Uncovering Enzyme-Specific Post-Translational Modifications: An Overview of Current Methods
Abstract
1. Introduction
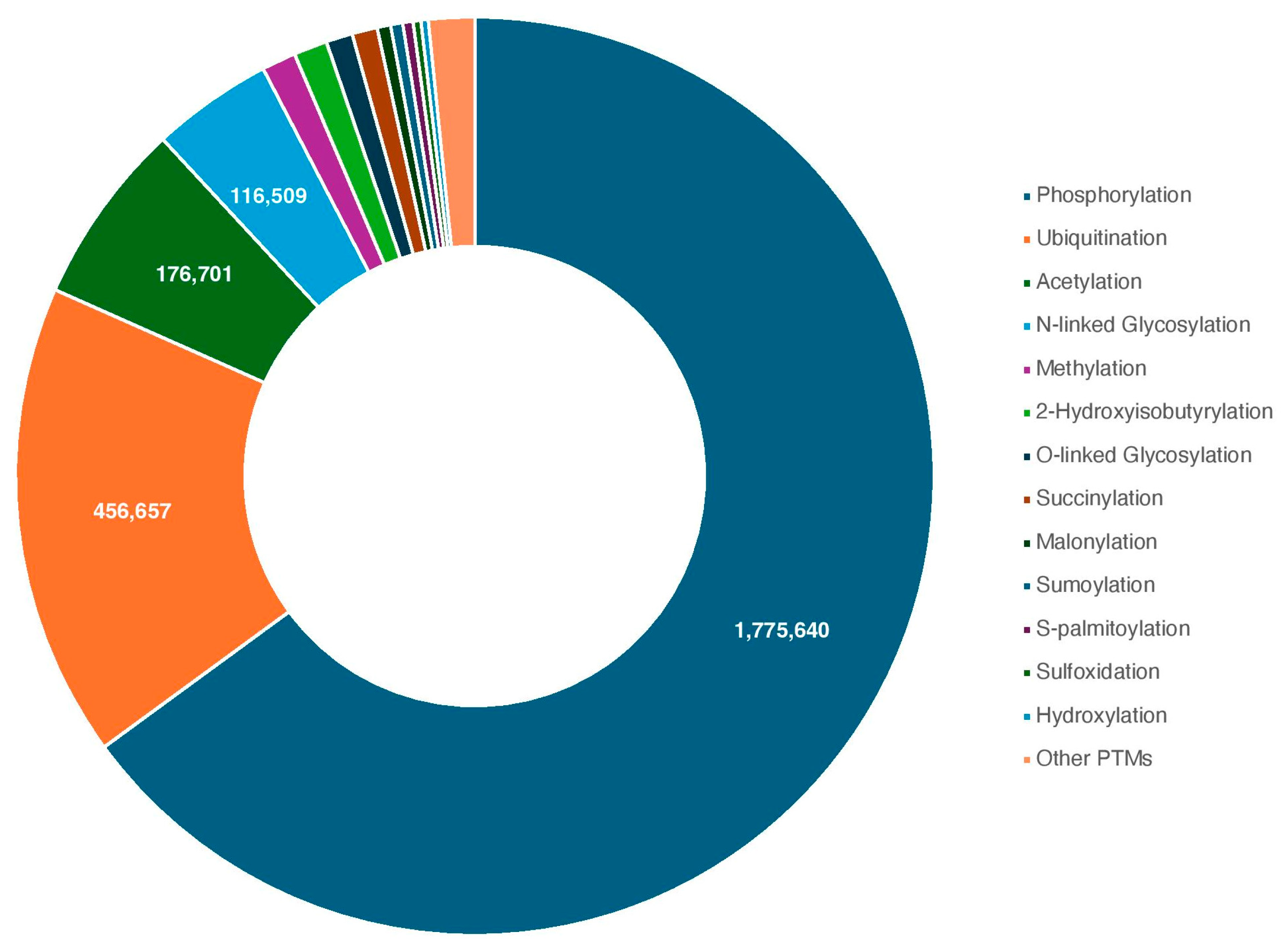
2. PTMs and the Enzymes Responsible
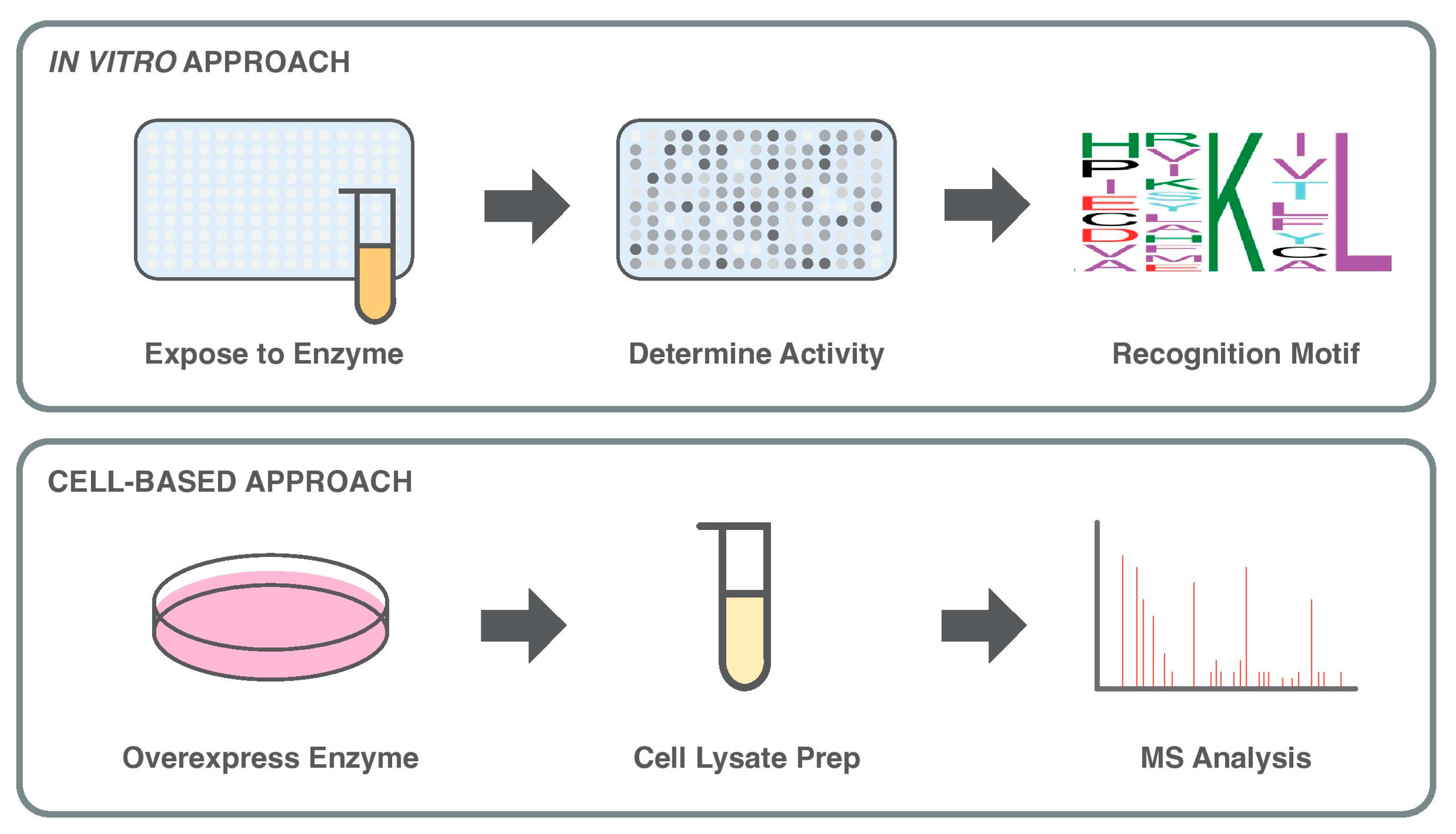
3. In Vitro Approaches to Enzyme–Substrate Discovery
4. Cell-Based Approaches to Explore Enzyme–Substrate Networks
5. Modern Computational Approaches to Guide Enzyme–Substrate Discovery
6. Limitations
7. Conclusions and Outlook
Funding
Conflicts of Interest
References
- Levene, P.A.; Alsberg, C.L. The Cleavage Products of Vitellin. J. Biol. Chem. 1906, 2, 127–133. [Google Scholar] [CrossRef]
- Keenan, E.K.; Zachman, D.K.; Hirschey, M.D. Discovering the Landscape of Protein Modifications. Mol. Cell 2021, 81, 1868–1878. [Google Scholar] [CrossRef] [PubMed]
- Ramazi, S.; Zahiri, J. Post-Translational Modifications in Proteins: Resources, Tools and Prediction Methods. Database 2021, 2021, baab012. [Google Scholar] [CrossRef] [PubMed]
- Bradley, D. The Evolution of Post-Translational Modifications. Curr. Opin. Genet. Dev. 2022, 76, 101956. [Google Scholar] [CrossRef]
- Harmel, R.; Fiedler, D. Features and Regulation of Non-Enzymatic Post-Translational Modifications. Nat. Chem. Biol. 2018, 14, 244–252. [Google Scholar] [CrossRef]
- Li, Z.; Li, S.; Luo, M.; Jhong, J.-H.; Li, W.; Yao, L.; Pang, Y.; Wang, Z.; Wang, R.; Ma, R.; et al. dbPTM in 2022: An Updated Database for Exploring Regulatory Networks and Functional Associations of Protein Post-Translational Modifications. Nucleic Acids Res. 2022, 50, D471–D479. [Google Scholar] [CrossRef]
- Leutert, M.; Entwisle, S.W.; Villén, J. Decoding Post-Translational Modification Crosstalk With Proteomics. Mol. Cell. Proteom. 2021, 20, 100129. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.A.; Cole, P.A. The Chemical Biology of Reversible Lysine Post-Translational Modifications. Cell Chem. Biol. 2020, 27, 953–969. [Google Scholar] [CrossRef]
- Duan, G.; Walther, D. The Roles of Post-Translational Modifications in the Context of Protein Interaction Networks. PLoS Comput. Biol. 2015, 11, e1004049. [Google Scholar] [CrossRef]
- Islam, K.; Zheng, W.; Yu, H.; Deng, H.; Luo, M. Expanding Cofactor Repertoire of Protein Lysine Methyltransferase for Substrate Labeling. ACS Chem. Biol. 2011, 6, 679–684. [Google Scholar] [CrossRef]
- Biggar, K.K.; Li, S.S.-C. Non-Histone Protein Methylation as a Regulator of Cellular Signalling and Function. Nat. Rev. Mol. Cell Biol. 2015, 16, 5–17. [Google Scholar] [CrossRef]
- Burnett, G.; Kennedy, E.P. The Enzymatic Phosphorylation of Proteins. J. Biol. Chem. 1954, 211, 969–980. [Google Scholar] [CrossRef]
- Sutherland, E.W.; Wosilait, W.D. Inactivation and Activation of Liver Phosphorylase. Nature 1955, 175, 169–170. [Google Scholar] [CrossRef]
