Proteomics Advances in the Understanding of Pollen–Pistil Interactions
Abstract
:1. Introduction
2. Proteomic Analysis of Pollen-Pistil Interaction with Successful Fertilization
2.1. Proteome Dynamics in Pollen at Different Points from Development to Germination
2.2. Protein Analysis on Pistils by Comparative Proteomics
3. Proteomic Analysis of Pollen-Pistil Interaction in SI Response
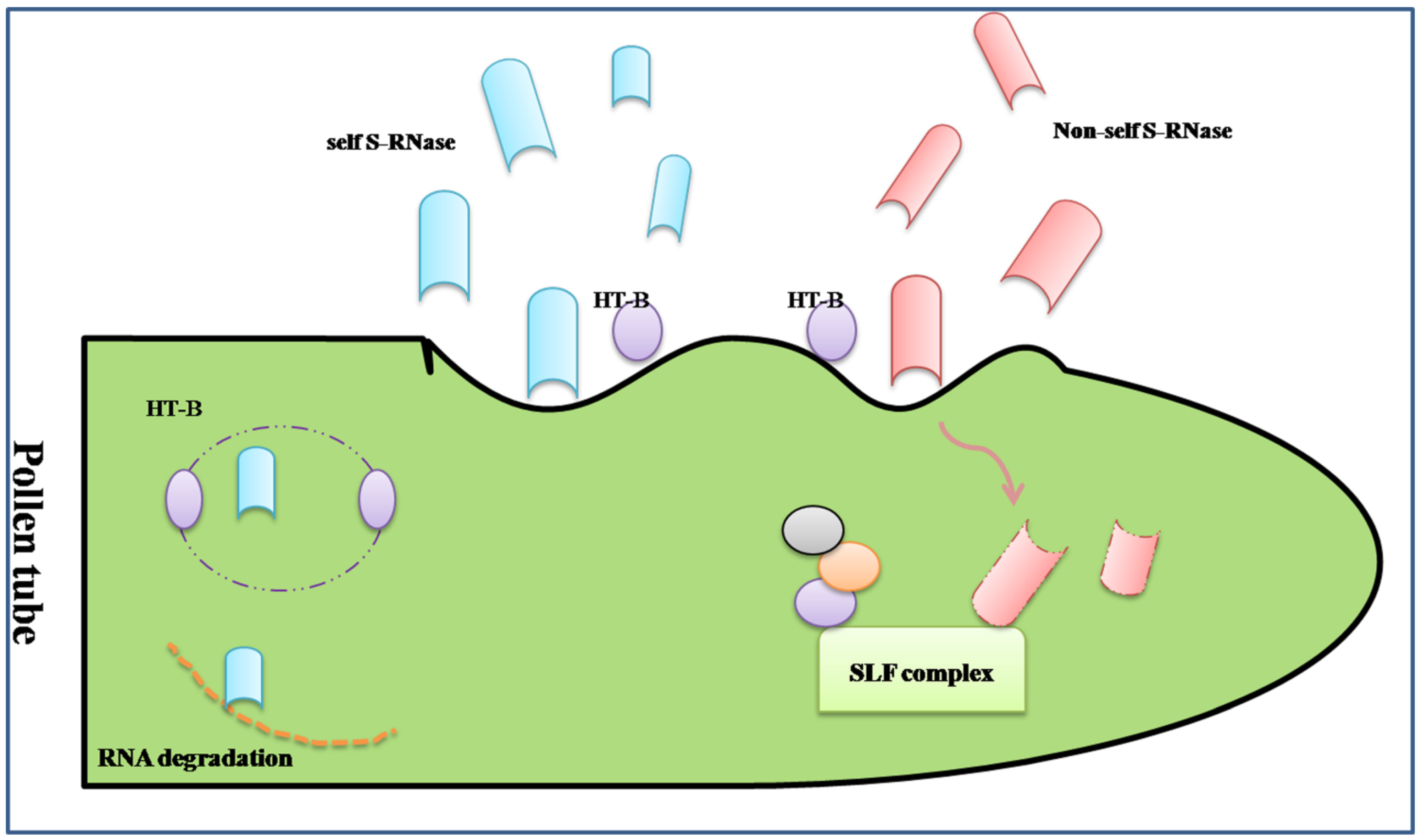
3.1. Proteomic Analysis of the Gametophytic Self-Incompatibility Response
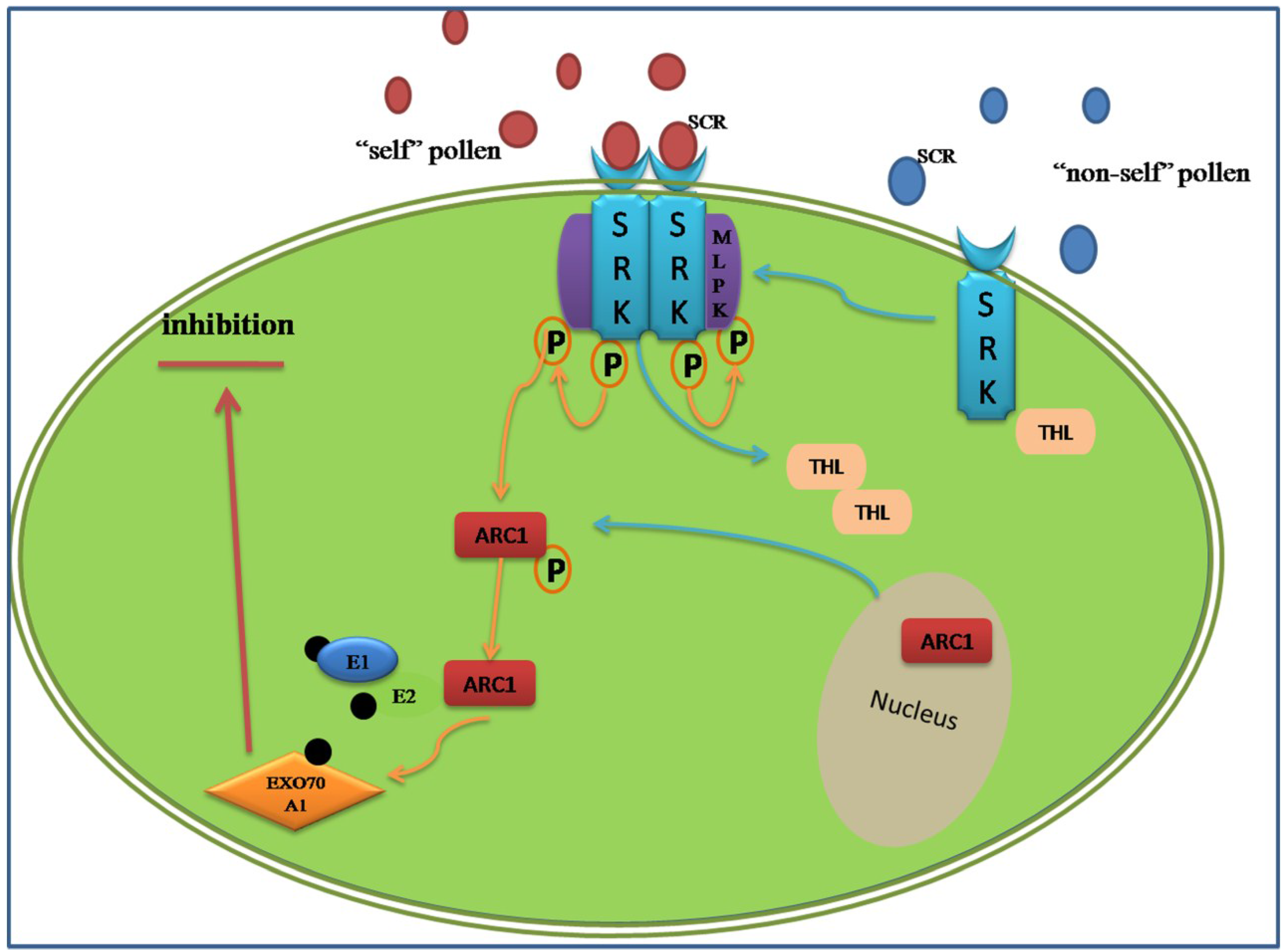
3.2. Proteomic Analysis of the Sporophytic Self-Incompatibility Response
4. Conclusions and Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kenrick, P.; Crane, P.R. The origin and early evolution of plants on land. Nature 1997, 389, 33–39. [Google Scholar]
- Hiscock, S.J.; Allen, A.M. Diverse cell signalling pathways regulate pollen-stigma interactions: The search for consensus. New Phytol. 2008, 179, 286–317. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.K.; Srivastav, M.; Rymbai, H.; Chaudhary, R.; Singh, A.K.; Dubey, A.K.; Lal, K. Pollen-pistil interaction studies in mango (Mangifera indica L.) cultivars. Sci. Horticult. 2013, 160, 213–221. [Google Scholar] [CrossRef]
- Takayama, S.; Shiba, H.; Iwano, M.; Shimosato, H.; Che, F.-S.; Kai, N.; Watanabe, M.; Suzuki, G.; Hinata, K.; Isogai, A. The pollen determinant of self-incompatibility in Brassica campestris. Proc. Natl. Acad. Sci. USA 2000, 97, 1920–1925. [Google Scholar] [CrossRef] [PubMed]
- Kao, T.-H.; Tsukamoto, T. The molecular and genetic bases of S-RNase-based self-incompatibility. Plant Cell Online 2004, 16, S72–S83. [Google Scholar]
- Ahmad, Y.; Lamond, A.I. A perspective on proteomics in cell biology. Trends Cell Biol. 2014, 24, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Sankaranarayanan, S.; Jamshed, M.; Samuel, M.A. Proteomics approaches advance our understanding of plant self-incompatibility response. J. Proteome Res. 2013, 12, 4717–4726. [Google Scholar] [CrossRef] [PubMed]
- Panchaud, A.; Affolter, M.; Moreillon, P.; Kussmann, M. Experimental and computational approaches to quantitative proteomics: Status quo and outlook. J. Proteomics 2008, 71, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Cho, J.-Y. Proteomics approaches for the studies of bone metabolism. BMB Rep. 2014, 47, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Wiese, S.; Reidegeld, K.A.; Meyer, H.E.; Warscheid, B. Protein labeling by iTRAQ: A new tool for quantitative mass spectrometry in proteome research. Proteomics 2007, 7, 340–350. [Google Scholar] [PubMed]
