S-Nitrosylation in Organs of Mice Exposed to Low or High Doses of γ-Rays: The Modulating Effect of Iodine Contrast Agent at a Low Radiation Dose
Abstract
:1. Introduction
2. Experimental Section
2.1. Animals and Radiation Treatment
2.2. Biotin Switch Analysis of Protein Nitrosylation
2.3. Immuno-Precipitation and Detection of S-Nitrosylated Proteins
2.4. Analysis of Nitrosylated Proteins and Peptides by Mass Spectrometry
2.5. Ingenuity Pathway Analysis
2.6. Protein Classification and Functional Analysis
2.7. Statistical Analysis
3. Results
3.1. The Effect of Low and High Doses of γ-Rays on Global Changes in S-Nitrosylation in Organs of Irradiated Mice: The Impact of Iodine Contrast Agent on the Low-Dose Radiation Response
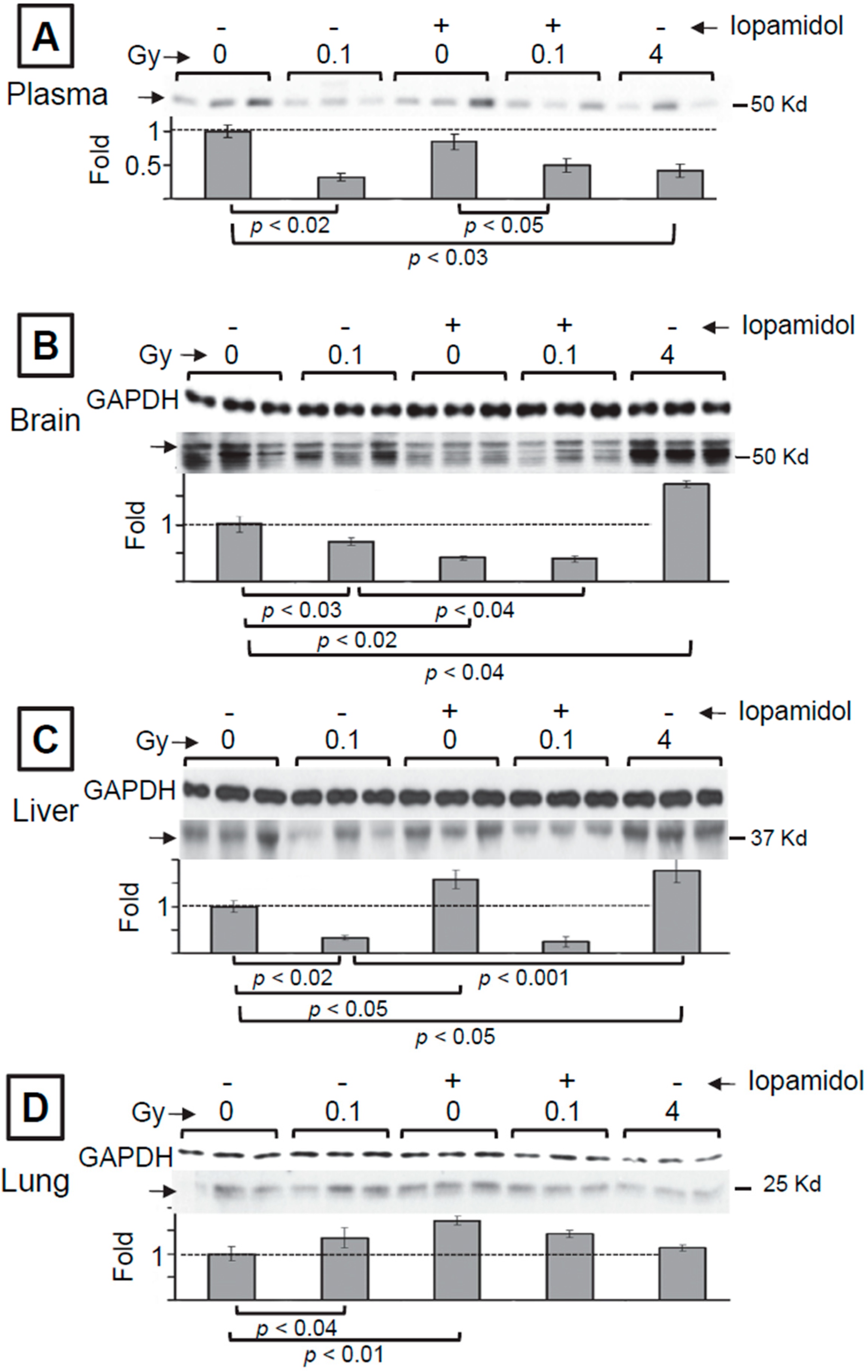
3.2. Mass Spectrometry Analysis of S-Nitrosylated Proteins in Brains of Irradiated Mice

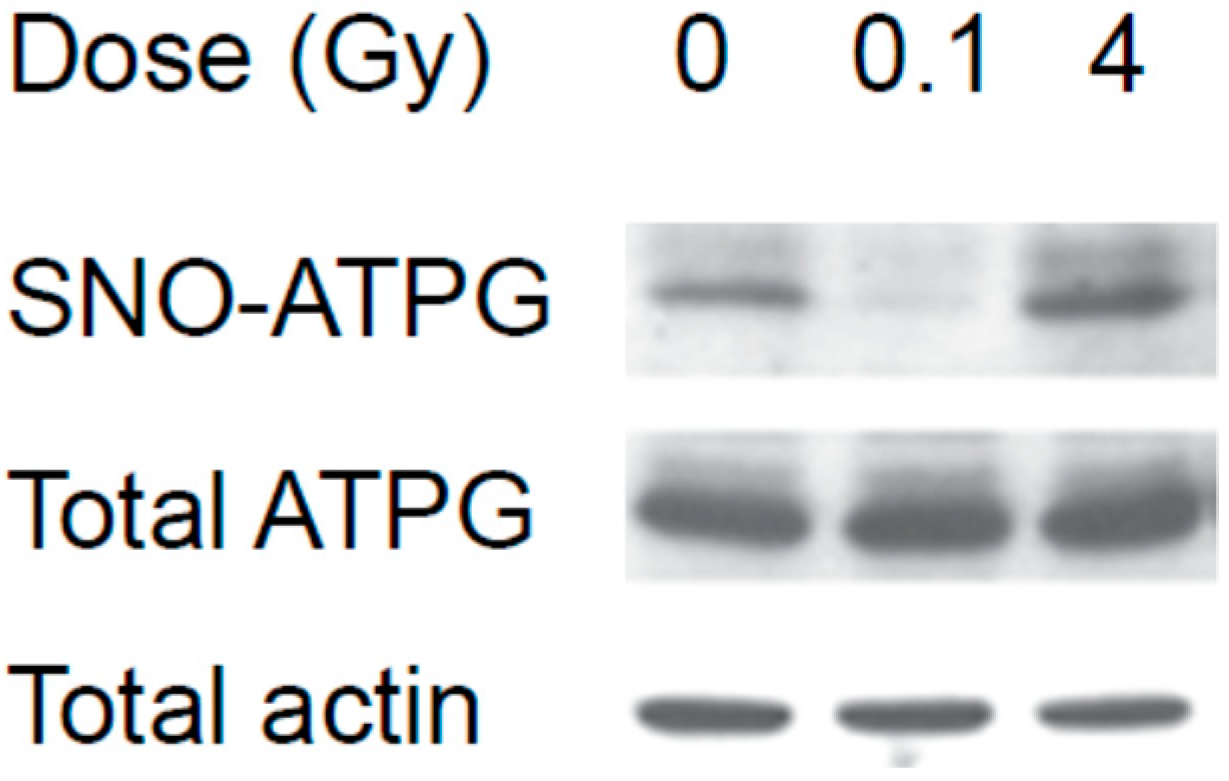
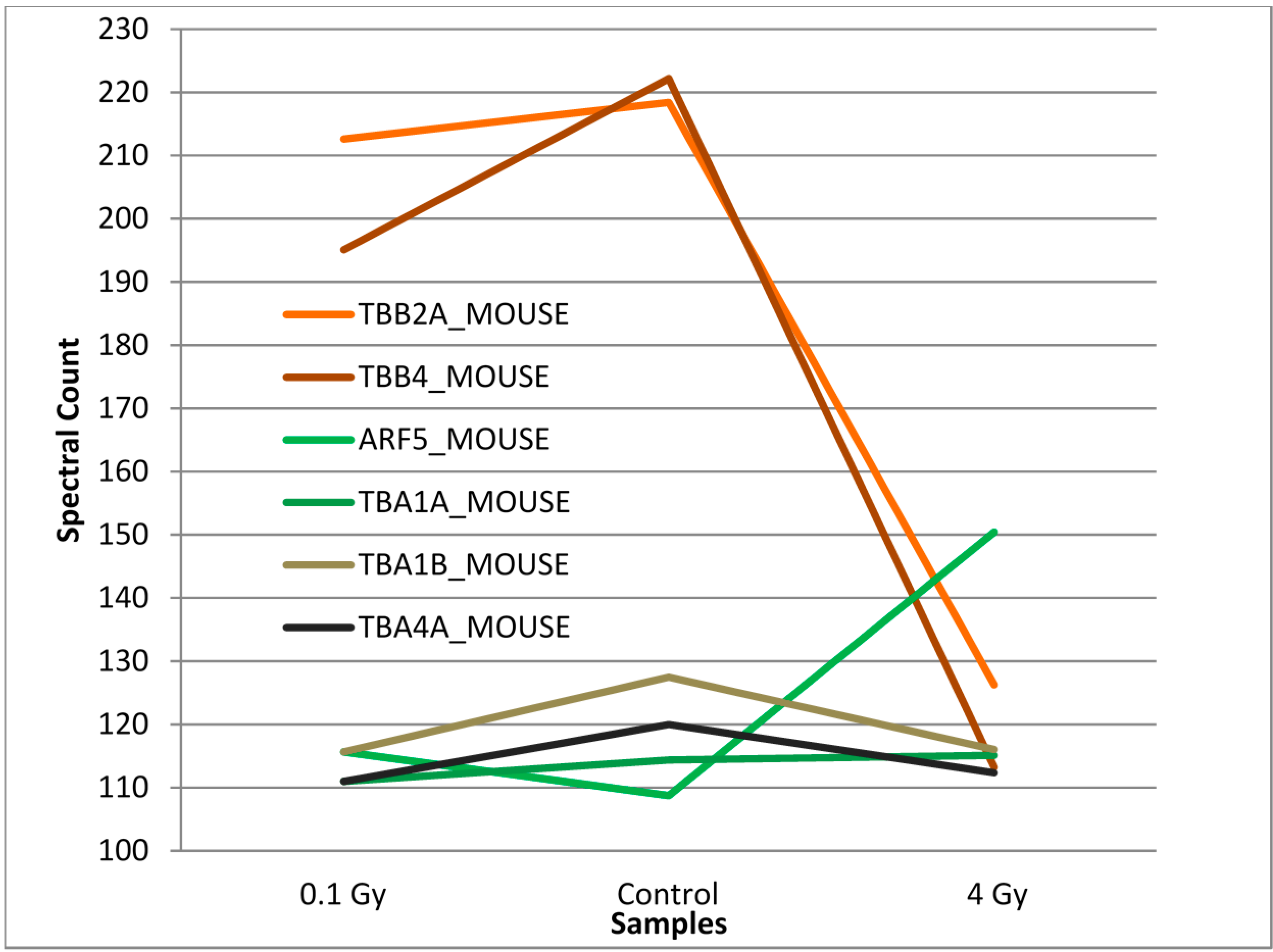
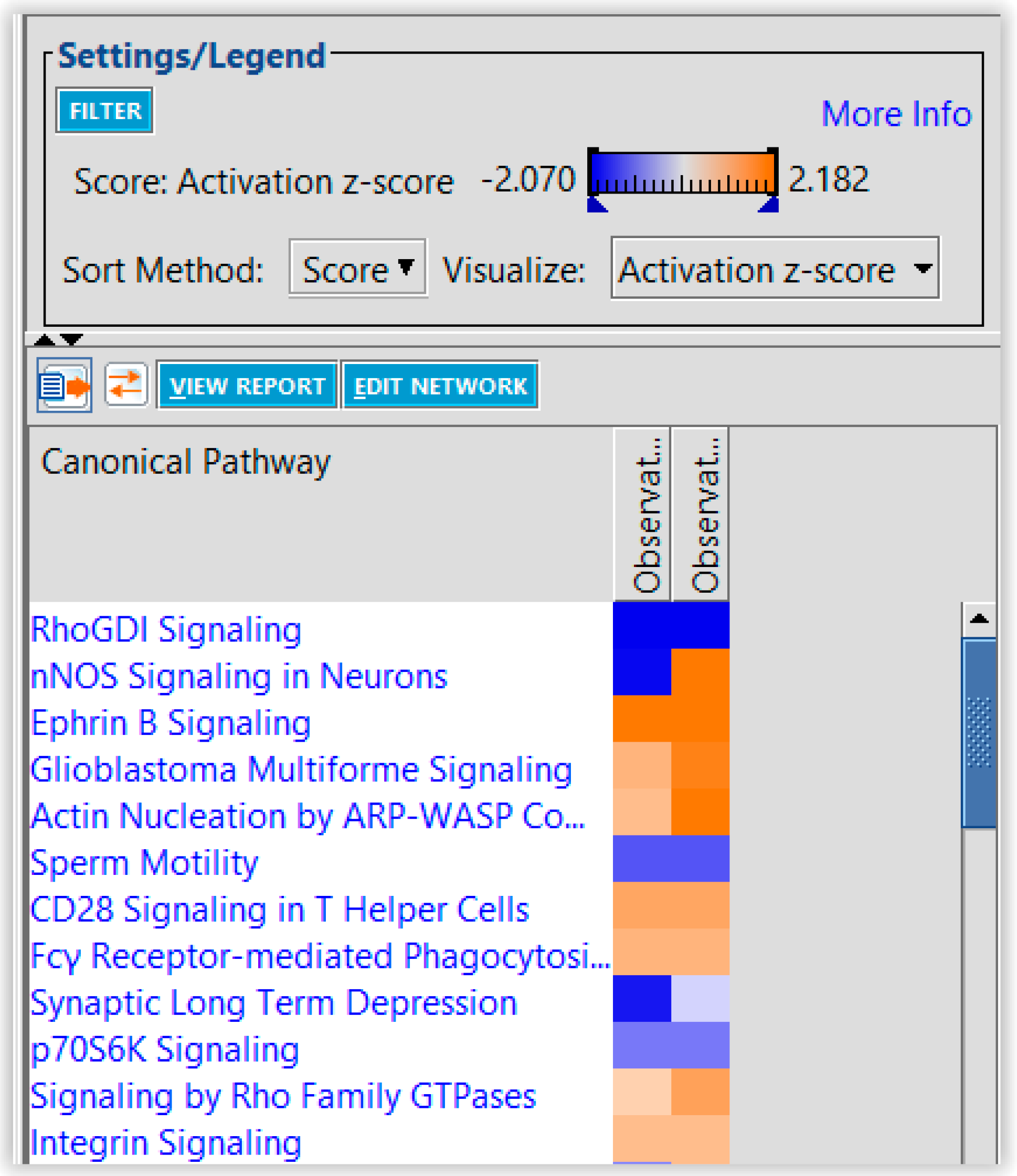
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflict of Interest
References
- NCRP. Ionizing Radiation Exposure of the Population of the United States; Report No. 160; National Council on Radiation Protection and Measurements: Bethesda, MD, USA, 3 March 2009. [Google Scholar]
- Azzam, E.I.; Jay-Gerin, J.P.; Pain, D. Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Lett 2012, 327, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Tomonaga, M. Leukaemia in nagasaki atomic bomb survivors from 1945 through 1959. Bull. World Health Organ. 1962, 26, 619–631. [Google Scholar] [PubMed]
- Little, J.B. Failla memorial lecture. Changing views of cellular radiosensitivity. Radiat. Res. 1994, 140, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Ron, E. Cancer risks from medical radiation. Health Phys. 2003, 85, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Tubiana, M.; Feinendegen, L.E.; Yang, C.; Kaminski, J.M. The linear no-threshold relationship is inconsistent with radiation biologic and experimental data. Radiology 2009, 251, 13–22. [Google Scholar] [CrossRef] [PubMed]
- BEIR-VII. In Health Risks from Exposure to Low Levels of Ionizing Radiation; National Research Council of the National Academies: Washington, DC, USA, 2005.