- Krebs, E.G.; Fischer, E.H. The Phosphorylase b to a Converting Enzyme of Rabbit Skeletal Muscle. Biochim. Biophys. Acta 1956, 20, 150–157. [Google Scholar] [CrossRef]
- Fischer, E.H. Phosphorylase and the Origin of Reversible Protein Phosphorylation. Biol. Chem. 2010, 391, 131–137. [Google Scholar] [CrossRef]
- Linn, T.C.; Pettit, F.H.; Reed, L.J. α-Keto Acid Dehydrogenase Complexes, X. Regulation of the Activity of the Pyruvate dehydrogenase Complex from Beef Kidney Mitochondria by Phosphorylation and Dephosphorylation. Proc. Natl. Acad. Sci. USA 1969, 62, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Allfrey, V.G.; Faulkner, R.; Mirsky, A.E. Acetylation and Methylation of Histones and Their Possible Role in the Regulation OF RNA Synthesis. Proc. Natl. Acad. Sci. USA 1964, 51, 786–794. [Google Scholar] [CrossRef] [PubMed]
- Sacco, F.; Perfetto, L.; Castagnoli, L.; Cesareni, G. The Human Phosphatase Interactome: An Intricate Family Portrait. FEBS Lett. 2012, 586, 2732–2739. [Google Scholar] [CrossRef] [PubMed]
- Esmaili, F.; Pourmirzaei, M.; Ramazi, S.; Shojaeilangari, S.; Yavari, E. A Review of Machine Learning and Algorithmic Methods for Protein Phosphorylation Sites Prediction. Genom. Proteom. Bioinform. 2023, 21, 1266–1285. [Google Scholar] [CrossRef]
- He, W.; Wei, L.; Zou, Q. Research Progress in Protein Posttranslational Modification Site Prediction. Brief. Funct. Genom. 2019, 18, 220–229. [Google Scholar] [CrossRef]
- Shi, X.; Kachirskaia, I.; Yamaguchi, H.; West, L.E.; Wen, H.; Wang, E.W.; Dutta, S.; Appella, E.; Gozani, O. Modulation of P53 Function by SET8-Mediated Methylation at Lysine 382. Mol. Cell 2007, 27, 636–646. [Google Scholar] [CrossRef]
- Skoge, R.H.; Dölle, C.; Ziegler, M. Regulation of SIRT2-Dependent α-Tubulin Deacetylation by Cellular NAD Levels. DNA Repair 2014, 23, 33–38. [Google Scholar] [CrossRef]
- Wu, X.; Xu, M.; Geng, M.; Chen, S.; Little, P.J.; Xu, S.; Weng, J. Targeting Protein Modifications in Metabolic Diseases: Molecular Mechanisms and Targeted Therapies. Signal Transduct. Target. Ther. 2023, 8, 220. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, I.A.; D’Souza, R.C.J.; Yang, B.; Verlaan-de Vries, M.; Mann, M.; Vertegaal, A.C.O. Uncovering Global SUMOylation Signaling Networks in a Site-Specific Manner. Nat. Struct. Mol. Biol. 2014, 21, 927–936. [Google Scholar] [CrossRef]
- Palacios, A.V.; Acharya, P.; Peidl, A.S.; Beck, M.R.; Blanco, E.; Mishra, A.; Bawa-Khalfe, T.; Pakhrin, S.C. SumoPred-PLM: Human SUMOylation and SUMO2/3 Sites Prediction Using Pre-Trained Protein Language Model. NAR Genom. Bioinform. 2024, 6, lqae011. [Google Scholar] [CrossRef] [PubMed]
- Buch-Larsen, S.C.; Hendriks, I.A.; Lodge, J.M.; Rykær, M.; Furtwängler, B.; Shishkova, E.; Westphall, M.S.; Coon, J.J.; Nielsen, M.L. Mapping Physiological ADP-Ribosylation Using Activated Ion Electron Transfer Dissociation. Cell Rep. 2020, 32, 108176. [Google Scholar] [CrossRef] [PubMed]