- Becker, C.H.; Bern, M. Recent developments in quantitative proteomics. Mutat. Res. Genet. Toxicol. Environ. Mutagenes. 2011, 722, 171–182. [Google Scholar] [CrossRef]
- McCormick, S. Male gametophyte development. Plant Cell 1993, 5, 1265–1275. [Google Scholar] [PubMed]
- McCormick, S. Control of male gametophyte development. Plant Cell 2004, 16, S142–S153. [Google Scholar] [PubMed]
- Taylor, L.P.; Hepler, P.K. Pollen germination and tube growth. Annu. Rev. Plant Biol. 1997, 48, 461–491. [Google Scholar] [CrossRef]
- Honys, D.; Twell, D. Comparative analysis of the arabidopsis pollen transcriptome. Plant Physiol. 2003, 132, 640–652. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.D.; Boavida, L.C.; Carneiro, J.; Haury, M.; Feijó, J.A. Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol. 2003, 133, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Holmes-Davis, R.; Tanaka, C.K.; Vensel, W.H.; Hurkman, W.J.; McCormick, S. Proteome mapping of mature pollen of Arabidopsis thaliana. Proteomics 2005, 5, 4864–4884. [Google Scholar] [PubMed]
- Grobei, M.A.; Qeli, E.; Brunner, E.; Rehrauer, H.; Zhang, R.; Roschitzki, B.; Basler, K.; Ahrens, C.H.; Grossniklaus, U. Deterministic protein inference for shotgun proteomics data provides new insights into Arabidopsis pollen development and function. Genome Res. 2009, 19, 1786–1800. [Google Scholar] [CrossRef] [PubMed]
- Mayfield, J.A.; Fiebig, A.; Johnstone, S.E.; Preuss, D. Gene families from the Arabidopsis thaliana pollen coat proteome. Science 2001, 292, 2482–2485. [Google Scholar] [PubMed]
- Dai, S.; Li, L.; Chen, T.; Chong, K.; Xue, Y.; Wang, T. Proteomic analyses of Oryza sativa mature pollen reveal novel proteins associated with pollen germination and tube growth. Proteomics 2006, 6, 2504–2529. [Google Scholar] [PubMed]
- Suen, D.F.; Wu, S.S.H.; Chang, H.C.; Dhugga, K.S.; Huang, A.H.C. Cell wall reactive proteins in the coat and wall of maize pollen: Potential role in pollen tube growth on the stigma and through the style. J. Biol. Chem. 2003, 278, 43672–43681. [Google Scholar] [CrossRef] [PubMed]
- Kerim, T.; Imin, N.; Weinman, J.J.; Rolfe, B.G. Proteome analysis of male gametophyte development in rice anthers. Proteomics 2003, 3, 738–751. [Google Scholar] [PubMed]
