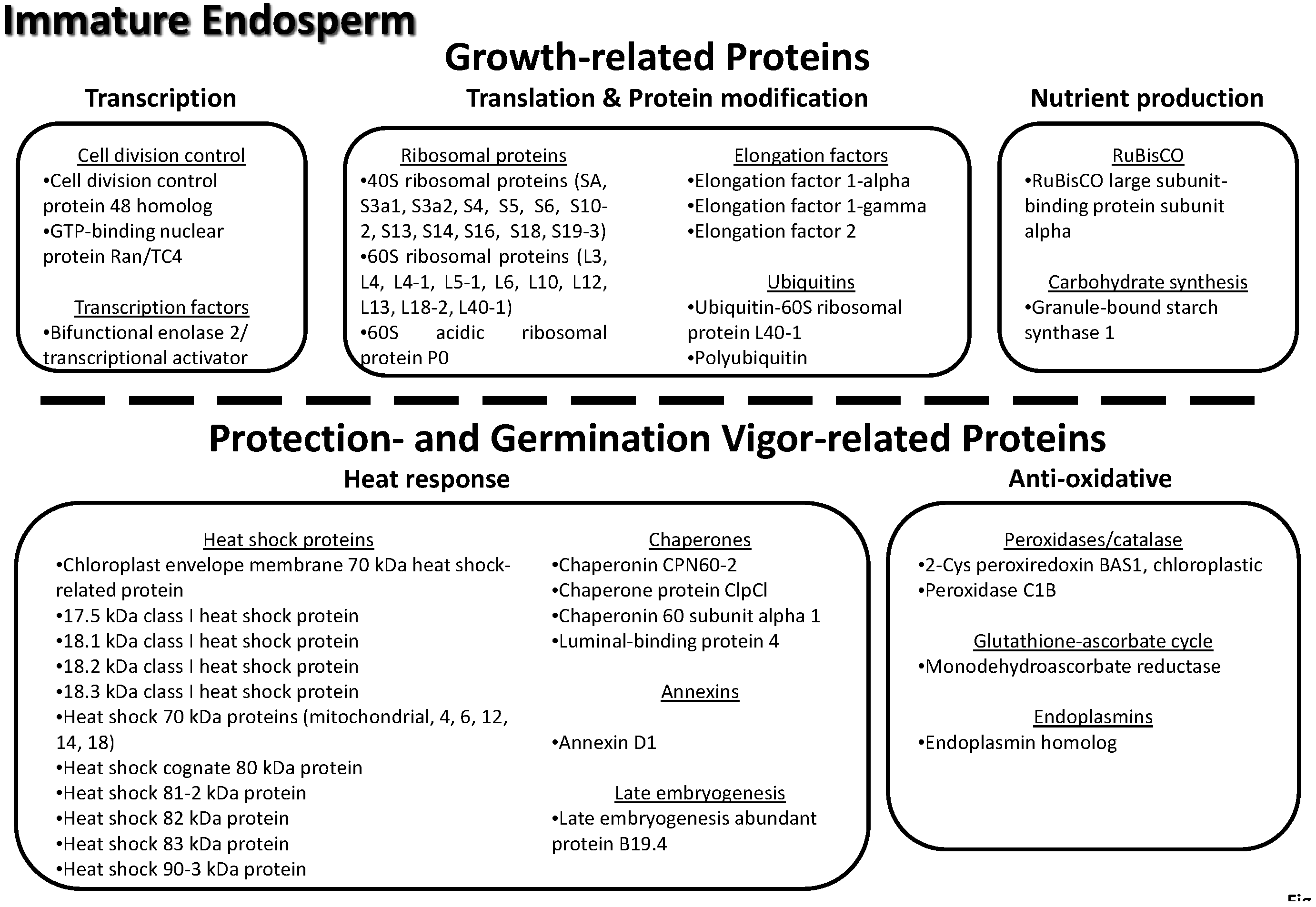
3.8. Key Proteins of the Lotus Immature Seed Endosperm
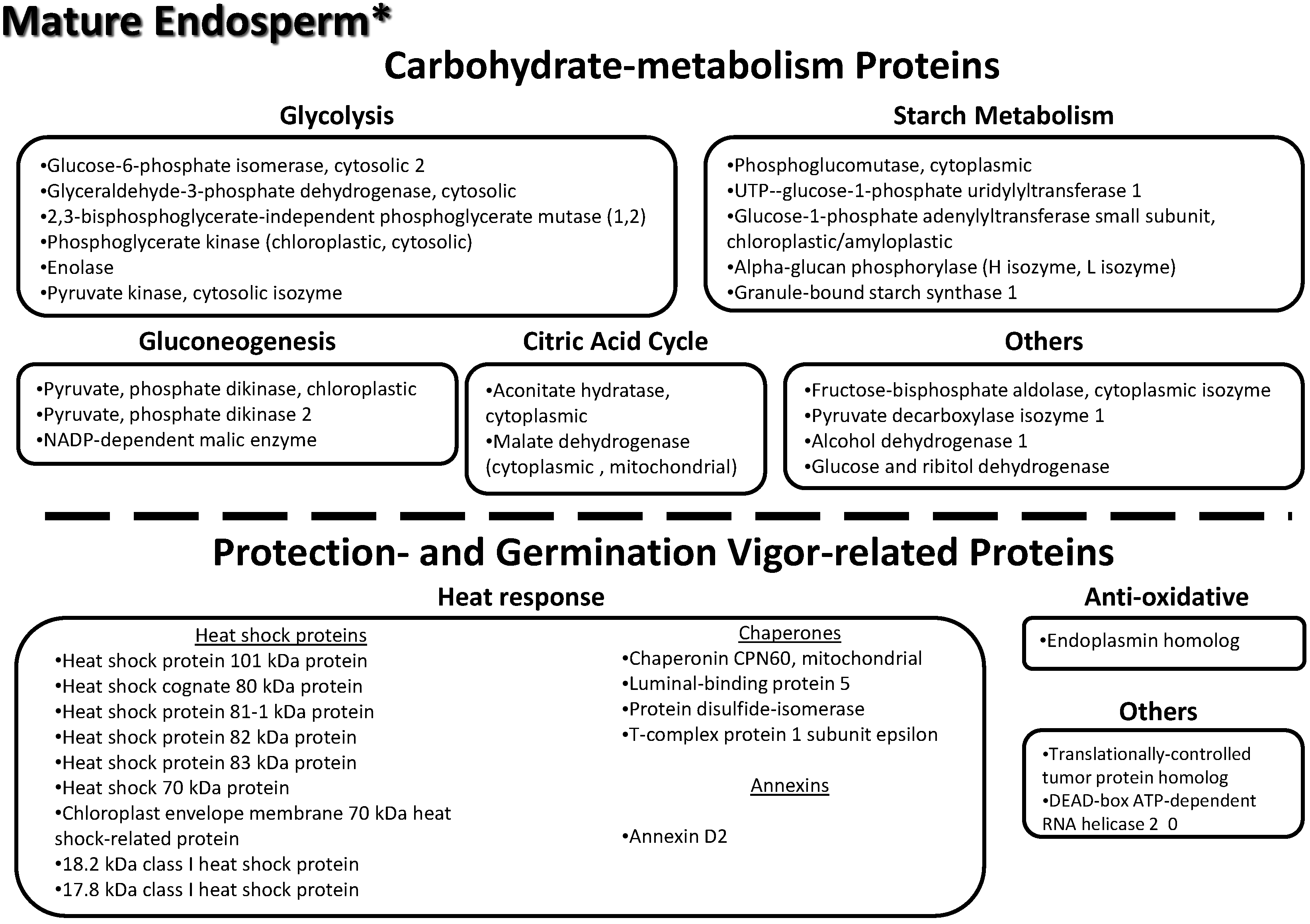
3.9. Key Proteins Previously Identified in the Lotus Mature Seed Endosperm
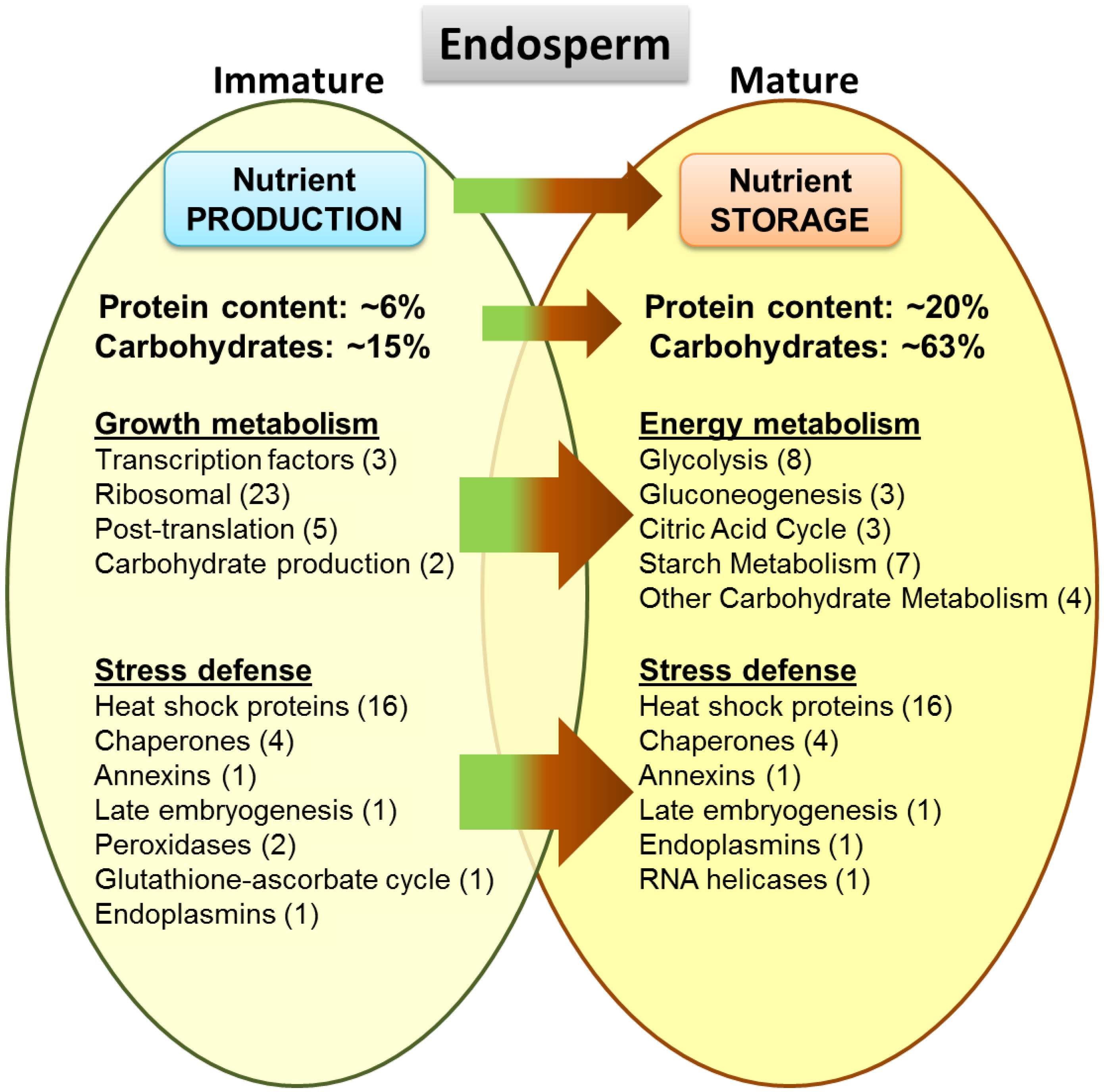
3.10. Proteome Changes between Mature and Immature Stages of the Endosperm
3.11. Key Proteins of the Lotus Seed Embryo
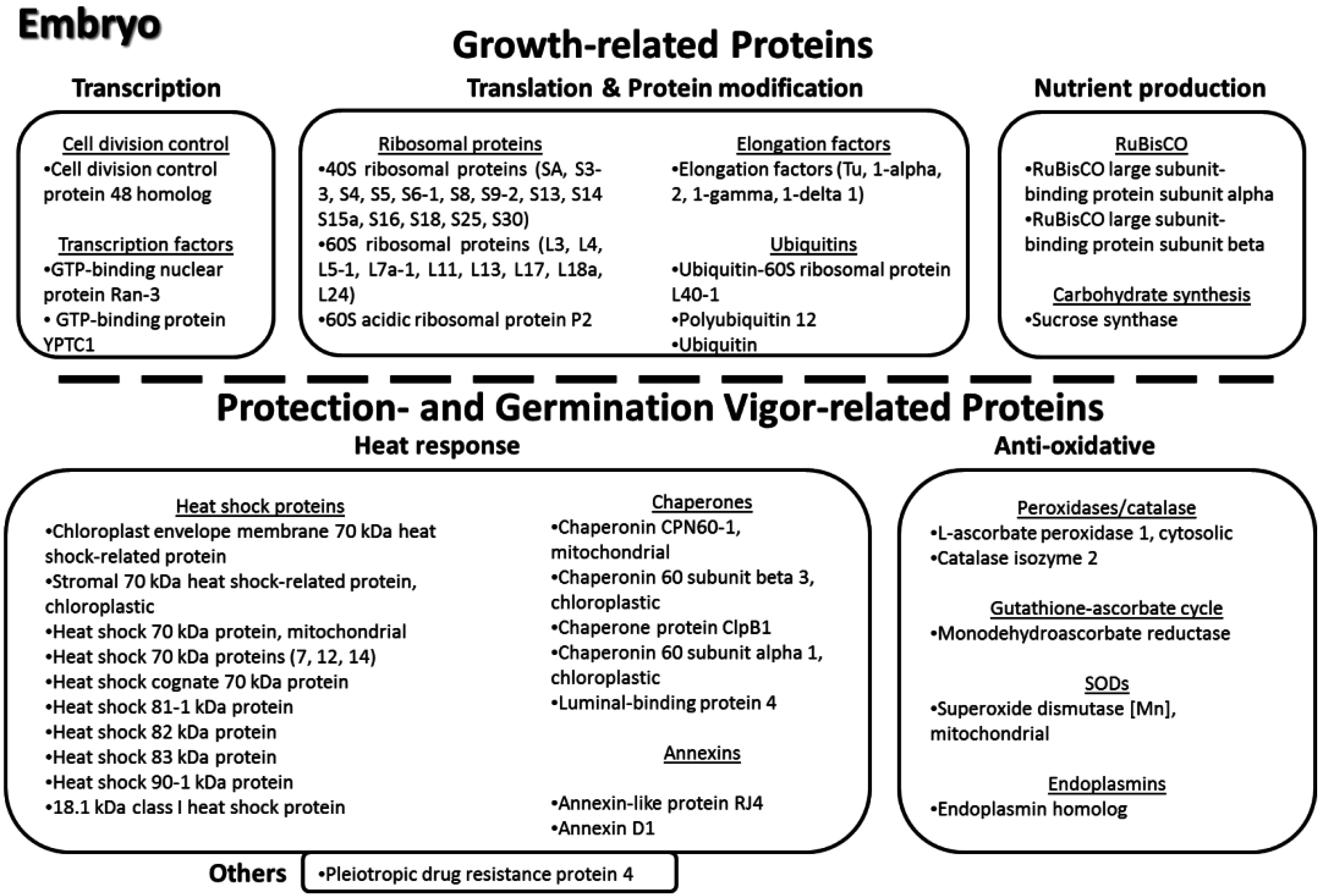
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sridhar, K.R.; Rajeev, B. Lotus—A potential nutraceutical source. J. Agric. Technol. 2007, 3, 143–155. [Google Scholar]
- Pandey, B.P. Economic Botany, 5th ed.; Chand (S.) & Co. Ltd.: New Delhi, India, 1999; p. 61. [Google Scholar]
- Loewer, H.P. Seeds: The Definitive Guide to Growing, History and Lore, 1st ed.; Timber Press: Cambridge, UK, 2005; p. 56. [Google Scholar]
- Shen-Miller, J.; Schopf, J.W.; Harbottle, G.; Cao, R.J.; Ouyang, S.; Zhou, K.S.; Southon, J.R.; Liu, G.H. Long-living lotus: germination and soil γ-irradiation of centuries-old fruits, and cultivation, growth, and phenotypic abnormalities of offspring. Am. J. Bot. 2002, 89, 236–247. [Google Scholar] [CrossRef] [PubMed]
- Shen-Miller, J.; Aung, L.H.; Turek, J.; Schopf, J.W.; Tholandi, M.; Yang, M.; Czaja, M. Centuries-old viable fruit of sacred lotus Nelumbo nucifera Gaertn var. China antique. Trop. Plant Biol. 2013, 6, 53–68. [Google Scholar] [CrossRef]