- Molendijk, J.; Yip, R.; Parker, B.L. urPTMdb/TeaProt: Upstream and Downstream Proteomics Analysis. J. Proteome Res. 2023, 22, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Dutta, H.; Jain, N. Post-Translational Modifications and Their Implications in Cancer. Front. Oncol. 2023, 13, 1240115. [Google Scholar] [CrossRef]
- Leibowitz, M.J.; Soffer, R.L. Enzymatic Modification of Proteins. J. Biol. Chem. 1971, 246, 5207–5212. [Google Scholar] [CrossRef]
- Zhu, H.; Klemic, J.F.; Chang, S.; Bertone, P.; Casamayor, A.; Klemic, K.G.; Smith, D.; Gerstein, M.; Reed, M.A.; Snyder, M. Analysis of Yeast Protein Kinases Using Protein Chips. Nat. Genet. 2000, 26, 283–289. [Google Scholar] [CrossRef]
- Ptacek, J.; Devgan, G.; Michaud, G.; Zhu, H.; Zhu, X.; Fasolo, J.; Guo, H.; Jona, G.; Breitkreutz, A.; Sopko, R.; et al. Global Analysis of Protein Phosphorylation in Yeast. Nature 2005, 438, 679–684. [Google Scholar] [CrossRef]
- Merbl, Y.; Kirschner, M.W. Large-Scale Detection of Ubiquitination Substrates Using Cell Extracts and Protein Microarrays. Proc. Natl. Acad. Sci. USA 2009, 106, 2543–2548. [Google Scholar] [CrossRef]
- Syu, G.-D.; Dunn, J.; Zhu, H. Developments and Applications of Functional Protein Microarrays. Mol. Cell. Proteom. 2020, 19, 916–927. [Google Scholar] [CrossRef]
- Levy, D.; Liu, C.L.; Yang, Z.; Newman, A.M.; Alizadeh, A.A.; Utz, P.J.; Gozani, O. A Proteomic Approach for the Identification of Novel Lysine Methyltransferase Substrates. Epigenetics Chromatin 2011, 4, 19. [Google Scholar] [CrossRef]
- Merrifield, R.B. Solid Phase Peptide Synthesis. I. The Synthesis of a Tetrapeptide. J. Am. Chem. Soc. 1963, 85, 2149–2154. [Google Scholar] [CrossRef]
- Geysen, H.M.; Meloen, R.H.; Barteling, S.J. Use of Peptide Synthesis to Probe Viral Antigens for Epitopes to a Resolution of a Single Amino Acid. Proc. Natl. Acad. Sci. USA 1984, 81, 3998–4002. [Google Scholar] [CrossRef] [PubMed]
- Houghten, R.A. General Method for the Rapid Solid-Phase Synthesis of Large Numbers of Peptides: Specificity of Antigen-Antibody Interaction at the Level of Individual Amino Acids. Proc. Natl. Acad. Sci. USA 1985, 82, 5131–5135. [Google Scholar] [CrossRef]
- Frank, R. The SPOT-Synthesis Technique. J. Immunol. Methods 2002, 267, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Szymczak, L.C.; Kuo, H.-Y.; Mrksich, M. Peptide Arrays: Development and Application. Anal. Chem. 2018, 90, 266–282. [Google Scholar] [CrossRef]
- Wu, C.; Li, S.S.-C. CelluSpotsTM: A Reproducible Means of Making Peptide Arrays for the Determination of SH2 Domain Binding Specificity. In Peptide Microarrays; Cretich, M., Chiari, M., Eds.; Humana Press: Totowa, NJ, USA, 2009; Volume 570, pp. 197–202. ISBN 978-1-60327-393-0. [Google Scholar]