- Dai, S.; Chen, T.; Chong, K.; Xue, Y.; Liu, S.; Wang, T. Proteomics identification of differentially expressed proteins associated with pollen germination and tube growth reveals characteristics of germinated Oryza sativa pollen. Mol. Cell. Proteomics 2007, 6, 207–230. [Google Scholar] [CrossRef] [PubMed]
- Sheoran, I.S.; Pedersen, E.J.; Ross, A.R.; Sawhney, V.K. Dynamics of protein expression during pollen germination in canola (Brassica napus). Planta 2009, 230, 779–793. [Google Scholar] [PubMed]
- Fernando, D.D. Characterization of pollen tube development in Pinus strobus (Eastern white pine) through proteomic analysis of differentially expressed proteins. Proteomics 2005, 5, 4917–4926. [Google Scholar] [PubMed]
- Chen, Y.; Chen, T.; Shen, S.; Zheng, M.; Guo, Y.; Lin, J.; Baluska, F.; Samaj, J. Differential display proteomic analysis of Picea meyeri pollen germination and pollen-tube growth after inhibition of actin polymerization by latrunculin B. Plant J. Cell Mol. Biol. 2006, 47, 174–195. [Google Scholar] [CrossRef]
- Zaidi, M.A.; O’Leary, S.; Wu, S.; Gleddie, S.; Eudes, F.; Laroche, A.; Robert, L.S. A molecular and proteomic investigation of proteins rapidly released from triticale pollen upon hydration. Plant Mol. Biol. 2012, 79, 101–121. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Chen, S.; Dai, S.; Yang, N.; Wang, T. Isobaric tags for relative and absolute quantification- based comparative proteomics reveals the features of plasma membrane-associated proteomes of pollen grains and pollen tubes from Lilium davidii. J. Integr. Plant Biol. 2010, 52, 1043–1058. [Google Scholar] [CrossRef] [PubMed]
- Gasser, C.S.; Robinson-Beers, K. Pistil development. Plant Cell 1993, 5, 1231. [Google Scholar] [PubMed]
- Faure, J.-E. Double fertilization in flowering plants: Discovery, study methods and mechanisms. Comptes Rendus de l’Académie des Sciences-Series III-Sciences de la Vie 2001, 324, 551–558. [Google Scholar] [CrossRef]
- Rea, A.C.; Nasrallah, J.B. Self-incompatibility systems: Barriers to self-fertilization in flowering plants. Int. J. Dev. Biol. 2008, 52, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Tung, C.W.; Dwyer, K.G.; Nasrallah, M.E.; Nasrallah, J.B. Genome-wide identification of genes expressed in Arabidopsis pistils specifically along the path of pollen tube growth. Plant Physiol. 2005, 138, 977–989. [Google Scholar] [CrossRef] [PubMed]
- Boavida, L.C.; Borges, F.; Becker, J.D.; Feijo, J.A. Whole genome analysis of gene expression reveals coordinated activation of signaling and metabolic pathways during pollen-pistil interactions in Arabidopsis. Plant Physiol. 2011, 155, 2066–2080. [Google Scholar] [CrossRef] [PubMed]
