Unraveling the Central Role of Global Regulator PprI in Deinococcus radiodurans Through Label-Free Quantitative Proteomics
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Chemical Reagents
2.2. Sample Preparation for Proteomics Analysis
2.3. LC-MS/MS Analysis
2.4. Data Analysis and Bioinformatics
3. Results
3.1. Growth Curves of the DR and ΔpprI Strains
3.2. Comparative Proteomic Analysis of Total Proteins in Wild-Type and pprI Knockout Strains
3.3. Gene Ontology (GO) and Clusters of Orthologous Groups (COG) Analysis
3.3.1. Gene Ontology Analysis
3.3.2. Clusters of Orthologous Groups Analysis
3.4. Protein–Protein Interactions and Biological Pathway Networks
4. Discussion
4.1. Dynamic Regulation of DNA Damage Repair
4.2. Molecular Basis of Antioxidant Defense
4.3. Metabolic Reprogramming
4.4. Limitations of This Present Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Makarova, K.S.; Aravind, L.; Wolf, Y.I.; Tatusov, R.L.; Minton, K.W.; Koonin, E.V.; Daly, M.J. Genome of the Extremely Radiation-Resistant Bacterium Deinococcus radiodurans Viewed from the Perspective of Comparative Genomics. Microbiol. Mol. Biol. Rev. 2001, 65, 44–79. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.M.; Battista, J.R. Deinococcus radiodurans—the consummate survivor. Nat. Rev. Microbiol. 2005, 3, 882–892. [Google Scholar] [CrossRef]
- White, O.; Eisen, J.A.; Heidelberg, J.F.; Hickey, E.K.; Peterson, J.D.; Dodson, R.J.; Haft, D.H.; Gwinn, M.L.; Nelson, W.C.; Richardson, D.L.; et al. Genome Sequence of the Radioresistant Bacterium Deinococcus radiodurans R1. Science 1999, 286, 1571–1577. [Google Scholar] [CrossRef]
- Yan, S.; Gao, S.S.; Zhou, P.K. Multi-Functions of Exonuclease 1 in DNA Damage Response and Cancer Susceptibility. Radiat. Med. Prot. 2021, 2, 146–154. [Google Scholar] [CrossRef]
- Nilsson, R.; Liu, N.A. Nuclear DNA Damages Generated by Reactive Oxygen Molecules (ROS) Under Oxidative Stress and Their Relevance to Human Cancers, including Ionizing Radiation-Induced Neoplasia Part I: Physical, Chemical and Molecular Biology Aspects. Radiat. Med. Prot. 2020, 1, 140–152. [Google Scholar] [CrossRef]
- Timmins, J.; Moe, E. A Decade of Biochemical and Structural Studies of the DNA Repair Machinery of Deinococcus radiodurans: Major Findings, Functional and Mechanistic Insight and Challenges. Comput. Struct. Biotechnol. J. 2016, 14, 168–176. [Google Scholar] [CrossRef]
- Hansler, A.; Chen, Q.; Ma, Y.; Gross, S.S. Untargeted Metabolite Profiling Reveals that Nitric Oxide Bioynthesis is an Endogenous Modulator of Carotenoid Biosynthesis in Deinococcus radiodurans and is Required for Extreme Ionizing Radiation Resistance. Arch. Biochem. Biophys. 2016, 589, 38–52. [Google Scholar] [CrossRef]
- Liu, F.; Li, N.M.; Zhang, Y.Q. The Radioresistant and Survival Mechanisms of Deinococcus radiodurans. Radiat. Med. Prot. 2023, 4, 70–79. [Google Scholar] [CrossRef]
