Oncogenic Proteomics Approaches for Translational Research and HIV-Associated Malignancy Mechanisms
Abstract
1. Introduction
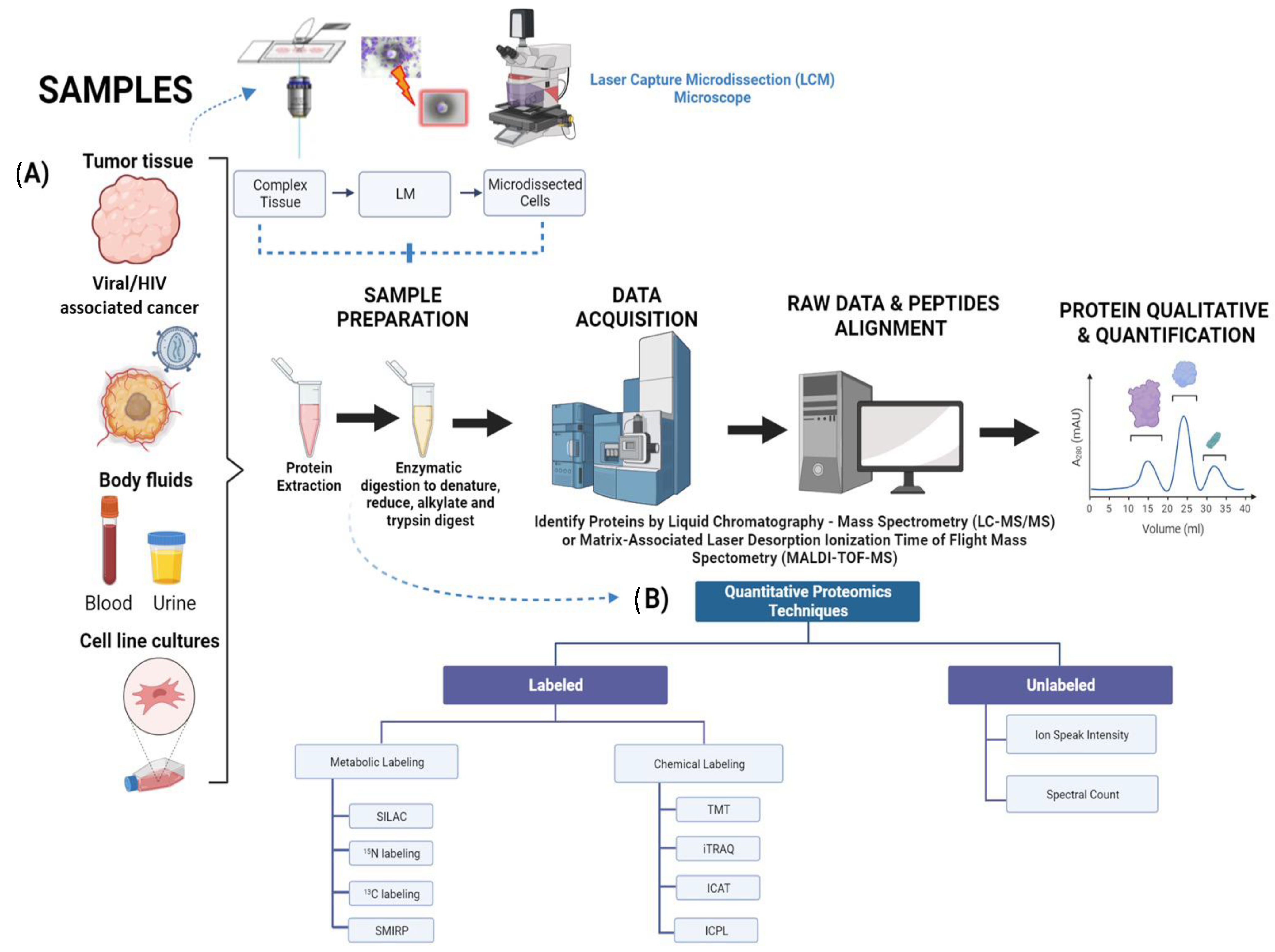
2. Biomarker Discovery in Cancer Proteomics
| Screening/Labeling Tools | Advantages | Drawbacks | Expenditures |
|---|---|---|---|
| MALDI-TOF-MS [40] | Easy to setup and analyzed | Poor sensitivity towards some bacteria specie such as Shigella and E-coli | Low cost |
| Relatively high sensitivity for detection of closely related microbial species | Not suitable for detecting small numbers of bacteria in pathogenic and sterile research | ||
| TMT [41,42] | Allows for identification of different samples with greater ease Increased sensitivity to phosphopeptides in reverse phase liquid chromatography Enable multiplexing of up to 18 samples simultaneously | Fragmentation of peptides may result in chimeric M/S spectra, which can lead to incorrect protein/peptide fold changes | High cost |
| iTRAQ [31,43] | Versatile tool that can be applied to a wide range of samples and organisms such as in vitro and in vivo. | High variations when implementing at the peptide level | Varies on sample numbers (High or low cost) |
| Fast method | Quantification tends to be rather poor due to small amounts of mass spectra involved (usually one or a few more) | ||
| Enables multiplexing of up to 8 samples simultaneously | Requires a mass spectrometer that can analyze the low regions of m/z | ||
| ICAT [31,44] | Can be applied to any sample type Fast method | Mild variations when implementing at the peptide level Only specific to Cys residues in peptides | High/Low cost |
| ICPL [25,31] | Can be applied to any sample type Fast method | Mild variations when implementing at the peptide level Only specific to Lys residues in peptides and protein N terminus | High/low cost |
| SILAC [31,33] | Can be implemented to an organism with relative ease and low variations High sensitivity and precision High preservation of proteins | Fails to be incorporated to samples relating to humans Slow method | High cost |
| 15N [31,34] | Low variation when implementing in organism Does not discriminate between peptides. Therefore, it can be incorporated to any sample | Fails to be incorporated to samples relating to humans Slow method Unable to identify molecular weight before each peptide be subjected to identification | High cost |