- Ohyanagi, H.; Sakata, K.; Komatsu, S. Soybean proteome database 2012: Update on the comprehensive data repository for soybean proteomics. Front. Plant Sci. 2012, 3, e110. [Google Scholar] [CrossRef]
- Haerizadeh, F.; Wong, C.E.; Bhalla, P.L.; Gresshoff, P.M.; Singh, M.B. Genomic expression profiling of mature soybean (Glycine max) pollen. BMC Plant Biol. 2009, 9, e25. [Google Scholar] [CrossRef]
- Li, M.; Sha, A.; Zhou, X.; Yang, P. Comparative proteomic analyses reveal the changes of metabolic features in soybean (Glycine max) pistils upon pollination. Sex. Plant Reprod. 2012, 25, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Verhoeven, T.; Feron, R.; Wolters-Arts, M.; Edqvist, J.; Gerats, T.; Derksen, J.; Mariani, C. STIG1 controls exudate secretion in the pistil of petunia and tobacco. Plant Physiol. 2005, 138, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Rejon, J.D.; Delalande, F.; Schaeffer-Reiss, C.; Carapito, C.; Zienkiewicz, K.; de Dios Alche, J.; Rodriguez-Garcia, M.I.; van Dorsselaer, A.; Castro, A.J. Proteomics profiling reveals novel proteins and functions of the plant stigma exudate. J. Exp. Bot. 2013, 64, 5695–5705. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, H.; Higashiyama, T. Attraction of tip-growing pollen tubes by the female gametophyte. Curr. Opin. Plant Biol. 2011, 14, 614–621. [Google Scholar] [CrossRef] [PubMed]
- Wagner, R.E.; Mugnaini, S.; Sniezko, R.; Hardie, D.; Poulis, B.; Nepi, M.; Pacini, E.; Aderkas, P. Proteomic evaluation of gymnosperm pollination drop proteins indicates highly conserved and complex biological functions. Sex. Plant Reprod. 2007, 20, 181–189. [Google Scholar] [CrossRef]
- Márton, M.-L.; Dresselhaus, T. Female gametophyte-controlled pollen tube guidance. Biochem. Soc. Trans. 2010, 38, 627–630. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, R.D.; Portereiko, M.F.; Sandaklie-Nikolova, L.; Rabiger, D.S.; Drews, G.N. MYB98 is required for pollen tube guidance and synergid cell differentiation in Arabidopsis. Plant Cell 2005, 17, 2981–2992. [Google Scholar] [PubMed]
- Kessler, S.A.; Grossniklaus, U. She’s the boss: Signaling in pollen tube reception. Curr. Opin. Plant Biol. 2011, 14, 622–627. [Google Scholar] [CrossRef]
- Takeuchi, H.; Higashiyama, T. A species-specific cluster of defensin-like genes encodes diffusible pollen tube attractants in Arabidopsis. PLoS Biol. 2012, 10, e1001449. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.M.; Bosch, M.; Bots, M.; Nieuwland, J.; Feron, R.; Mariani, C. Pistil factors controlling pollination. Plant Cell 2004, 16, S98–S106. [Google Scholar] [PubMed]