- Agrawal, D.; Satpathy, S.; Samal, D. Deinococcus Radiodurans: The World’s Toughest Bacterium. A Review. Glob. J. Med. Res. 2020, 20, 1–4. [Google Scholar] [CrossRef]
- Chen, Z.J.; Tang, Y.Y.; Hua, Y.; Zhao, Y. Structural Features and Functional Implications of Proteins Enabling the Robustness of Deinococcus radiodurans. Comput. Struct. Biotechnol. J. 2020, 18, 2810–2817. [Google Scholar] [CrossRef]
- Battista, J.R.; Cox, M.M.; Daly, M.J.; Narumi, I.; Radman, M.; Sommer, S. The Structure of D. radiodurans. Science 2003, 302, 567–568. [Google Scholar] [CrossRef]
- Dai, S.; Xie, Z.M.; Wang, B.; Yu, N.; Zhao, J.; Zhou, Y.; Hua, Y.; Tian, B. Dynamic Polyphosphate Metabolism Coordinating with Manganese Ions Defends against Oxidative Stress in the Extreme Bacterium Deinococcus radiodurans. Appl. Environ. Microbiol. 2021, 87, e02785-20. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Zhou, Z.; Chen, X.; Zhang, W.; Lin, M.; Chen, M. Comparative Proteomics Analysis Reveals New Features of the Oxidative Stress Response in the Polyextremophilic Bacterium Deinococcus radiodurans. Microorganisms 2020, 8, 451. [Google Scholar] [CrossRef] [PubMed]
- Daly, M.J.; Gaidamakova, E.K.; Matrosova, V.Y.; Vasilenko, A.; Zhai, M.; Venkateswaran, A.; Hess, M.; Omelchenko, M.V.; Kostandarithes, H.M.; Makarova, K.S.; et al. Accumulation of Mn(II) in Deinococcus radiodurans Facilitates Gamma-Radiation Resistance. Science 2004, 306, 1025–1028. [Google Scholar] [CrossRef]
- Daly, M.J.; Gaidamakova, E.K.; Matrosova, V.Y.; Vasilenko, A.; Zhai, M.; Leapman, R.D.; Lai, B.; Ravel, B.; Li, S.-M.W.; Kemner, K.M. Protein Oxidation Implicated as the Primary Determinant of Bacterial Radioresistance. PLoS Biol. 2007, 5, e92. [Google Scholar] [CrossRef]
- Slade, D.; Radman, M. Oxidative Stress Resistance in Deinococcus radiodurans. Microbiol. Mol. Biol. Rev. 2011, 75, 133–191. [Google Scholar] [CrossRef]
- Cox, M.M.; Keck, J.L.; Battista, J.R. Rising From the Ashes: DNA Repair in Deinococcus radiodurans. PLOS Genet. 2010, 6, e1000815. [Google Scholar] [CrossRef]
- Gao, G.; Tian, B.; Liu, L.; Sheng, D.; Shen, B.; Hua, Y. Expression of Deinococcus radiodurans PprI Enhances the Radioresistance of Escherichia coli. DNA Repair 2003, 2, 1419–1427. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Gao, G.; Xu, G.; Fan, L.; Yin, L.; Shen, B.; Hua, Y. Deinococcus radiodurans PprI Switches on DNA Damage Response and Cellular Survival Networks after Radiation Damage. Mol. Cell. Proteom. 2009, 8, 481–494. [Google Scholar] [CrossRef]
- Lu, H.; Hua, Y. PprI: The Key Protein in Response to DNA Damage in Deinococcus. Front. Cell Dev. Biol. 2021, 8, 609714. [Google Scholar] [CrossRef]