| 13C [31,34] | Low variation when implementing in organism Does not discriminate between peptides. Therefore, it can be incorporated to any sample | Fails to be incorporated to samples relating to humans Slow method Isotopes can inhibit identification and quantification | High cost |
| SMIRP [31,35] | Low variation when implementing in organism Can be incorporated to a wide variety of in-vivo organisms Does not discriminate between peptides. Therefore, it can be incorporated to any sample | Fails to be incorporated to samples relating to humans Slow method | High/low cost |
3. Prospects of Diagnostics and Therapeutics of Proteomic Research in Predominant Cancers including HIV-Associated Malignancies
4. HIV/AIDS Malignancies in CD4+/CXCR4/CCR5-Infected Cells including GBM
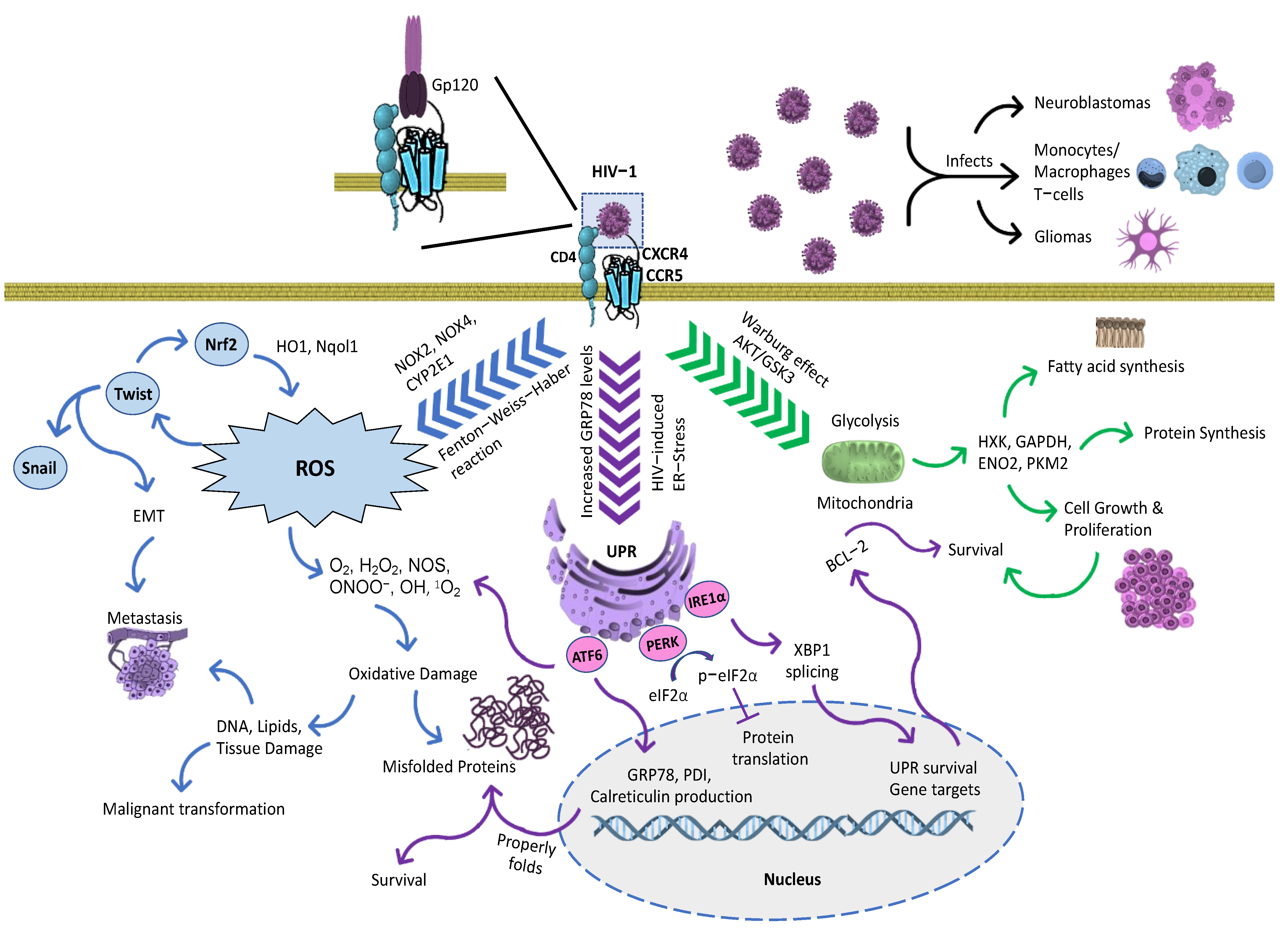
5. Signaling Pathways in Cancer: Mutual Crosstalk between ER Stress, Survival, Proliferation, Cell Cycle, and Migration


6. The Role of Proteomics in Personalized Medicine
7. The Critical Role of Bioinformatics
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Al-Amrani, S.; Al-Jabri, Z.; Al-Zaabi, A.; Alshekaili, J.; Al-Khabori, M. Proteomics: Concepts and Applications in Human Medicine. World J. Biol. Chem. 2021, 12, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Al Shweiki, M.R.; Oeckl, P.; Steinacker, P.; Barschke, P.; Dorner-Ciossek, C.; Hengerer, B.; Schönfeldt-Lecuona, C.; Otto, M. Proteomic Analysis Reveals a Biosignature of Decreased Synaptic Protein in Cerebrospinal Fluid of Major Depressive Disorder. Transl. Psychiatry 2020, 10, 144. [Google Scholar] [CrossRef]
- Hermann, J.; Schurgers, L.; Jankowski, V. Identification and Characterization of Post-Translational Modifications: Clinical Implications. Mol. Asp. Med. 2022, 86, 101066. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.W.; Jo, H.-S.; Bae, S.; Seo, Y.; Song, P.; Song, M.; Yoon, J.H. Application of Proteomics in Cancer: Recent Trends and Approaches for Biomarkers Discovery. Front. Med. 2021, 8, 1644. [Google Scholar] [CrossRef]
- Yang, X.-L.; Shi, Y.; Zhang, D.-D.; Xin, R.; Deng, J.; Wu, T.-M.; Wang, H.-M.; Wang, P.-Y.; Liu, J.-B.; Li, W.; et al. Quantitative Proteomics Characterization of Cancer Biomarkers and Treatment. Mol. Ther. Oncolytics 2021, 21, 255–263. [Google Scholar] [CrossRef]