- MEXT—Ministry of Education, Culture, Sports, Science and Technology. Standard Tables of Food Composition in Japan, 5th ed.; MEXT: Tokyo, Japan, 2000. [Google Scholar]
- Guo, H.B. Cultivation of lotus (Nelumbo nucifera Gaertn. ssp. nucifera) and its utilization in China. Gen. Res. Crop Evol. 2009, 56, 323–330. [Google Scholar] [CrossRef]
- Komatsu, E.; Tsukahara, A.; Amagaya, H.; Okazawa, N.; Noguchi, T.; Okuyama, T. Lotus. In The Cultivation and Management in Aquatic Vegetables; Izaki, M., Ed.; Ie-No-Hikari Kyokai Press: Tokyo, Japan, 1975; Volume 1, pp. 9–94. [Google Scholar]
- Ling, Z.Q.; Xie, B.J.; Yang, E.L. Isolation, characterization, and determination of antioxidative activity of oligomeric procyanidins from the seedpod of Nelumbo nucifera Gaertn. J. Agric. Food Chem. 2005, 53, 2441–2445. [Google Scholar] [CrossRef] [PubMed]
- Ou, M. Chinese-English Manual of Commonly-Used in Traditional Chinese Medicine; Joint Publishing Co. Ltd.: Hong Kong, China, 1989. [Google Scholar]
- Moro, C.F.; Yonekura, M.; Kouzuma, Y.; Agrawal, G.K.; Rakwal, R. Lotus—A source of food and medicine: Current status and future perspectives in context of the seed proteomics. Int. J. Life Sci. 2013, 7, 1–5. [Google Scholar] [CrossRef]