- Wang, L.; Tan, Y.S.; Chen, K.; Ntakirutimana, S.; Liu, Z.H.; Li, B.Z.; Yuan, Y.J. Global Regulator IrrE on Stress Tolerance: A Review. Crit. Rev. Biotechnol. 2024, 44, 1439–1459. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Lu, H.; Huang, L.; Hua, Y. Construction of DNA Damage Response Gene pprI Function-Deficient and Function-Complementary Mutants in Deinococcus radiodurans. Chin. Sci. Bull. 2005, 50, 311–316. [Google Scholar] [CrossRef]
- Wang, L.; Yin, L.; Xu, G.; Li, M.; Zhang, H.; Tian, B.; Hua, Y. Cooperation of PprI and DrRRA in Response to Extreme Ionizing Radiation in Deinococcus radiodurans. Chin. Sci. Bull. 2012, 57, 98–104. [Google Scholar] [CrossRef]
- Narumi, I.; Satoh, K.; Kikuchi, M.; Funayama, T.; Ikeuchi, M.; Watanabe, H. PprI protein regulates RecA protein expression and DNA repair in Deinococcus radiodurans. J. Bacteriol. 2004, 186, 35–41. [Google Scholar]
- Adachi, M.; Shimizu, R.; Shibazaki, C.; Satoh, K.; Fujiwara, S.; Arai, S.; Narumi, I.; Kuroki, R. Extended Structure of Pleiotropic DNA Repair-Promoting Protein PprA from Deinococcus radiodurans. FASEB J. 2019, 33, 3647–3658. [Google Scholar] [CrossRef]
- Rajpurohit, Y.S.; Sharma, D.K.; Misra, H.S. PprA Protein Inhibits DNA Strand Exchange and ATP Hydrolysis of Deinococcus RecA and Regulates the Recombination in Gamma-Irradiated Cells. Front. Cell Dev. Biol. 2021, 9, 636178. [Google Scholar] [CrossRef]
- Tian, B.; Zhang, S.W.; Xu, Z.J.; Sheng, D.H.; Hua, Y.J. Effects of PprI and RecX on Antioxidant Activity of Deinococcus radiodurans. Acta Microbiol. Sin. 2006, 46, 238–242. [Google Scholar]
- Maurya, G.K.; Chaudhary, R.; Pandey, N.; Misra, H.S. Molecular Insights into Replication Initiation in a Multipartite Genome Harboring Bacterium Deinococcus radiodurans. J. Biol. Chem. 2021, 296, 100451. [Google Scholar] [CrossRef]
- Jeong, S.W.; Seo, H.S.; Kim, M.K.; Choi, J.; Lim, H.M.; Lim, S.Y. PprM is Necessary for Up-Regulation of katE1, Encoding the Major Catalase of Deinococcus radiodurans, under Unstressed Culture Conditions. J. Microbiol. 2016, 54, 426–431. [Google Scholar] [CrossRef]
- Lu, H.; Wang, L.; Li, S.; Pan, C.; Cheng, K.; Luo, Y.; Xu, H.; Tian, B.; Zhao, Y.; Hua, Y. Structure and DNA Damage-Dependent Derepression Mechanism for the XRE Family Member DG-DdrO. Nucleic Acids Res. 2019, 47, 9925–9933. [Google Scholar] [CrossRef]
- Lu, H.; Chen, H.; Xu, G.; Shah, A.M.; Hua, Y. DNA Binding is Essential for PprI Function in Response to Radiation Damage in Deinococcus radiodurans. DNA Repair 2012, 11, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.G.; Zhang, C.C.; Hua, Y.J. Proteomic Analysis of pprI mutant of Deinococcus radiodurans. J. Zhejiang Univ. 2005, 31, 27–31. (In Chinese) [Google Scholar]
- Matrosova, V.Y.; Gaidamakova, E.K.; Makarova, K.S.; Grichenko, O.; Klimenkova, P.; Volpe, R.P.; Tkavc, R.; Ertem, G.; Conze, I.H.; Brambilla, E.; et al. High-quality genome sequence of the radioresistant bacterium Deinococcus ficus KS 0460. Stand. Genomic Sci. 2017, 12, 46. [Google Scholar] [CrossRef]