- Cozzolino, F.; Landolfi, A.; Iacobucci, I.; Monaco, V.; Caterino, M.; Celentano, S.; Zuccato, C.; Cattaneo, E.; Monti, M. New Label-Free Methods for Protein Relative Quantification Applied to the Investigation of an Animal Model of Huntington Disease. PLoS ONE 2020, 15, e0238037. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.M.; Wilding, K.M.; Calve, S.; Bundy, B.C.; Kinzer-Ursem, T.L. Non-Canonical Amino Acid Labeling in Proteomics and Biotechnology. J. Biol. Eng. 2019, 13, 43. [Google Scholar] [CrossRef]
- Čuklina, J.; Lee, C.H.; Williams, E.G.; Sajic, T.; Collins, B.C.; Rodríguez Martínez, M.; Sharma, V.S.; Wendt, F.; Goetze, S.; Keele, G.R.; et al. Diagnostics and Correction of Batch Effects in Large-Scale Proteomic Studies: A Tutorial. Mol. Syst. Biol. 2021, 17, e10240. [Google Scholar] [CrossRef]
- Miles, H.N.; Delafield, D.G.; Li, L. Recent Developments and Applications of Quantitative Proteomics Strategies for High-Throughput Biomolecular Analyses in Cancer Research. RSC Chem. Biol. 2021, 2, 1050–1072. [Google Scholar] [CrossRef]
- Koziol, J.; Griffin, N.; Long, F.; Li, Y.; Latterich, M.; Schnitzer, J. On Protein Abundance Distributions in Complex Mixtures. Proteome Sci. 2013, 11, 5. [Google Scholar] [CrossRef]
- Zhang, W.; Sakashita, S.; Taylor, P.; Tsao, M.S.; Moran, M.F. Comprehensive Proteome Analysis of Fresh Frozen and Optimal Cutting Temperature (OCT) Embedded Primary Non-Small Cell Lung Carcinoma by LC–MS/MS. Methods 2015, 81, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Dapic, I.; Uwugiaren, N.; Jansen, P.; Corthals, G.L. Fast and Simple Protocols for Mass Spectrometry-Based Proteomics of Small Fresh Frozen Uterine Tissue Sections. Anal. Chem. 2017, 89, 10769–10775. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Huffman, K.E.; Fujimoto, J.; Canales, J.R.; Girard, L.; Nie, G.; Heymach, J.V.; Wistuba, I.I.; Minna, J.D.; Yu, Y. Quantitative Proteomic Analysis of Optimal Cutting Temperature (OCT) Embedded Core-Needle Biopsy of Lung Cancer. J. Am. Soc. Spectrom. 2017, 28, 2078–2089. [Google Scholar] [CrossRef] [PubMed]
- Sprung, R.W.; Brock, J.W.C.; Tanksley, J.P.; Li, M.; Washington, M.K.; Slebos, R.J.C.; Liebler, D.C. Equivalence of Protein Inventories Obtained from Formalin-Fixed Paraffin-Embedded and Frozen Tissue in Multidimensional Liquid Chromatography-Tandem Mass Spectrometry Shotgun Proteomic Analysis. Mol. Cell. Proteom. 2009, 8, 1988–1998. [Google Scholar] [CrossRef]