- Ming, R.; Vanburen, R.; Liu, Y.; Yang, M.; Han, Y.; Li, L.T.; Zhang, Q.; Kim, M.J.; Schatz, M.C.; Campbell, M.; et al. Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.). Genome Bio. 2013, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, C.; Zheng, X.; Li, G.; Zhu, H.; Zhou, M.; Hu, Z. Molecular cloning and expression of two cytosolic copper–zinc superoxide dismutases genes from Nelumbo nucifera. Appl. Biochem. Biotechnol. 2010, 163, 679–691. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Chen, H.; Chu, P.; Li, Y.; Tan, B.; Ding, Y.; Tsang, E.W.T.; Jiang, L.; Wu, K.; Huang, S. NnHSP17.5, a cytosolic class II small heat shock protein gene from Nelumbo nucifera, contributes to seed germination vigor and seedling thermotolerance in transgenic Arabidopsis. Plant Cell Rep. 2012, 31, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gu, G.; Chen, F.; Yang, D.; Wu, K.; Chen, S.; Jiang, J.; Zhang, Z. Heterologous expression of a Nelumbo nucifera phytochelatin synthase gene enhances cadmium tolerance in Arabidopsis thaliana. Appl. Biochem. Biotechnol. 2012, 166, 722–734. [Google Scholar] [CrossRef] [PubMed]
- Chu, P.; Chen, H.; Zhou, Y.; Li, Y.; Ding, Y.; Jiang, L.; Tsang, E.W.; Wu, K.; Huang, S. 2012. Proteomic and functional analyses of Nelumbo nucifera annexins involved in seed thermotolerance and germination vigor. Planta 2012, 235, 1271–1288. [Google Scholar] [CrossRef] [PubMed]