- Gao, Y.; Li, N.; Zhou, Y.; Zhang, Z.; Zhang, Y.; Fan, P.; Zhou, H.; Zhang, T.; Chang, L.; Gao, H.; et al. iTRAQ-based proteomic analysis of Deinococcus radiodurans in response to 12C6+ heavy ion irradiation. BMC Microbiol. 2022, 22, 264. [Google Scholar] [CrossRef]
- Chi, H.; Liu, C.; Yang, H.; Zeng, W.-F.; Wu, L.; Zhou, W.-J.; Wang, R.-M.; Niu, X.-N.; Ding, Y.-H.; Zhang, Y.; et al. Comprehensive identification of peptides in tandem mass spectra using an efficient open search engine. Nat. Biotechnol. 2018, 36, 1059–1061. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Song, C.Q.; Yuan, Z.F.; Fu, Y.; Chi, H.; Wang, L.-H.; Fan, S.-B.; Zhang, K.; Zeng, W.-F.; He, S.-M.; et al. pQuant Improves Quantitation by Keeping out Interfering Signals and Evaluating the Accuracy of Calculated Ratios. Anal. Chem. 2014, 86, 5286–5294. [Google Scholar] [CrossRef]
- Qi, H.; Wang, W.; He, J.; Ma, Y.; Xiao, F.; He, S. Antioxidative system of Deinococcus radiodurans. Res. Microbiol. 2020, 171, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Wu, W.; Qiao, H.; Yue, L.; Ren, L.; Zhang, S.; Yang, W.; Yang, Z. The protein PprI provides protection against radiation injury in human and mouse cells. Sci. Rep. 2016, 6, 26664. [Google Scholar] [CrossRef]
- Villa, J.K.; Han, R.; Tsai, C.H.; Chen, A.; Sweet, P.; Franco, G.; Vaezian, R.; Tkavc, R.; Daly, M.J.; Contreras, L.M. A small RNA regulates pprM, a modulator of pleiotropic proteins promoting DNA repair, in Deinococcus radiodurans under ionizing radiation. Sci. Rep. 2021, 11, 12949. [Google Scholar] [CrossRef]
- Cheng, K.Y.; Xu, Y.; Chen, X.Y.; Lu, H.Z.; He, Y.; Wang, L.Y.; Hua, Y.J. Participation of RecJ in the base excision repair pathway of Deinococcus radiodurans. Nucleic Acids Res. 2020, 48, 9859–9871. [Google Scholar] [CrossRef]
- Vujičić-Žagar, A.; Serre, L.; Dulermo, R.; Le Gorrec, M.; Vannier, F.; Servant, P.; Sommer, S.; de Groot, A. Crystal Structure of the IrrE Protein, a Central Regulator of DNA Damage Repair in Deinococcaceae. J. Mol. Biol. 2009, 386, 704–716. [Google Scholar] [CrossRef]
- Lu, H.; Chen, Z.; Xie, T.; Zhong, S.; Suo, S.; Song, S.; Wang, L.; Xu, H.; Tian, B.; Zhao, Y.; et al. The Deinococcus protease PprI senses DNA damage by directly interacting with single-stranded DNA. Nat. Commun. 2024, 15, 1892. [Google Scholar] [CrossRef]
- Dai, S.; Wang, B.; Ye, R.; Zhang, D.; Xie, Z.; Yu, N.; Cai, C.; Huang, C.; Zhao, J.; Zhang, F.; et al. Structural Evolution of Bacterial Polyphosphate Degradation Enzyme for Phosphorus Cycling. Adv.Sci. 2024, 11, 2309602. [Google Scholar] [CrossRef]
- Hua, X.T.; Tian, B.; Hua, Y.J. Research Progress on Homologous Recombination Repair Mechanism in Deinococcus radiodurans. J. Nucl. Agric. Sci. 2010, 24, 1192–1197. [Google Scholar]