- Averbeck, D. Does scientific evidence support a change from the lnt model for low-dose radiation risk extrapolation? Health Phys. 2009, 97, 493–504. [Google Scholar] [CrossRef] [PubMed]
- Mullenders, L.; Atkinson, M.; Paretzke, H.; Sabatier, L.; Bouffler, S. Assessing cancer risks of low-dose radiation. Nat. Rev. Cancer 2009, 9, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Amundson, S.A.; Fornace, A.J., Jr. Gene expression profiles for monitoring radiation exposure. Radiat. Prot. Dosim. 2001, 97, 11–16. [Google Scholar] [CrossRef]
- Azzam, E.I.; de Toledo, S.M.; Little, J.B. Expression of connexin43 is highly sensitive to ionizing radiation and environmental stresses. Cancer Res. 2003, 63, 7128–7135. [Google Scholar] [PubMed]
- Coleman, M.A.; Yin, E.; Peterson, L.E.; Nelson, D.; Sorensen, K.; Tucker, J.D.; Wyrobek, A.J. Low-dose irradiation alters the transcript profiles of human lymphoblastoid cells including genes associated with cytogenetic radioadaptive response. Radiat. Res. 2005, 164, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Amundson, S.A. Development of gene expression signatures for practical radiation biodosimetry. Int. J. Radiat. Oncol. Biol. Phys. 2008, 71, 1236–1244. [Google Scholar] [CrossRef] [PubMed]
- Barjaktarovic, Z.; Anastasov, N.; Azimzadeh, O.; Sriharshan, A.; Sarioglu, H.; Ueffing, M.; Tammio, H.; Hakanen, A.; Leszczynski, D.; Atkinson, M.J.; et al. Integrative proteomic and microrna analysis of primary human coronary artery endothelial cells exposed to low-dose γ radiation. Radiat. Environ. Biophys. 2013, 52, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, M.A.; Omaruddin, R.A.; Kreger, B.; de Toledo, S.M.; Azzam, E.I. Micro RNA responses to chronic or acute exposures to low dose ionizing radiation. Mol. Biol. Rep. 2012, 39, 7549–7558. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; de Toledo, S.M.; Pandey, B.N.; Guo, G.; Pain, D.; Li, H.; Azzam, E.I. Role of the translationally controlled tumor protein in DNA damage sensing and repair. Proc. Natl. Acad. Sci. USA 2012, 109, E926–E933. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, M.V.; Azimzadeh, O.; Barjaktarovic, Z.; Kempf, S.J.; Merl-Pham, J.; Hauck, S.M.; Buratovic, S.; Eriksson, P.; Atkinson, M.J.; Tapio, S. Total body exposure to low-dose ionizing radiation induces long-term alterations to the liver proteome of neonatally exposed mice. J. Proteome Res. 2014, 14, 366–373. [Google Scholar] [CrossRef]
- Zhang, Q.; Matzke, M.; Schepmoes, A.A.; Moore, R.J.; Webb-Robertson, B.J.; Hu, Z.; Monroe, M.E.; Qian, W.J.; Smith, R.D.; Morgan, W.F. High and low doses of ionizing radiation induce different secretome profiles in a human skin model. PloS ONE 2014, 9, e92332. [Google Scholar] [CrossRef] [PubMed]
- Chatgilialoglu, C.; Ferreri, C.; Torreggiani, A.; Salzano, A.M.; Renzone, G.; Scaloni, A. Radiation-induced reductive modifications of sulfur-containing amino acids within peptides and proteins. J. Proteomics 2011, 74, 2264–2273. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, G.; Bodycote, J.; Wolff, S. Adaptive response of human lymphocytes to low concentrations of radioactive thymidine. Science 1984, 223, 594–597. [Google Scholar] [CrossRef] [PubMed]
- ICRP. Recommendations of the International Commission on Radiological Protection. Pergamon Press: Oxford, UK, 1990. Available online: http://www.icrp.org/publication.asp?id=ICRP%20Publication%2060 (accessed on 4 March 2015).