- Moro, C.F.; Fukao, Y.; Shibato, J.; Rakwal, R.; Timperio, A.M.; Zolla, L.; Agrawal, G.K.; Shioda, S.; Kouzuma, Y.; Yonekura, M. Unraveling the seed endosperm proteome of the lotus (Nelumbo nucifera Gaertn.) utilizing 1DE and 2DE separation in conjunction with tandem mass spectrometry. Proteomics 2015, 15, 1717–1735. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Wang, X.; Li, X.; Li, Y. A modified Coomassie Brilliant Blue staining method at nanogram sensitivity compatible with proteomic analysis. Biotech. Lett. 2007, 29, 1599–1603. [Google Scholar]
- Horie, K.; Rakwal, R.; Hirano, M.; Shibato, J.; Nam, H.W.; Kim, Y.S.; Kouzuma, Y.; Agrawal, G.K.; Masuo, Y.; Yonekura, M. Proteomics of two cultivated mushrooms Sparassis crispa and Hericium erinaceum provides insight into their numerous functional protein components and diversity. J. Proteome Res. 2008, 7, 1819–1835. [Google Scholar] [CrossRef] [PubMed]
- Yates, J.R.; Ruse, C.I.; Nakorchevsky, A. Proteomics by mass spectrometry: Approaches, advances, and applications. Ann. Rev. Biomed. Eng. 2009, 11, 49–79. [Google Scholar] [CrossRef] [PubMed]
- Bevan, M.; Bancroft, I.; Bent, E.; Love, K.; Goodman, H.; Dean, C.; Bergkamp, R.; Dirkse, W.; van Staveren, M.; Stiekema, W.; et al. Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 1998, 391, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yang, Z.; Yang, M; Shen, S. The differential proteome of endosperm and embryo from mature seed of Jatropha curcas. Plant Sci. 2011, 181, 660–666. [Google Scholar] [CrossRef] [PubMed]