- Liu, J.; Zhong, S.; Guo, X.; Hao, L.; Wei, X.; Huang, Q.; Hou, Y.; Shi, J.; Wang, C.; Gu, H.; et al. Membrane-bound RLCKs LIP1 and LIP2 are essential male factors controlling male-female attraction in Arabidopsis. Curr. Biol. 2013, 23, 993–998. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Lu, J.; Xu, J.; McClure, B.; Zhang, S. Two Mitogen-activated protein kinases, MPK3 and MPK6, are required for funicular guidance of pollen tubes in Arabidopsis. Plant Physiol. 2014, 165, 528–533. [Google Scholar] [CrossRef] [PubMed]
- Swanson, R.; Edlund, A.F.; Preuss, D. Species specificity in pollen-pistil interactions. Annu. Rev. Genet. 2004, 38, 793–818. [Google Scholar] [CrossRef] [PubMed]
- Swanson, W.J.; Vacquier, V.D. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 2002, 3, 137–144. [Google Scholar] [CrossRef]
- Showalter, A. Arabinogalactan-proteins: Structure, expression and function. Cell. Mol. Life Sci. 2001, 58, 1399–1417. [Google Scholar] [CrossRef] [PubMed]
- Suárez, C.; Zienkiewicz, A.; Castro, A.J.; Zienkiewicz, K.; Majewska-Sawka, A.; Rodríguez-García, M.I. Cellular localization and levels of pectins and arabinogalactan proteins in olive (Olea europaea L.) pistil tissues during development: Implications for pollen-pistil interaction. Planta 2012, 237, 305–319. [Google Scholar] [PubMed]
- Cruz-Garcia, F.; Hancock, C.N.; McClure, B. S-RNase complexes and pollen rejection. J. Exp. Bot. 2003, 54, 123–130. [Google Scholar] [CrossRef] [PubMed]
- De Nettancourt, D. Incompatibility in angiosperms. Sex. Plant Reprod. 1997, 10, 185–199. [Google Scholar]
- McCubbin, A.G.; Kao, T.-H. Molecular recognition and response in pollen and pistil interactions. Annu. Rev. Cell Dev. Biol. 2000, 16, 333–364. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Sun, P.; Kao, T.H. S-RNase-based self-incompatibility in Petunia inflata. Ann. Bot. 2011, 108, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, S.; Gu, Y.; Zhang, S.; Publicover, S.J.; Franklin-Tong, V.E. Self-Incompatibility in Papaver rhoeas activates nonspecific cation conductance permeable to Ca2+ and K+. Plant Physiol. 2010, 155, 963–973. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, M.J.; de Graaf, B.H.; Hadjiosif, N.; Perry, R.M.; Poulter, N.S.; Osman, K.; Vatovec, S.; Harper, A.; Franklin, F.C.; Franklin-Tong, V.E. Identification of the pollen self-incompatibility determinant in Papaver rhoeas. Nature 2009, 459, 992–995. [Google Scholar] [PubMed]