- Heubach, Y.; Planatscher, H.; Sommersdorf, C.; Maisch, D.; Maier, J.; Joos, T.O.; Templin, M.F.; Poetz, O. From Spots to Beads-PTM-peptide Bead Arrays for the Characterization of Anti-histone Antibodies. Proteomics 2013, 13, 1010–1015. [Google Scholar] [CrossRef]
- Beyer, M.; Nesterov, A.; Block, I.; König, K.; Felgenhauer, T.; Fernandez, S.; Leibe, K.; Torralba, G.; Hausmann, M.; Trunk, U.; et al. Combinatorial Synthesis of Peptide Arrays onto a Microchip. Science 2007, 318, 1888. [Google Scholar] [CrossRef]
- Fodor, S.P.A.; Read, J.L.; Pirrung, M.C.; Stryer, L.; Lu, A.T.; Solas, D. Light-Directed, Spatially Addressable Parallel Chemical Synthesis. Science 1991, 251, 767–773. [Google Scholar] [CrossRef]
- Legutki, J.B.; Zhao, Z.-G.; Greving, M.; Woodbury, N.; Johnston, S.A.; Stafford, P. Scalable High-Density Peptide Arrays for Comprehensive Health Monitoring. Nat. Commun. 2014, 5, 4785. [Google Scholar] [CrossRef]
- Krystkowiak, I.; Manguy, J.; Davey, N.E. PSSMSearch: A Server for Modeling, Visualization, Proteome-Wide Discovery and Annotation of Protein Motif Specificity Determinants. Nucleic Acids Res. 2018, 46, W235–W241. [Google Scholar] [CrossRef] [PubMed]
- Hilpert, K.; Hansen, G.; Wessner, H.; Schneider-Mergener, J.; Hohne, W. Characterizing and Optimizing Protease/Peptide Inhibitor Interactions, a New Application for Spot Synthesis. J. Biochem. 2000, 128, 1051–1057. [Google Scholar] [CrossRef] [PubMed]
- Rathert, P.; Zhang, X.; Freund, C.; Cheng, X.; Jeltsch, A. Analysis of the Substrate Specificity of the Dim-5 Histone Lysine Methyltransferase Using Peptide Arrays. Chem. Biol. 2008, 15, 5–11. [Google Scholar] [CrossRef][Green Version]
- Rathert, P.; Dhayalan, A.; Murakami, M.; Zhang, X.; Tamas, R.; Jurkowska, R.; Komatsu, Y.; Shinkai, Y.; Cheng, X.; Jeltsch, A. Protein Lysine Methyltransferase G9a Acts on Non-Histone Targets. Nat. Chem. Biol. 2008, 4, 344–346. [Google Scholar] [CrossRef] [PubMed]
- Kudithipudi, S.; Lungu, C.; Rathert, P.; Happel, N.; Jeltsch, A. Substrate Specificity Analysis and Novel Substrates of the Protein Lysine Methyltransferase NSD1. Chem. Biol. 2014, 21, 226–237. [Google Scholar] [CrossRef]
- Rodriguez, M.; Li, S.S.-C.; Harper, J.W.; Songyang, Z. An Oriented Peptide Array Library (OPAL) Strategy to Study Protein-Protein Interactions. J. Biol. Chem. 2004, 279, 8802–8807. [Google Scholar] [CrossRef]
- Huang, H.; Li, L.; Wu, C.; Schibli, D.; Colwill, K.; Ma, S.; Li, C.; Roy, P.; Ho, K.; Songyang, Z.; et al. Defining the Specificity Space of the Human Src Homology 2 Domain. Mol. Cell. Proteom. 2008, 7, 768–784. [Google Scholar] [CrossRef]
- Gayatri, S.; Cowles, M.W.; Vemulapalli, V.; Cheng, D.; Sun, Z.-W.; Bedford, M.T. Using Oriented Peptide Array Libraries to Evaluate Methylarginine-Specific Antibodies and Arginine Methyltransferase Substrate Motifs. Sci. Rep. 2016, 6, 28718. [Google Scholar] [CrossRef] [PubMed]