- Zhang, C.Y.; Qiu, Q.T.; Ma, B.G. Transcriptomic Analysis of Deinococcus radiodurans During the Early Recovery Stage from Ultraviolet Irradiation. Prog. Biochem. Biophys. 2023, 50, 1701–1715. [Google Scholar]
- Xu, G.; Wang, L.; Chen, H.; Lu, H.; Ying, N.; Tian, B.; Hua, Y. RecO Is Essential for DNA Damage Repair in Deinococcus radiodurans. J. Bacteriol. 2008, 190, 2624–2628. [Google Scholar] [CrossRef]
- Nivedita, P.K.; Vidya, A.K.; Hari, S.M. RecBC enzyme overproduction affects UV and gamma radiation survival of Deinococcus radiodurans. DNA Repair 2008, 7, 40–47. [Google Scholar]
- Zhou, Q.; Zhang, X.; Xu, H.; Xu, B.; Hua, Y.J. A new role of Deinococcus radiodurans RecD in antioxidant pathway. FEMS Microbiol. Lett. 2007, 271, 118–125. [Google Scholar] [CrossRef]
- Servinsky, M.D.; Julin, D.A. Effect of a recD Mutation on DNA Damage Resistance and Transformation in Deinococcus radiodurans. J. Bacteriol. 2007, 189, 5101–5107. [Google Scholar] [CrossRef]
- Huang, L.; Hua, X.; Lu, H.; Gao, G.; Tian, B.; Shen, B.; Hua, Y. Three tandem HRDC domains have synergistic effect on the RecQ functions in Deinococcus radiodurans. DNA Repair 2007, 6, 167–176. [Google Scholar] [CrossRef]
- Cheng, K.; Xu, H.; Chen, X.; Wang, L.; Tian, B.; Zhao, Y.; Hua, Y. Structural basis for DNA 5’-end resection by RecJ. eLife 2016, 5, e14294. [Google Scholar] [CrossRef]
- Kim, J.I.; Cox, M.M. The RecA proteins of Deinococcus radiodurans and Escherichia coli promote DNA strand exchange via inverse pathways. Proc. Natl. Acad. Sci. USA 2002, 99, 7917–7921. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Chen, W.; Zhao, Y.; Hua, Y. Involvement of RecG in H2O2-induced damage repair in Deinococcus radiodurans. Can. J. Microbiol. 2009, 55, 841–848. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.W.; Kim, M.K.; Zhao, L.; Yang, S.-K.; Jung, J.-H.; Lim, H.-M.; Lim, S. Effects of Conserved Wedge Domain Residues on DNA Binding Activity of Deinococcus radiodurans RecG Helicase. Front. Genet. 2021, 12, 634615. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Narumi, I.; Funayama, T.; Kikuchi, M.; Watanabe, H.; Matsunaga, T.; Nikaido, O.; Yamamoto, K. Characterization of pathways dependent on the uvsE, uvrA1, or uvrA2 gene product for UV resistance in Deinococcus radiodurans. J. Bacteriol. 2005, 187, 3693–3697. [Google Scholar] [CrossRef] [PubMed]
- Santos, S.P.; Yang, Y.; Rosa, M.T.G.; Rodrigues, M.A.A.; De La Tour, C.B.; Sommer, S.; Teixeira, M.; Carrondo, M.A.; Cloetens, P.; Abreu, I.A. The interplay between Mn and Fe in Deinococcus radiodurans triggers cellular protection during paraquat-induced oxidative stress. Sci. Rep. 2019, 9, 17217. [Google Scholar] [CrossRef]
- Chen, H.; Wu, R.; Xu, G.; Fang, X.; Qiu, X.; Guo, H.; Tian, B.; Hua, Y. DR2539 is a novel DtxR-like regulator of Mn/Fe ion homeostasis and antioxidant enzyme in Deinococcus radiodurans. Biochem. Biophys. Res. Commun. 2010, 396, 413–418. [Google Scholar] [CrossRef]