- Macklin, A.; Khan, S.; Kislinger, T. Recent Advances in Mass Spectrometry Based Clinical Proteomics: Applications to Cancer Research. Clin. Proteom. 2020, 17, 17. [Google Scholar] [CrossRef]
- Liotta, L.A.; Pappalardo, P.A.; Carpino, A.; Haymond, A.; Howard, M.; Espina, V.; Wulfkuhle, J.; Petricoin, E. Laser Capture Proteomics: Spatial Tissue Molecular Profiling from the Bench to Personalized Medicine. Expert Rev. Proteom. 2021, 18, 845–861. [Google Scholar] [CrossRef]
- Alghanem, B.; Ali, R.; Nehdi, A.; Al Zahrani, H.; Altolayyan, A.; Shaibah, H.; Baz, O.; Alhallaj, A.; Moresco, J.J.; Diedrich, J.K.; et al. Proteomics Profiling of KAIMRC1 in Comparison to MDA-MB231 and MCF-7. Int. J. Mol. Sci. 2020, 21, 4328. [Google Scholar] [CrossRef]
- Curran, S. Laser Capture Microscopy. Mol. Pathol. 2000, 53, 64–68. [Google Scholar] [CrossRef]
- Dapic, I.; Uwugiaren, N.; Kers, J.; Mohammed, Y.; Goodlett, D.R.; Corthals, G. Evaluation of Fast and Sensitive Proteome Profiling of FF and FFPE Kidney Patient Tissues. Molecules 2022, 27, 1137. [Google Scholar] [CrossRef]
- Wimmer, I.; Tröscher, A.R.; Brunner, F.; Rubino, S.J.; Bien, C.G.; Weiner, H.L.; Lassmann, H.; Bauer, J. Systematic Evaluation of RNA Quality, Microarray Data Reliability and Pathway Analysis in Fresh, Fresh Frozen and Formalin-Fixed Paraffin-Embedded Tissue Samples. Sci. Rep. 2018, 8, 6351. [Google Scholar] [CrossRef]
- Pais, R.J.; Jardine, C.; Zmuidinaite, R.; Lacey, J.; Butler, S.; Iles, R. Rapid, Affordable and Efficient Screening of Multiple Blood Abnormalities Made Possible Using an Automated Tool for MALDI-ToF Spectrometry Analysis. Appl. Sci. 2019, 9, 4999. [Google Scholar] [CrossRef]
- Sun, H.; Poudel, S.; Vanderwall, D.; Lee, D.-G.; Li, Y.; Peng, J. 29-Plex Tandem Mass Tag Mass Spectrometry Enabling Accurate Quantification by Interference Correction. Proteomics 2022, 22, 2100243. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Li, R.; Di, X.; Jin, X.; Wang, Y.; Lai, B.; Shi, C.; Mingxin, J.; Zhu, X.-R.; Wang, K. ITRAQ-Based Proteomic Analysis Reveals Possible Target-Related Proteins in Human Adrenocortical Adenomas. BMC Genom. 2019, 20, 655. [Google Scholar] [CrossRef]
- Isotope Coded Affinity Tags (ICATTM) (Institute for Systems Biology) | Innovative Molecular Analysis Technologies (IMAT). imat.cancer.gov. Available online: https://imat.cancer.gov/about-imat/outputs-and-achievements/individual-technologies-and-platforms/isotope-coded-affinity (accessed on 28 June 2023).