- Hanquier, J.N.; Sanders, K.; Berryhill, C.A.; Sahoo, F.K.; Hudmon, A.; Vilseck, J.Z.; Cornett, E.M. Identification of Nonhistone Substrates of the Lysine Methyltransferase PRDM9. J. Biol. Chem. 2023, 299, 104651. [Google Scholar] [CrossRef] [PubMed]
- Tegge, W.; Frank, R.; Hofmann, F.; Dostmann, W.R.G. Determination of Cyclic Nucleotide-Dependent Protein Kinase Substrate Specificity by the Use of Peptide Libraries on Cellulose Paper. Biochemistry 1995, 34, 10569–10577. [Google Scholar] [CrossRef]
- Luo, K.; Zhou, P.; Lodish, H.F. The Specificity of the Transforming Growth Factor Beta Receptor Kinases Determined by a Spatially Addressable Peptide Library. Proc. Natl. Acad. Sci. USA 1995, 92, 11761–11765. [Google Scholar] [CrossRef] [PubMed]
- Rauh, D.; Fischer, F.; Gertz, M.; Lakshminarasimhan, M.; Bergbrede, T.; Aladini, F.; Kambach, C.; Becker, C.F.W.; Zerweck, J.; Schutkowski, M.; et al. An Acetylome Peptide Microarray Reveals Specificities and Deacetylation Substrates for All Human Sirtuin Isoforms. Nat. Commun. 2013, 4, 2327. [Google Scholar] [CrossRef]
- Rychlewski, L.; Kschischo, M.; Dong, L.; Schutkowski, M.; Reimer, U. Target Specificity Analysis of the Abl Kinase Using Peptide Microarray Data. J. Mol. Biol. 2004, 336, 307–311. [Google Scholar] [CrossRef]
- Panse, S.; Dong, L.; Burian, A.; Carus, R.; Schutkowski, M.; Reimer, U.; Schneider-Mergener, J. Profiling of Generic Anti-Phosphopeptide Antibodies and Kinases with Peptide Microarrays Using Radioactive and Fluorescence-Based Assays. Mol. Divers. 2004, 8, 291–299. [Google Scholar] [CrossRef]
- Schutkowski, M.; Reimer, U.; Panse, S.; Dong, L.; Lizcano, J.M.; Alessi, D.R.; Schneider-Mergener, J. High-Content Peptide Microarrays for Deciphering Kinase Specificity and Biology. Angew. Chem. 2004, 116, 2725–2728. [Google Scholar] [CrossRef]
- Thiele, A.; Stangl, G.I.; Schutkowski, M. Deciphering Enzyme Function Using Peptide Arrays. Mol. Biotechnol. 2011, 49, 283–305. [Google Scholar] [CrossRef]
- Ho, C.S.; Lam, C.W.K.; Chan, M.H.M.; Cheung, R.C.K.; Law, L.K.; Lit, L.C.W.; Ng, K.F.; Suen, M.W.M.; Tai, H.L. Electrospray Ionisation Mass Spectrometry: Principles and Clinical Applications. Clin. Biochem. Rev. 2003, 24, 3–12. [Google Scholar]
- Chen, W.; Ji, G.; Wu, R.; Fang, C.; Lu, H. Mass Spectrometry-Based Candidate Substrate and Site Identification of PTM Enzymes. TrAC Trends Anal. Chem. 2023, 160, 116991. [Google Scholar] [CrossRef]
- Černý, M.; Skalák, J.; Cerna, H.; Brzobohatý, B. Advances in Purification and Separation of Posttranslationally Modified Proteins. J. Proteom. 2013, 92, 2–27. [Google Scholar] [CrossRef] [PubMed]
- Ubersax, J.A.; Woodbury, E.L.; Quang, P.N.; Paraz, M.; Blethrow, J.D.; Shah, K.; Shokat, K.M.; Morgan, D.O. Targets of the Cyclin-Dependent Kinase Cdk1. Nature 2003, 425, 859–864. [Google Scholar] [CrossRef] [PubMed]