- Sun, H.; Li, M.; Xu, G.; Chen, H.; Jiao, J.; Tian, B.; Wang, L.; Hua, Y. Regulation of MntH by a Dual Mn(II)- and Fe(II)-Dependent Transcriptional Repressor (DR2539) in Deinococcus radiodurans. PLoS ONE 2012, 7, e35057. [Google Scholar] [CrossRef]
- Bing, T.; Xu, Z.; Sun, Z.; Lin, J.; Hua, Y. Evaluation of the antioxidant effects of carotenoids from Deinococcus radiodurans through targeted mutagenesis, chemiluminescence, and DNA damage analyses. BBA-Gen. Subj. 2007, 1770, 902–911. [Google Scholar]
- Jeong, S.W.; Kim, J.H.; Kim, J.W.; Kim, C.Y.; Kim, S.Y.; Choi, Y.J. Metabolic Engineering of Extremophilic Bacterium Deinococcus radiodurans for the Production of the Novel Carotenoid Deinoxanthin. Microorganisms 2021, 9, 44. [Google Scholar] [CrossRef]
- Xu, Z.J.; Tian, B.; Xu, G.Z.; Hua, Y. Construction and functional analysis of the crtI gene disruptant in Deinococcus radiodurans. Acta Microbiol. Sin. 2006, 46, 210–213. [Google Scholar]
- Li, S.F.; Xie, J.Y.; Qiu, S.; Xu, S.Y.; Cheng, F.; Wang, Y.J.; Zheng, Y.G. Semirational engineering of an aldo-keto reductase KmAKR for overcoming trade-offs between catalytic activity and thermostability. Biotechnol. Bioeng. 2021, 118, 4441–4452. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Morar, M.; Ealick, S.E. Structural biology of the purine biosynthetic pathway. Cell. Mol. Life Sci. 2008, 65, 3699–3724. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, S.P.; Grove, A. HucR, a Novel Uric Acid-responsive Member of the MarR Family of Transcriptional Regulators from Deinococcus radiodurans. J. Biol. Chem. 2004, 279, 51442–51450. [Google Scholar] [CrossRef]
- Holbrook, S.R. The Nudix Hydrolase Family: Structural, Functional and Evolutionary Relationships; LBNL Report LBNL-52014 Abs; Lawrence Berkeley National Laboratory (LBNL): Berkeley, CA, USA, 2003. [Google Scholar]
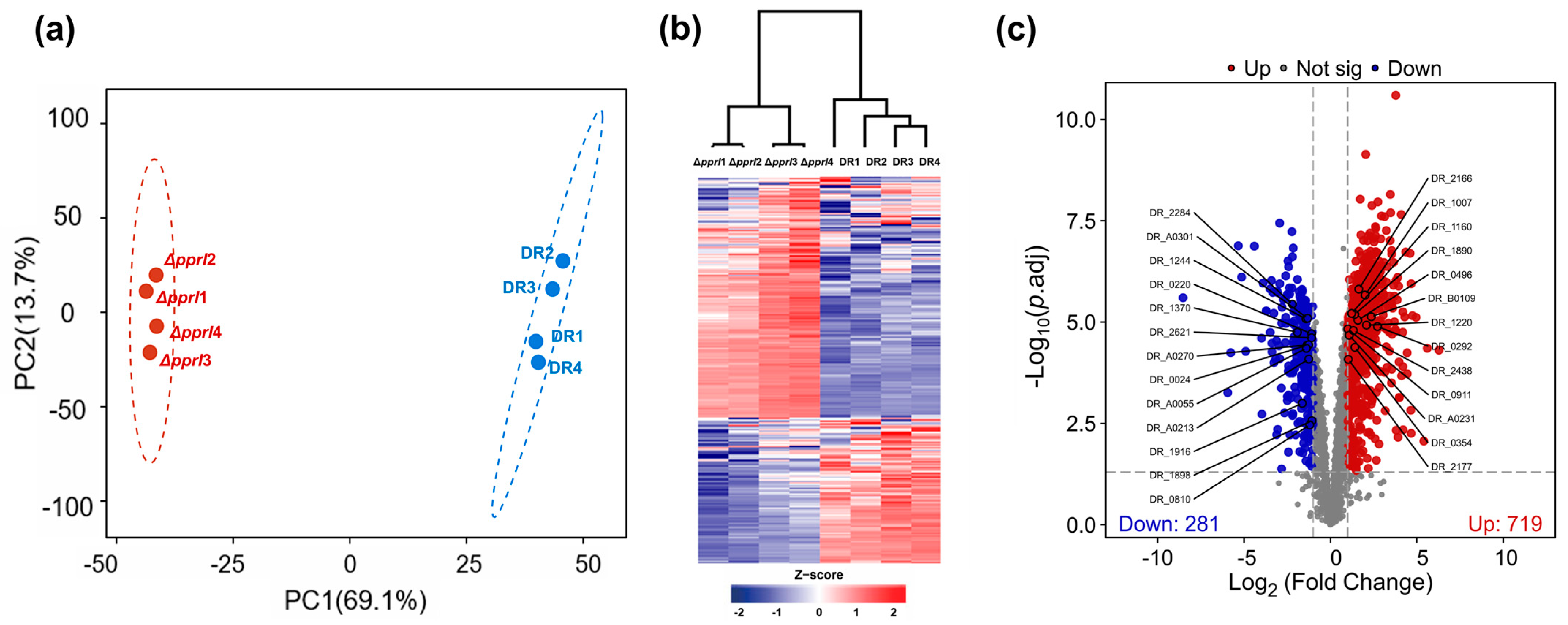
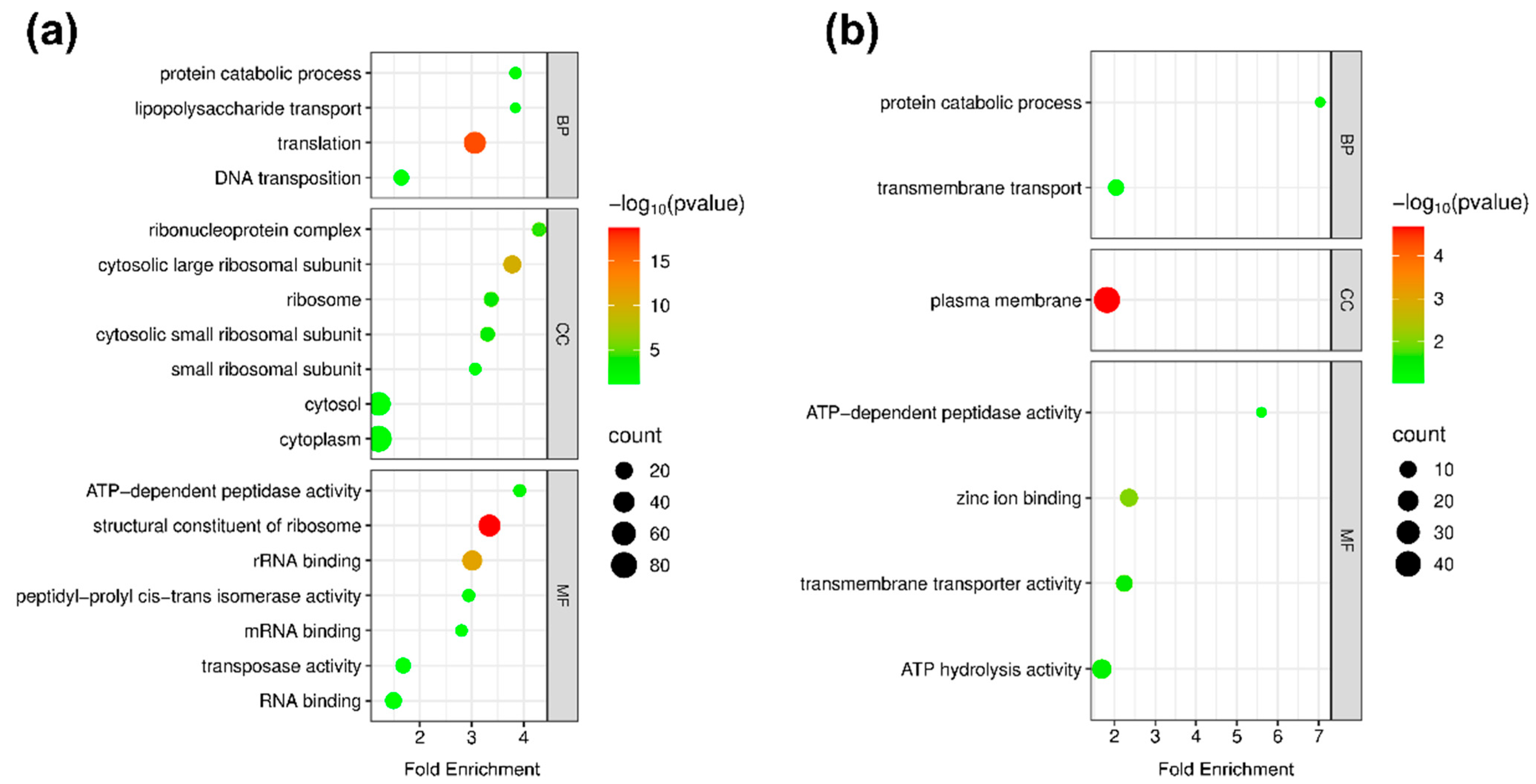
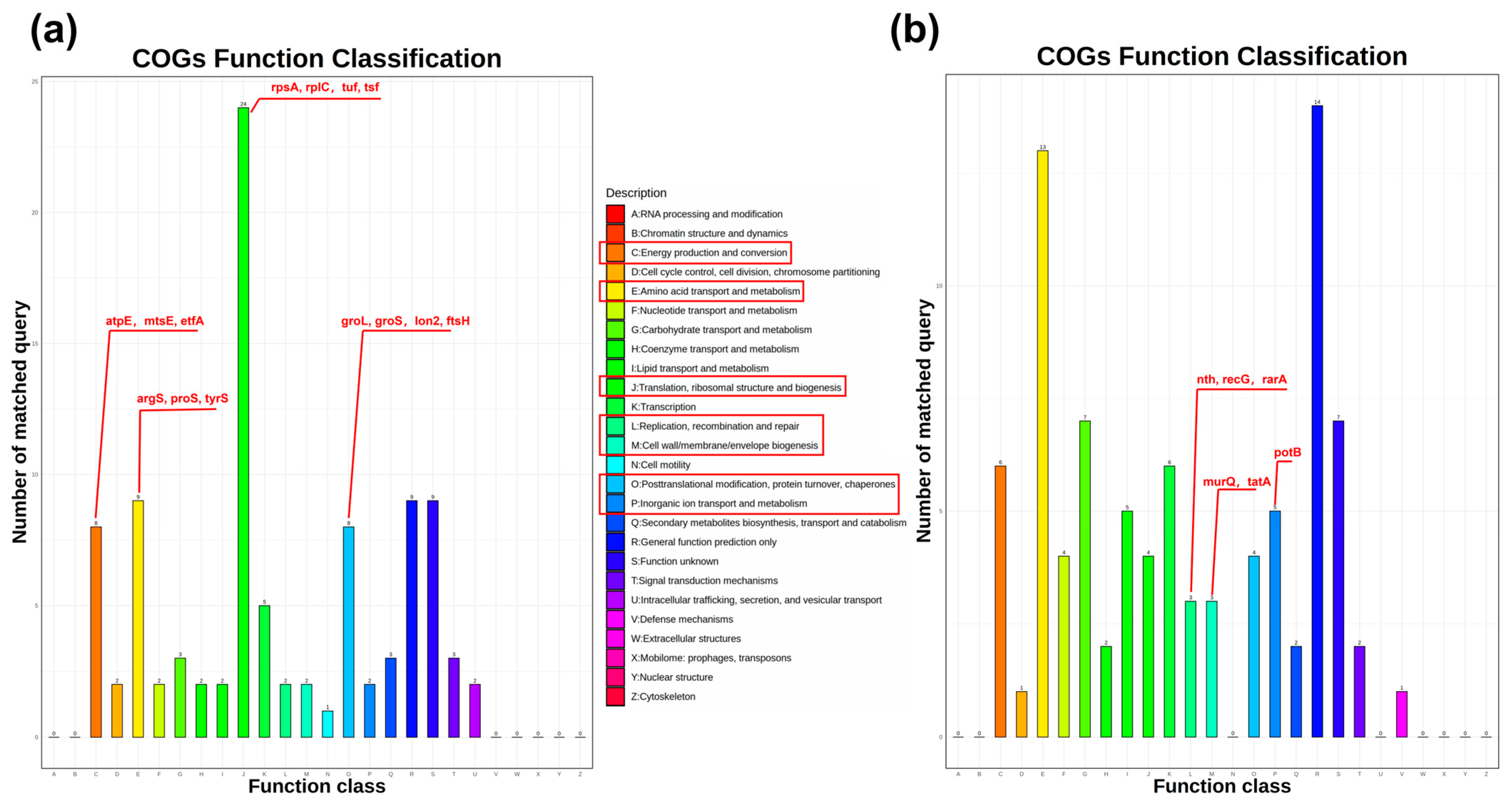
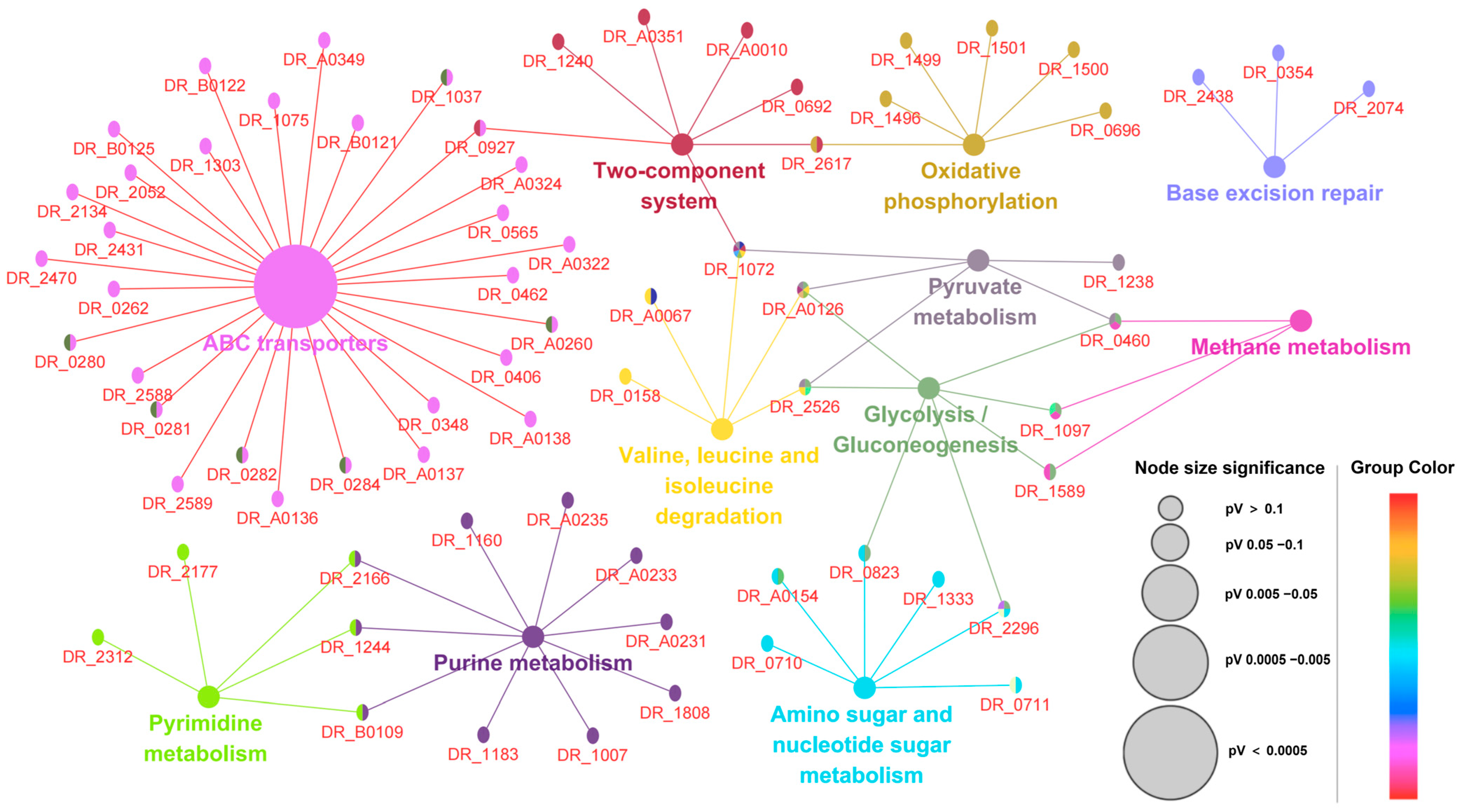

| Accession ID | Protein Name | Gene Name | Fold Change | COG Category | Protein Score |
|---|---|---|---|---|---|
| Q9RXH8 | RNA helicase | DR_0335 | 3.217 | LKJ | 875.0 |
| Q9RXF9 | Exodeoxyribonuclease III | DR_0354 | 2.669 | L | 577.0 |
| Q9RZ89 | DNA-binding protein HU | DR_A0065 | 2.642 | L | 186.0 |
| Q9RRQ0 | Endonuclease III | DR_2438 | 2.513 | L | 540.0 |
| Q9RSJ6 | DNA-directed RNA polymerase subunit alpha | DR_2128 | 2.187 | K | 662.0 |
| Q9RYM2 | ATP-dependent zinc metalloprotease FtsH | DR_A0290 | 2.141 | O | 1270.0 |
| Q9RXG4 | Lon protease | DR_0349 | 2.108 | O | 1573.0 |
| Q9RVW0 | DNA-directed RNA polymerase subunit beta’ | DR_0911 | 2.011 | K | 3033.0 |
| Q9RSC5 | Orotidine-5′-phosphate decarboxylase | DR_2200 | 0.498 | F | 520.0 |