- Lottspeich, F.; Kellermann, J. ICPL Labeling Strategies for Proteome Research. Methods Mol. Biol. 2011, 753, 55–64. [Google Scholar] [CrossRef]
- Thompson, A.; Schäfer, J.; Kuhn, K.; Kienle, S.; Schwarz, J.; Schmidt, G.; Neumann, T.; Hamon, C. Tandem Mass Tags: A Novel Quantification Strategy for Comparative Analysis of Complex Protein Mixtures by MS/MS. Anal. Chem. 2003, 75, 1895–1904. [Google Scholar] [CrossRef] [PubMed]
- Bathla, S.; Sindhu, A.; Kumar, S.; Dubey, S.K.; Pattnaik, S.; Rawat, P.; Chopra, A.; Dang, A.; Kaushik, J.K.; Mohanty, A.K. Tandem Mass Tag (TMT)-Based Quantitative Proteomics Reveals Potential Targets Associated with Onset of Sub-Clinical Mastitis in Cows. Sci. Rep. 2020, 10, 9321. [Google Scholar] [CrossRef]
- Kelstrup, C.D.; Aizikov, K.; Batth, T.S.; Kreutzman, A.; Grinfeld, D.; Lange, O.; Mourad, D.; Makarov, A.A.; Olsen, J.V. Limits for Resolving Isobaric Tandem Mass Tag Reporter Ions Using Phase-Constrained Spectrum Deconvolution. J. Proteome Res. 2018, 17, 4008–4016. [Google Scholar] [CrossRef]
- Pottiez, G.; Wiederin, J.; Fox, H.S.; Ciborowski, P. Comparison of 4-Plex to 8-Plex ITRAQ Quantitative Measurements of Proteins in Human Plasma Samples. J. Proteome Res. 2012, 11, 3774–3781. [Google Scholar] [CrossRef]
- Grabowska, K.; Harwood, E.; Ciborowski, P. HIV and Proteomics: What We Have Learned from High Throughput Studies. PROTEOMICS Clin. Appl. 2021, 15, 2000040. [Google Scholar] [CrossRef]
- Gouw, J.W.; Krijgsveld, J.; Heck, A.J.R. Quantitative Proteomics by Metabolic Labeling of Model Organisms. Mol. Cell. Proteom. 2010, 9, 11–24. [Google Scholar] [CrossRef]
- Koomen, J.M.; Haura, E.B.; Bepler, G.; Sutphen, R.; Remily-Wood, E.R.; Benson, K.; Hussein, M.; Hazlehurst, L.A.; Yeatman, T.J.; Hildreth, L.T.; et al. Proteomic Contributions to Personalized Cancer Care. Mol. Cell. Proteom. 2008, 7, 1780–1794. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wei, S.; Ji, Y.; Guo, X.; Yang, F. Quantitative Proteomics Using SILAC: Principles, Applications, and Developments. Proteomics 2015, 15, 3175–3192. [Google Scholar] [CrossRef] [PubMed]
- Blair; Chen, S.-W.; Drechsler, R.; Gafken, P.R.; Olsen, C.P. 13C- and 15N-Labeling Strategies Combined with Mass Spectrometry Comprehensively Quantify Phospholipid Dynamics in C. Elegans. PLoS ONE 2015, 10, e0141850. [Google Scholar] [CrossRef]