- Roux, K.J.; Kim, D.I.; Raida, M.; Burke, B. A Promiscuous Biotin Ligase Fusion Protein Identifies Proximal and Interacting Proteins in Mammalian Cells. J. Cell Biol. 2012, 196, 801–810. [Google Scholar] [CrossRef]
- Branon, T.C.; Bosch, J.A.; Sanchez, A.D.; Udeshi, N.D.; Svinkina, T.; Carr, S.A.; Feldman, J.L.; Perrimon, N.; Ting, A.Y. Efficient Proximity Labeling in Living Cells and Organisms with TurboID. Nat. Biotechnol. 2018, 36, 880–887. [Google Scholar] [CrossRef] [PubMed]
- Hung, V.; Zou, P.; Rhee, H.-W.; Udeshi, N.D.; Cracan, V.; Svinkina, T.; Carr, S.A.; Mootha, V.K.; Ting, A.Y. Proteomic Mapping of the Human Mitochondrial Intermembrane Space in Live Cells via Ratiometric APEX Tagging. Mol. Cell 2014, 55, 332–341. [Google Scholar] [CrossRef]
- Kim, M.; Zhong, J.; Pandey, A. Common Errors in Mass Spectrometry-based Analysis of Post-translational Modifications. Proteomics 2016, 16, 700–714. [Google Scholar] [CrossRef]
- Xue, L.; Tao, W.A. Current Technologies to Identify Protein Kinase Substrates in High Throughput. Front. Biol. 2013, 8, 216–227. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Kornhauser, J.M.; Tkachev, S.; Zhang, B.; Skrzypek, E.; Murray, B.; Latham, V.; Sullivan, M. PhosphoSitePlus: A Comprehensive Resource for Investigating the Structure and Function of Experimentally Determined Post-Translational Modifications in Man and Mouse. Nucleic Acids Res. 2012, 40, D261–D270. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and Recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID Database: A Comprehensive Biomedical Resource of Curated Protein, Genetic, and Chemical Interactions. Protein Sci. 2021, 30, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Blom, N.; Gammeltoft, S.; Brunak, S. Sequence and Structure-Based Prediction of Eukaryotic Protein Phosphorylation Sites. J. Mol. Biol. 1999, 294, 1351–1362. [Google Scholar] [CrossRef] [PubMed]
- Biggar, K.K.; Charih, F.; Liu, H.; Ruiz-Blanco, Y.B.; Stalker, L.; Chopra, A.; Connolly, J.; Adhikary, H.; Frensemier, K.; Hoekstra, M.; et al. Proteome-Wide Prediction of Lysine Methylation Leads to Identification of H2BK43 Methylation and Outlines the Potential Methyllysine Proteome. Cell Rep. 2020, 32, 107896. [Google Scholar] [CrossRef]
- Wang, D.; Liu, D.; Yuchi, J.; He, F.; Jiang, Y.; Cai, S.; Li, J.; Xu, D. MusiteDeep: A Deep-Learning Based Webserver for Protein Post-Translational Modification Site Prediction and Visualization. Nucleic Acids Res. 2020, 48, W140–W146. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Lee, J.; Oh, B.; Kimm, K.; Koh, I. Prediction of Phosphorylation Sites Using SVMs. Bioinformatics 2004, 20, 3179–3184. [Google Scholar] [CrossRef]