| Q9RW54 | GntR family transcriptional regulator | DR_0815 | 0.494 | K | 508.0 |
| Q9RZD9 | DNA-binding response regulator | DR_A0010 | 0.493 | T | 396.0 |
| Q9RT67 | AAA + ATPase domain-containing protein | DR_1898 | 0.476 | L | 843.0 |
| Q9RV39 | A-adding tRNA nucleotidyltransferase | DR_1191 | 0.385 | J | 859.0 |
| Q9RZ99 | Ribokinase | DR_A0055 | 0.380 | H | 573.0 |
| Q9RUY6 | DNA-directed DNA polymerase | DR_1244 | 0.375 | L | 575.0 |
| Q9RT50 | ATP-dependent DNA helicase RecG | DR_1916 | 0.323 | LK | 1517.0 |
| Q9RSQ0 | Putative 3-methyladenine DNA glycosylase | DR_2074 | 0.183 | L | 392.0 |
| Accession ID | Protein Name | Gene Name | Fold Change | COG Category | Protein Score |
|---|---|---|---|---|---|
| Q9RV10 | Ferrous iron transport protein A | DR_1220 | 4.245 | P | 152.0 |
| Q9RX19 | Ferredoxin/Ferredoxin--NADP reductase | DR_0496 | 3.009 | ER | 939.0 |
| Q9RT75 | Oxidoreductase | DR_1890 | 2.653 | C | 650.0 |
| Q9RYS7 | Oxidoreductase | DR_A0231 | 2.122 | C | 1455.0 |
| Q9RVS5 | Isochorismatase family protein | DR_0947 | 0.489 | Q | 228.0 |
| Q9RW59 | Dehydrogenase | DR_0810 | 0.436 | Q | 901.0 |
| Q9RU93 | NADH-quinone oxidoreductase | DR_1499 | 0.422 | C | 1398.0 |
| Q9RYL1 | Cytochrome-c peroxidase | DR_A0301 | 0.400 | P | 687.0 |
| Q9RS43 | Manganese ABC transporter, ATP-binding protein | DR_2284 | 0.217 | P | 482.0 |
| Accession ID | Protein Name | Gene Name | Fold Change | COG Category | Protein Score |
|---|---|---|---|---|---|
| Q9RW75 | [LysW]-aminoadipate semialdehyde transaminase | DR_0794 | 5.494 | E | 835.0 |
| Q9RZL6 | Ribonucleotide–diphosphate reductase subunit beta | DR_B0109 | 5.144 | F | 603.0 |
| Q9RVM0 | MutT/nudix family protein | DR_1007 | 4.027 | L | 324.0 |
| Q9RU24 | ABC transporter substrate-binding protein | DR_1571 | 3.378 | E | 1194.0 |
| Q9RSW3 | Aspartate-semialdehyde dehydrogenase | DR_2008 | 3.351 | E | 660.0 |
| Q9RWQ9 | Chaperonin GroEL | DR_0607 | 3.219 | O | 1016.0 |
| Q9RUB7 | Phage shock protein A homolog | DR_1473 | 3.137 | KT | 403.0 |
| Q9RSF8 | Uridine phosphorylase | DR_2166 | 3.123 | F | 612.0 |
| Q9RW01 | Bifunctional purine biosynthesis protein PurH | DR_0868 | 3.068 | F | 1002.0 |
| Q9RUW4 | Proline--tRNA ligase | DR_1266 | 3.004 | J | 1021.0 |
| Q9RRC4 | Arginine--tRNA ligase | DR_2568 | 2.867 | J | 1194.0 |
| Q9RXK2 | Large ribosomal subunit protein uL3 | DR_0311 | 2.733 | J | 419.0 |
| Q9RY06 | Valine--tRNA ligase | DR_0148 | 2.596 | J | 1840.0 |
| Q9RUP2 | Phosphoglycerate kinase | DR_1342 | 2.563 | G | 805.0 |
| Q9RR70 | Fumarate hydratase class II | DR_2627 | 2.552 | C | 919.0 |
| Q9RSL8 | Fe-S cluster assembly protein SufB | DR_2106 | 2.525 | O | 942.0 |
| Q9RY67 | 2-oxoglutarate dehydrogenase complex dihydrolipoyllysine-residue Succinyltransferase(odhB) | DR_0083 | 2.489 | C | 783.0 |