- Sheoran, I.S.; Olson, D.J.; Ross, A.R.; Sawhney, V.K. Proteome analysis of embryo and endosperm from germinating tomato seeds. Proteomics 2005, 5, 3752–3764. [Google Scholar] [CrossRef] [PubMed]
- Catusse, J.; Strub, J.M.; Job, C.; Dorsselaer, A.; Job, D. Proteome-wide characterization of sugarbeet seed vigor and its tissue specific expression. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 10262–10267. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Gao, Y.; Xu, H.; Zhang, Y.; Wang, J. A proteomic analysis of seed development in Brassica campestri L. PLoS ONE 2012, 7, e50290. [Google Scholar] [CrossRef] [PubMed]
- Moro, C.F. Study of the Lotus (Nelumbo nucifera Gaertn.) Seed Proteome. Ph.D. Thesis, Tokyo University of Agriculture and Technology, Tokyo, Japan, March 2015. [Google Scholar]
- Ravanel, S.; Gakière, B.; Job, D.; Douce, R. The specific features of methionine biosynthesis and metabolism in plants. Proc. Natl. Acad. Sci. U.S.A. 1998, 95, 7805–7812. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moro, C.F.; Fukao, Y.; Shibato, J.; Rakwal, R.; Agrawal, G.K.; Shioda, S.; Kouzuma, Y.; Yonekura, M. Immature Seed Endosperm and Embryo Proteomics of the Lotus (Nelumbo Nucifera Gaertn.) by One-Dimensional Gel-Based Tandem Mass Spectrometry and a Comparison with the Mature Endosperm Proteome. Proteomes 2015, 3, 184-235. https://doi.org/10.3390/proteomes3030184