- Li, F.; Li, C.; Marquez-Lago, T.T.; Leier, A.; Akutsu, T.; Purcell, A.W.; Ian Smith, A.; Lithgow, T.; Daly, R.J.; Song, J.; et al. Quokka: A Comprehensive Tool for Rapid and Accurate Prediction of Kinase Family-Specific Phosphorylation Sites in the Human Proteome. Bioinformatics 2018, 34, 4223–4231. [Google Scholar] [CrossRef]
- Wang, L.; Du, Y.; Lu, M.; Li, T. ASEB: A Web Server for KAT-Specific Acetylation Site Prediction. Nucleic Acids Res. 2012, 40, W376–W379. [Google Scholar] [CrossRef]
- Deng, W.; Wang, C.; Zhang, Y.; Xu, Y.; Zhang, S.; Liu, Z. GPS-PAIL: Prediction of Lysine Acetyltransferase-Specific Modification Sites from Protein Sequences. Sci. Rep. 2016, 22, 39787. [Google Scholar] [CrossRef]
- Dang, T.H.; Van Leemput, K.; Verschoren, A.; Laukens, K. Prediction of Kinase-Specific Phosphorylation Sites Using Conditional Random Fields. Bioinformatics 2008, 24, 2857–2864. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Wang, H.; Wang, J.; Leier, A.; Marquez-Lago, T.; Yang, B.; Zhang, Z.; Akutsu, T.; Webb, G.I.; Daly, R.J. PhosphoPredict: A Bioinformatics Tool for Prediction of Human Kinase-Specific Phosphorylation Substrates and Sites by Integrating Heterogeneous Feature Selection. Sci. Rep. 2017, 7, 6862. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Zuo, Z.; Zhu, Q.; Hong, A.; Zhou, X.; Gao, X.; Li, T. Qualitative and Quantitative Analysis of Peptide Microarray Binding Experiments Using SVM-PEPARRAY. In Peptide Microarrays; Cretich, M., Chiari, M., Eds.; Humana Press: Totowa, NJ, USA, 2009; Volume 570, pp. 403–411. ISBN 978-1-60327-393-0. [Google Scholar]
- Tallorin, L.; Wang, J.; Kim, W.E.; Sahu, S.; Kosa, N.M.; Yang, P.; Thompson, M.; Gilson, M.K.; Frazier, P.I.; Burkart, M.D.; et al. Discovering de Novo Peptide Substrates for Enzymes Using Machine Learning. Nat. Commun. 2018, 9, 5253. [Google Scholar] [CrossRef]
- Vinogradov, A.A.; Chang, J.S.; Onaka, H.; Goto, Y.; Suga, H. Accurate Models of Substrate Preferences of Post-Translational Modification Enzymes from a Combination of mRNA Display and Deep Learning. ACS Cent. Sci. 2022, 8, 814–824. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Du, Y.; Wang, L.; Huang, L.; Li, W.; Lu, M.; Zhang, X.; Zhu, W.-G. Characterization and Prediction of Lysine (K)-Acetyl-Transferase Specific Acetylation Sites. Mol. Cell. Proteom. 2012, 11, M111.011080. [Google Scholar] [CrossRef]
- Yan, Y.; Jiang, J.-Y.; Fu, M.; Wang, D.; Pelletier, A.R.; Sigdel, D.; Ng, D.C.M.; Wang, W.; Ping, P. MIND-S Is a Deep-Learning Prediction Model for Elucidating Protein Post-Translational Modifications in Human Diseases. Cell Rep. Methods 2023, 3, 100430. [Google Scholar] [CrossRef]
- Shrestha, P.; Kandel, J.; Tayara, H.; Chong, K.T. Post-Translational Modification Prediction via Prompt-Based Fine-Tuning of a GPT-2 Model. Nat. Commun. 2024, 15, 6699. [Google Scholar] [CrossRef]