- Ushijima, K.; Sassa, H.; Dandekar, A.M.; Gradziel, T.M.; Tao, R.; Hirano, H. Structural and transcriptional analysis of the self-incompatibility locus of almond: Identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell Online 2003, 15, 771–781. [Google Scholar]
- Bredemeijer, G.; Blaas, J. S-Specific proteins in styles of self-incompatible Nicotiana alata. Theor. Appl. Genet. 1981, 59, 185–190. [Google Scholar] [CrossRef] [PubMed]
- McClure, B.A.; Haring, V.; Ebert, P.R.; Anderson, M.A.; Simpson, R.J.; Sakiyama, F.; Clarke, A.E. Style self-incompatibility gene products of Nicotlana alata are ribonucleases. Nature 1989, 342, 955–957. [Google Scholar] [PubMed]
- Broothaerts, W.J.; van Laere, A.; Witters, R.; Préaux, G.; Decock, B.; van Damme, J.; Vendrig, J.C. Purification and N-terminal sequencing of style glycoproteins associated with self-incompatibility in Petunia hybrida. Plant Mol. Biol. 1990, 14, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Ai, Y.; Kao, T.-H. Characterization of ribonuclease activity of three S-allele-associated proteins of Petunia inflata. Plant Physiol. 1991, 96, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Hiratsuka, S. Characterization of an S-allele associated protein in Japanese pear. Euphytica 1992, 62, 103–110. [Google Scholar]
- Ishimizu, T.; Sato, Y.; Saito, T.; Yoshimura, Y.; Norioka, S.; Nakanishi, T.; Sakiyama, F. Identification and partial amino acid sequences of seven S-RNases associated with self-incompatibility of Japanese pear, Pyrus pyrifolia Nakai. J. Biochem. 1996, 120, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Ma, W.; Han, B.; Liang, L.; Zhang, Y.; Hong, G.; Xue, Y. An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol. Biol. 2002, 50, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Entani, T.; Iwano, M.; Shiba, H.; Che, F.S.; Isogai, A.; Takayama, S. Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: Identification of a pollen-expressed F-box gene with allelic diversity. Genes Cells 2003, 8, 203–213. [Google Scholar] [PubMed]
- Sijacic, P.; Wang, X.; Skirpan, A.L.; Wang, Y.; Dowd, P.E.; McCubbin, A.G.; Huang, S.; Kao, T.-H. Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature 2004, 429, 302–305. [Google Scholar] [PubMed]
- Hua, Z.; Kao, T.-H. Identification and characterization of components of a putative Petunia S-locus F-box–containing E3 ligase complex involved in S-RNase-based self-incompatibility. Plant Cell Online 2006, 18, 2531–2553. [Google Scholar]