- Ozawa, K.; Whalen, E.J.; Nelson, C.D.; Mu, Y.; Hess, D.T.; Lefkowitz, R.J.; Stamler, J.S. S-nitrosylation of β-arrestin regulates β-adrenergic receptor trafficking. Mol. Cell 2008, 31, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Foster, M.W.; McMahon, T.J.; Stamler, J.S. S-nitrosylation in health and disease. Trends Mol. Med. 2003, 9, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Switzer, C.H.; Cheng, R.Y.; Ridnour, L.A.; Glynn, S.A.; Ambs, S.; Wink, D.A. Ets-1 is a transcriptional mediator of oncogenic nitric oxide signaling in estrogen receptor-negative breast cancer. Breast Cancer Res. 2012, 14, R125. [Google Scholar] [CrossRef]
- Nakamura, T.; Tu, S.; Akhtar, M.W.; Sunico, C.R.; Okamoto, S.; Lipton, S.A. Aberrant protein S-nitrosylation in neurodegenerative diseases. Neuron 2013, 78, 596–614. [Google Scholar] [CrossRef]
- Li, F.; Sonveaux, P.; Rabbani, Z.N.; Liu, S.; Yan, B.; Huang, Q.; Vujaskovic, Z.; Dewhirst, M.W.; Li, C.Y. Regulation of HIF-1α stability through S-nitrosylation. Mol. Cell 2007, 26, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, H.; Hayashi, S.; Hatashita, M.; Shioura, H.; Ohtsubo, T.; Kitai, R.; Ohnishi, T.; Yukawa, O.; Furusawa, Y.; Kano, E. Induction of radioresistance to accelerated carbon-ion beams in recipient cells by nitric oxide excreted from irradiated donor cells of human glioblastoma. Int. J. Radiat. Biol. 2000, 76, 1649–1657. [Google Scholar] [CrossRef] [PubMed]
- Leach, J.K.; Van Tuyle, G.; Lin, P.S.; Schmidt-Ullrich, R.; Mikkelsen, R.B. Ionizing radiation-induced, mitochondria-dependent generation of reactive oxygen/nitrogen. Cancer Res. 2001, 61, 3894–3901. [Google Scholar] [PubMed]
- Ames, B.N. Endogenous oxidative DNA damage, aging, and cancer. Free Radic. Res. Commun. 1989, 7, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Yuhas, J.M.; Storer, J.B. On mouse strain differences in radiation resistance: Hematopoietic death and the endogenous colony-forming unit. Radiat. Res. 1969, 39, 608–622. [Google Scholar] [CrossRef] [PubMed]
- Hall, E.J.; Giaccia, A.J. Radiobiology for the Radiologist, 6th ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2006. [Google Scholar]
- Wu, C.; Parrott, A.M.; Liu, T.; Beuve, A.; Li, H. Functional proteomics approaches for the identification of transnitrosylase and denitrosylase targets. Methods 2013, 62, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Elias, J.E.; Thoreen, C.C.; Licklider, L.J.; Gygi, S.P. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: The yeast proteome. J. Proteome Res. 2003, 2, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Ingenuity Pathway Analysis (IPA), QIAGEN, Redwood City, CA. Available online: http://www.qiagen.com/ingenuity (accessed on 4 March 2015).
- PANTHER (Protein ANalysis THrough Evolutionary Relationships) classification system. Available online: http://pantherdb.org/ (accessed on 4 March 2015).
- Halloran, M.; Parakh, S.; Atkin, J.D. The role of S-nitrosylation and S-glutathionylation of protein disulphide isomerase in protein misfolding and neurodegeneration. Int. J. Cell Biol. 2013, 2013, 797914. [Google Scholar] [CrossRef]
- Little, M.P. A review of non-cancer effects, especially circulatory and ocular diseases. Radiat. Environ. Biophys. 2013, 52, 435–449. [Google Scholar] [CrossRef] [PubMed]
- Kempf, S.J.; Casciati, A.; Buratovic, S.; Janik, D.; von Toerne, C.; Ueffing, M.; Neff, F.; Moertl, S.; Stenerlow, B.; Saran, A.; et al. The cognitive defects of neonatally irradiated mice are accompanied by changed synaptic plasticity, adult neurogenesis and neuroinflammation. Mol. Neurodegener. 2014, 9, 57. [Google Scholar] [CrossRef]