- Ferrari, E.; Tinti, M.; Costa, S.; Corallino, S.; Nardozza, A.P.; Chatraryamontri, A.; Ceol, A.; Cesareni, G.; Castagnoli, L. Identification of New Substrates of the Protein-Tyrosine Phosphatase PTP1B by Bayesian Integration of Proteome Evidence. J. Biol. Chem. 2011, 286, 4173–4185. [Google Scholar] [CrossRef] [PubMed]
- Lanouette, S.; Davey, J.A.; Elisma, F.; Ning, Z.; Figeys, D.; Chica, R.A.; Couture, J.-F. Discovery of Substrates for a SET Domain Lysine Methyltransferase Predicted by Multistate Computational Protein Design. Structure 2015, 23, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Piovesan, D.; Hatos, A.; Minervini, G.; Quaglia, F.; Monzon, A.M.; Tosatto, S.C.E. Assessing Predictors for New Post Translational Modification Sites: A Case Study on Hydroxylation. PLoS Comput. Biol. 2020, 16, e1007967. [Google Scholar] [CrossRef]
- Newton, M.S.; Cabezas-Perusse, Y.; Tong, C.L.; Seelig, B. In Vitro Selection of Peptides and Proteins—Advantages of mRNA Display. ACS Synth. Biol. 2020, 9, 181–190. [Google Scholar] [CrossRef]

| Type of Feature | Tool-Specific Details | Tool Name |
|---|---|---|
| Sequence-Based | Peptide or fragment of protein, surrounding PTM site | NetPhos [73], MethylSight [74], MusiteDeep [75], PredPhospho [76], Quokka [77], ASEB [78], GPS-PAIL [79], CRPhos [80], PhosphoPredict [81], SVM-PEPARRAY [82], POOL [83], mRNA Display [84] |
| Sequence-Based | Physicochemical feature vectors | MethylSight [74], CRPhos [85], PhosphoPredict [81], mRNA Display [84] |
| Sequence-Based | Full protein sequence, PTM site(s) indicated | MIND-S [86], PTMGPT2 [87] |
| Sequence-Based | Substitution matrix: BLOSUM or PSSM | Quokka [77], ASEB [78], GPS-PAIL [79], PTP1B Bayesian Modelling [88] |
| Sequence-Based | Recognition motif | Quokka [77] |
| Sequence-Based | Protein interaction network | PhosphoPredict [81] |
| Sequence-Based | Predicted secondary structure | MIND-S [86], PhosphoPredict [81] |
| Structure-Based | Structure of enzyme–substrate complex | CPD [89] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ridgeway, N.H.; Biggar, K.K. Uncovering Enzyme-Specific Post-Translational Modifications: An Overview of Current Methods. Proteomes 2025, 13, 37. https://doi.org/10.3390/proteomes13030037
Ridgeway NH, Biggar KK. Uncovering Enzyme-Specific Post-Translational Modifications: An Overview of Current Methods. Proteomes. 2025; 13(3):37. https://doi.org/10.3390/proteomes13030037
Chicago/Turabian StyleRidgeway, Nashira H., and Kyle K. Biggar. 2025. "Uncovering Enzyme-Specific Post-Translational Modifications: An Overview of Current Methods" Proteomes 13, no. 3: 37. https://doi.org/10.3390/proteomes13030037
APA StyleRidgeway, N. H., & Biggar, K. K. (2025). Uncovering Enzyme-Specific Post-Translational Modifications: An Overview of Current Methods. Proteomes, 13(3), 37. https://doi.org/10.3390/proteomes13030037