- Chen, G.; Zhang, B.; Liu, L.; Li, Q.; Zhang, Y.; Xie, Q.; Xue, Y. Identification of a ubiquitin-binding structure in the S-locus F-box protein controlling S-RNase-based self-incompatibility. J. Genet. Genomics (Yi Chuan Xue Bao) 2012, 39, 93–102. [Google Scholar] [CrossRef]
- Sun, P.; Kao, T.H. Self-Incompatibility in Petunia inflata: The relationship between a self-incompatibility locus F-box protein and its non-self S-RNases. Plant Cell 2013, 25, 470–485. [Google Scholar] [PubMed]
- McClure, B.; Cruz-Garcia, F.; Romero, C. Compatibility and incompatibility in S-RNase-based systems. Ann. Bot. 2011, 108, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Soulard, J.; Boivin, N.; Morse, D.; Cappadocia, M. eEF1A Is an S-RNase binding factor in self-incompatible solanum chacoense. PLoS One 2014, 9, e90206. [Google Scholar] [PubMed]
- McClure, B. Darwin’s foundation for investigating self-incompatibility and the progress toward a physiological model for S-RNase-based SI. J. Exp. Bot. 2009, 60, 1069–1081. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Chen, X.; Yuan, Z.; He, T.; Zhang, L.; Wu, Y.; Liu, W.; Liang, Q. Proteome comparison following self- and across-pollination in self-incompatible apricot (Prunus armeniaca L.). Protein J. 2006, 25, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Chen, X.; Yuan, Z.; Zhang, L.; Ci, Z.; Liu, X.; Zhang, C. Primary molecular features of self-incompatible and self-compatible F1 seedling from apricot (Prunus armeniaca L.) Katy× Xinshiji. Mol. Biol. Rep. 2009, 36, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Uchida, A.; Takenaka, S.; Sakakibara, Y.; Kurogi, S. Comprehensive analysis of expressed proteins in the different stages of the style development of self-incompatible “Hyuganatsu” (Citrus tamurana hort. ex Tanaka). J. Jpn. Soc. Horticult. Sci. 2012, 81, 150–158. [Google Scholar] [CrossRef]
- Tantikanjana, T.; Nasrallah, M.E.; Nasrallah, J.B. Complex networks of self-incompatibility signaling in the Brassicaceae. Curr. Opin. Plant Biol. 2010, 13, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Gu, T.; Mazzurco, M.; Sulaman, W.; Matias, D.D.; Goring, D.R. Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc. Natl. Acad. Sci. USA 1998, 95, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L.; Anderson, E.M.; Mullen, R.T.; Goring, D.R. ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell Online 2003, 15, 885–898. [Google Scholar]