Moro CF, Fukao Y, Shibato J, Rakwal R, Agrawal GK, Shioda S, Kouzuma Y, Yonekura M. Immature Seed Endosperm and Embryo Proteomics of the Lotus (Nelumbo Nucifera Gaertn.) by One-Dimensional Gel-Based Tandem Mass Spectrometry and a Comparison with the Mature Endosperm Proteome. Proteomes. 2015; 3(3):184-235. https://doi.org/10.3390/proteomes3030184
Chicago/Turabian StyleMoro, Carlo F., Yoichiro Fukao, Junko Shibato, Randeep Rakwal, Ganesh Kumar Agrawal, Seiji Shioda, Yoshiaki Kouzuma, and Masami Yonekura. 2015. "Immature Seed Endosperm and Embryo Proteomics of the Lotus (Nelumbo Nucifera Gaertn.) by One-Dimensional Gel-Based Tandem Mass Spectrometry and a Comparison with the Mature Endosperm Proteome" Proteomes 3, no. 3: 184-235. https://doi.org/10.3390/proteomes3030184
APA StyleMoro, C. F., Fukao, Y., Shibato, J., Rakwal, R., Agrawal, G. K., Shioda, S., Kouzuma, Y., & Yonekura, M. (2015). Immature Seed Endosperm and Embryo Proteomics of the Lotus (Nelumbo Nucifera Gaertn.) by One-Dimensional Gel-Based Tandem Mass Spectrometry and a Comparison with the Mature Endosperm Proteome. Proteomes, 3(3), 184-235. https://doi.org/10.3390/proteomes3030184