- Pearce, M.S.; Salotti, J.A.; Little, M.P.; McHugh, K.; Lee, C.; Kim, K.P.; Howe, N.L.; Ronckers, C.M.; Rajaraman, P.; Sir Craft, A.W.; et al. Radiation exposure from ct scans in childhood and subsequent risk of leukaemia and brain tumours: A retrospective cohort study. Lancet 2012, 380, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Bredt, D.S.; Snyder, S.H. Isolation of nitric oxide synthetase, a calmodulin-requiring enzyme. Proc. Natl. Acad. Sci. USA 1990, 87, 682–685. [Google Scholar] [CrossRef] [PubMed]
- Knowles, R.G.; Moncada, S. Nitric oxide synthases in mammals. Biochem. J. 1994, 298, 249–258. [Google Scholar] [PubMed]
- Huttlin, E.L.; Jedrychowski, M.P.; Elias, J.E.; Goswami, T.; Rad, R.; Beausoleil, S.A.; Villen, J.; Haas, W.; Sowa, M.E.; Gygi, S.P. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 2010, 143, 1174–1189. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.J.; Seto, E. Lysine acetylation: Codified crosstalk with other posttranslational modifications. Mol Cell 2008, 31, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Hess, D.T.; Stamler, J.S. Regulation by S-nitrosylation of protein post-translational modification. J. Biol. Chem. 2012, 287, 4411–4418. [Google Scholar] [CrossRef] [PubMed]
- Harapanhalli, R.S.; Yaghmai, V.; Patel, Y.D.; Baker, S.R.; Rao, D.V. Assay of radiographic contrast agents in mice plasma and testes by high performance liquid chromatography. Anal. Chem. 1993, 65, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Yaghmai, V.; Harapanhalli, R.S.; Patel, Y.D.; Baker, S.R.; Rao, D.V. Effects of diatrizoate and iopamidol on spermatogenesis. Invest. Radiol. 1993, 28, 1160–1164. [Google Scholar] [CrossRef] [PubMed]
- Almog, T.; Naor, Z. The role of mitogen activated protein kinase (MAPK) in sperm functions. Mol. Cell Endocrinol. 2010, 314, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Numajiri, N.; Takasawa, K.; Nishiya, T.; Tanaka, H.; Ohno, K.; Hayakawa, W.; Asada, M.; Matsuda, H.; Azumi, K.; Kamata, H.; et al. On-off system for pi3-kinase-akt signaling through S-nitrosylation of phosphatase with sequence homology to tensin (PTEN). Proc. Natl. Acad. Sci. USA 2011, 108, 10349–10354. [Google Scholar] [CrossRef] [PubMed]
- Downward, J. Pi 3-kinase, akt and cell survival. Semin. Cell Dev. Biol. 2004, 15, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Jaffrey, S.R.; Erdjument-Bromage, H.; Ferris, C.D.; Tempst, P.; Snyder, S.H. Protein S-nitrosylation: A physiological signal for neuronal nitric oxide. Nat. Cell Biol. 2001, 3, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Hao, G.; Derakhshan, B.; Shi, L.; Campagne, F.; Gross, S.S. Snosid, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc. Natl. Acad. Sci. USA 2006, 103, 1012–1017. [Google Scholar] [CrossRef] [PubMed]
- Landino, L.M.; Koumas, M.T.; Mason, C.E.; Alston, J.A. Modification of tubulin cysteines by nitric oxide and nitroxyl donors alters tubulin polymerization activity. Chem. Res. Toxicol. 2007, 20, 1693–1700. [Google Scholar] [CrossRef] [PubMed]
- McMurray, C.T. Neurodegeneration: Diseases of the cytoskeleton? Cell Death Differ. 2000, 7, 861–865. [Google Scholar] [CrossRef] [PubMed]
- D’Souza-Schorey, C.; Chavrier, P. Arf proteins: Roles in membrane traffic and beyond. Nat. Rev. 2006, 7, 347–358. [Google Scholar] [CrossRef]
- Takatsu, H.; Yoshino, K.; Toda, K.; Nakayama, K. GGA proteins associate with golgi membranes through interaction between their ggah domains and ADP-ribosylation factors. Biochem. J. 2002, 365, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, P.L. Mechanism of the Na+, K+ pump. Protein structure and conformations of the pure (Na+ + K+)-atpase. Biochim. Biophys. Acta 1982, 694, 27–68. [Google Scholar] [CrossRef] [PubMed]