| Q9RYB2 | Serine hydroxymethyltransferase | DR_0038 | 2.462 | E | 810.0 |
| O32507 | Succinate-semialdehyde dehydrogenase [NADP(+)] | DR_A0343 | 2.455 | C | 936.0 |
| Q9RUF5 | Phosphoribosylamine--glycine ligase | DR_1431 | 2.396 | F | 811.0 |
| Q9RV70 | Uricase | DR_1160 | 2.339 | Q | 617.0 |
| Q9RWB2 | Citrate synthase | DR_0757 | 2.322 | C | 746.0 |
| Q9RVQ2 | Electron transfer flavoprotein, alpha subunit | DR_0970 | 2.307 | C | 597.0 |
| Q9RRA0 | Magnesium protoporphyrin chelatase | DR_2594 | 2.272 | H | 949.0 |
| Q9RWH2 | V-type ATP synthase, K subunit | DR_0696 | 2.268 | C | 170.0 |
| Q9RS27 | Alanine--tRNA ligase | DR_2300 | 2.231 | J | 1748.0 |
| Q9RR63 | Tyrosine--tRNA ligase | DR_2634 | 2.070 | J | 807.0 |
| Q9R342 | Elongation factor Tu | DR_0309 | 2.064 | J | 797.0 |
| Q9RSE7 | Cytidine deaminase | DR_2177 | 2.044 | F | 317.0 |
| Q9RVK7 | ATP-dependent zinc metalloprotease FtsH | DR_1020 | 0.498 | O | 1201.0 |
| Q9RYY2 | Hydroxymethylpyrimidine kinase | DR_A0171 | 0.491 | H | 486.0 |
| Q9RXT6 | Amidophosphoribosyltransferase | DR_0220 | 0.469 | F | 959.0 |
| Q9RR76 | Glycerol-3-phosphate dehydrogenase [NAD(P)+] | DR_2621 | 0.469 | I | 625.0 |
| Q9RVF3 | Cell wall synthesis protein | DR_1076 | 0.454 | M | 813.0 |
| Q9RYU5 | N-acetylmuramic acid 6-phosphate etherase | DR_A0213 | 0.420 | S | 565.0 |
| Q9RXL8 | Sec-independent protein translocase protein TatA | DR_0292 | 0.419 | U | 211.0 |
| Q9RYP0 | Adenine deaminase | DR_A0270 | 0.417 | F | 1031.0 |
| Q9RYC6 | N5-carboxyaminoimidazole ribonucleotide synthase | DR_0024 | 0.387 | F | 727.0 |
| Q9RVJ1 | Branched-chain amino acid ABC transporter, permease protein | DR_1037 | 0.361 | E | 655.0 |
| Q9RX55 | Acetate--CoA ligase | DR_0460 | 0.360 | I | 1301.0 |
| Q9RUL4 | Outer membrane lipoprotein-sorting protein | DR_1370 | 0.266 | M | 414.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, S.; Liu, F.; Wang, H.; Zhang, Y. Unraveling the Central Role of Global Regulator PprI in Deinococcus radiodurans Through Label-Free Quantitative Proteomics. Proteomes 2025, 13, 19. https://doi.org/10.3390/proteomes13020019
Zhu S, Liu F, Wang H, Zhang Y. Unraveling the Central Role of Global Regulator PprI in Deinococcus radiodurans Through Label-Free Quantitative Proteomics. Proteomes. 2025; 13(2):19. https://doi.org/10.3390/proteomes13020019
Chicago/Turabian StyleZhu, Siyu, Feng Liu, Hao Wang, and Yongqian Zhang. 2025. "Unraveling the Central Role of Global Regulator PprI in Deinococcus radiodurans Through Label-Free Quantitative Proteomics" Proteomes 13, no. 2: 19. https://doi.org/10.3390/proteomes13020019
APA StyleZhu, S., Liu, F., Wang, H., & Zhang, Y. (2025). Unraveling the Central Role of Global Regulator PprI in Deinococcus radiodurans Through Label-Free Quantitative Proteomics. Proteomes, 13(2), 19. https://doi.org/10.3390/proteomes13020019