- Samuel, M.A.; Chong, Y.T.; Haasen, K.E.; Aldea-Brydges, M.G.; Stone, S.L.; Goring, D.R. Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. Plant Cell Online 2009, 21, 2655–2671. [Google Scholar]
- Bower, M.S.; Matias, D.D.; Fernandes-Carvalho, E.; Mazzurco, M.; Gu, T.; Rothstein, S.J.; Goring, D.R. Two members of the thioredoxin-h family interact with the kinase domain of a Brassica S locus receptor kinase. Plant Cell Online 1996, 8, 1641–1650. [Google Scholar]
- Kakita, M.; Murase, K.; Iwano, M.; Matsumoto, T.; Watanabe, M.; Shiba, H.; Isogai, A.; Takayama, S. Two distinct forms of M-locus protein kinase localize to the plasma membrane and interact directly with S-locus receptor kinase to transduce self-incompatibility signaling in Brassica rapa. Plant Cell Online 2007, 19, 3961–3973. [Google Scholar]
- Samuel, M.A.; Tang, W.; Jamshed, M.; Northey, J.; Patel, D.; Smith, D.; Siu, K.M.; Muench, D.G.; Wang, Z.-Y.; Goring, D.R. Proteomic analysis of Brassica stigmatic proteins following the self-incompatibility reaction reveals a role for microtubule dynamics during pollen responses. Mol. Cell. Proteomics 2011, 10. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Peng, H.; Ge, T.; Liu, T.; Hou, X.; Li, Y. Identification of differentially accumulating pistil proteins associated with self-incompatibility of non-heading Chinese cabbage. Plant Biol. 2013, 16, 49–57. [Google Scholar] [CrossRef]
- Sankaranarayanan, S.; Jamshed, M.; Deb, S.; Chatfield-Reed, K.; Kwon, E.-J.G.; Chua, G.; Samuel, M.A. Deciphering the stigmatic transcriptional landscape of compatible and self-incompatible pollinations in Brassica napus reveals a rapid stigma senescence response following compatible pollination. Mol. Plant 2013. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, Z.; Yang, P. Proteomics Advances in the Understanding of Pollen–Pistil Interactions. Proteomes 2014, 2, 468-484. https://doi.org/10.3390/proteomes2040468
Fu Z, Yang P. Proteomics Advances in the Understanding of Pollen–Pistil Interactions. Proteomes. 2014; 2(4):468-484. https://doi.org/10.3390/proteomes2040468
Chicago/Turabian StyleFu, Ziyang, and Pingfang Yang. 2014. "Proteomics Advances in the Understanding of Pollen–Pistil Interactions" Proteomes 2, no. 4: 468-484. https://doi.org/10.3390/proteomes2040468
APA StyleFu, Z., & Yang, P. (2014). Proteomics Advances in the Understanding of Pollen–Pistil Interactions. Proteomes, 2(4), 468-484. https://doi.org/10.3390/proteomes2040468