- Clausen, M.J.; Poulsen, H. Sodium/potassium homeostasis in the cell. Met. Ions Life Sci. 2013, 12, 41–67. [Google Scholar] [PubMed]
- Ginsberg, M.D. Neuroprotection for ischemic stroke: Past, present and future. Neuropharmacology 2008, 55, 363–389. [Google Scholar] [CrossRef] [PubMed]
- Gisone, P.; Boveris, A.D.; Dubner, D.; Perez, M.R.; Robello, E.; Puntarulo, S. Early neuroprotective effect of nitric oxide in developing rat brain irradiated in utero. Neurotoxicology 2003, 24, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Russel, M.C.; Delman, K.A. Comparative effectiveness in melanoma. Cancer Treat Res. 2015, 164, 31–49. [Google Scholar] [PubMed]
- Lea, D.E. The inactivation of viruses by radiations. Br. J. Radiol. 1946, 19, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Barker, S.; Weinfeld, M.; Zheng, J.; Li, L.; Murray, D. Identification of mammalian proteins cross-linked to DNA by ionizing radiation. J. Biol. Chem. 2005, 280, 33826–33838. [Google Scholar] [CrossRef] [PubMed]
- Berovic, N.; Pratontep, S.; Bryant, A.; Montouris, A.; Green, R.G. The kinetics of radiation damage to the protein luciferase and recovery of enzyme activity after irradiation. Radiat. Res. 2002, 157, 122–127. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, P.; Wardman, P. Radiation chemistry comes before radiation biology. Int. J. Radiat. Biol. 2009, 85, 9–25. [Google Scholar] [CrossRef] [PubMed]
- Tsang, A.H.; Lee, Y.I.; Ko, H.S.; Savitt, J.M.; Pletnikova, O.; Troncoso, J.C.; Dawson, V.L.; Dawson, T.M.; Chung, K.K. S-nitrosylation of xiap compromises neuronal survival in Parkinson’s disease. Proc. Natl. Acad. Sci. USA 2009, 106, 4900–4905. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.; Campbell, S.L. Mechanism of p21Ras S-nitrosylation and kinetics of nitric oxide-mediated guanine nucleotide exchange. Biochemistry 2004, 43, 2314–2322. [Google Scholar] [CrossRef] [PubMed]
- Carrier, S.; Hricak, H.; Lee, S.S.; Baba, K.; Morgan, D.M.; Nunes, L.; Ross, G.Y.; Phillips, T.L.; Lue, T.F. Radiation-induced decrease in nitric oxide synthase—Containing nerves in the rat penis. Radiology 1995, 195, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, H.; Tomita, M.; Otsuka, K.; Hatashita, M.; Hamada, N. Nitric oxide is a key molecule serving as a bridge between radiation-induced bystander and adaptive responses. Curr. Mol. Pharmacol. 2011, 4, 126–134. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nicolas, F.; Wu, C.; Bukhari, S.; De Toledo, S.M.; Li, H.; Shibata, M.; Azzam, E.I. S-Nitrosylation in Organs of Mice Exposed to Low or High Doses of γ-Rays: The Modulating Effect of Iodine Contrast Agent at a Low Radiation Dose. Proteomes 2015, 3, 56-73. https://doi.org/10.3390/proteomes3020056
Nicolas F, Wu C, Bukhari S, De Toledo SM, Li H, Shibata M, Azzam EI. S-Nitrosylation in Organs of Mice Exposed to Low or High Doses of γ-Rays: The Modulating Effect of Iodine Contrast Agent at a Low Radiation Dose. Proteomes. 2015; 3(2):56-73. https://doi.org/10.3390/proteomes3020056
Chicago/Turabian StyleNicolas, Fadia, Changgong Wu, Salwa Bukhari, Sonia M. De Toledo, Hong Li, Masayuki Shibata, and Edouard I. Azzam. 2015. "S-Nitrosylation in Organs of Mice Exposed to Low or High Doses of γ-Rays: The Modulating Effect of Iodine Contrast Agent at a Low Radiation Dose" Proteomes 3, no. 2: 56-73. https://doi.org/10.3390/proteomes3020056
APA StyleNicolas, F., Wu, C., Bukhari, S., De Toledo, S. M., Li, H., Shibata, M., & Azzam, E. I. (2015). S-Nitrosylation in Organs of Mice Exposed to Low or High Doses of γ-Rays: The Modulating Effect of Iodine Contrast Agent at a Low Radiation Dose. Proteomes, 3(2), 56-73. https://doi.org/10.3390/proteomes3020056