- Whitelegge, J.P.; Katz, J.E.; Pihakari, K.A.; Hale, R.; Aguilera, R.; Gómez, S.M.; Faull, K.F.; Vavilin, D.; Vermaas, W. Subtle Modification of Isotope Ratio Proteomics; an Integrated Strategy for Expression Proteomics. Phytochemistry 2004, 65, 1507–1515. [Google Scholar] [CrossRef] [PubMed]
- Lau, H.-T.; Suh, H.W.; Golkowski, M.; Ong, S.-E. Comparing SILAC- and Stable Isotope Dimethyl-Labeling Approaches for Quantitative Proteomics. J. Proteome Res. 2014, 13, 4164–4174. [Google Scholar] [CrossRef]
- Luo, H.; Ge, H. Application of Proteomics in the Discovery of Radiosensitive Cancer Biomarkers. Front. Oncol. 2022, 12, 628. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate Proteome-Wide Label-Free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- He, B.; Shi, J.; Wang, X.; Jiang, H.; Zhu, H.-J. Label-Free Absolute Protein Quantification with Data-Independent Acquisition. J. Proteom. 2019, 200, 51–59. [Google Scholar] [CrossRef]
- Rychert, J. Benefits and Limitations of MALDI-TOF Mass Spectrometry for the Identification of Microorganisms. Available online: https://www.infectiologyjournal.com/articles/benefits-and-limitations-of-malditof-mass-spectrometry-for-the-identification-of-microorganisms.html (accessed on 28 June 2023).
- Thermo Fisher Scientific. Tandem Mass Tag (TMT) Multiplexing Approach to Protein Quantitation: Q&A. Available online: https://www.analyteguru.com/t5/Blog/Tandem-Mass-Tag-TMT-Multiplexing-Approach-to-Protein/ba-p/21253 (accessed on 5 June 2023).
- Sturm, R.M.; Lietz, C.B.; Li, L. Improved Isobaric Tandem Mass Tag Quantification by Ion Mobility Mass Spectrometry. Rapid Commun. Mass Spectrom. 2014, 28, 1051–1060. [Google Scholar] [CrossRef]
- Trinh, H.V.; Grossmann, J.; Gehrig, P.; Roschitzki, B.; Schlapbach, R.; Greber, U.F.; Hemmi, S. ITRAQ-Based and Label-Free Proteomics Approaches for Studies of Human Adenovirus Infections. Int. J. Proteom. 2013, 2013, 581862. [Google Scholar] [CrossRef]
- Zhao, Y.; Lu, Q.-Y.; Xiao, G.G. Quantitative Proteomics and Biomarker Discovery in Human Cancer. Expert Rev. Proteomics 2009, 6, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Melby, J.A.; Roberts, D.S.; Larson, E.J.; Brown, K.A.; Bayne, E.F.; Jin, S.; Ge, Y. Novel Strategies to Address the Challenges in Top-down Proteomics. J. Am. Soc. Mass Spectrom. 2021, 32, 1278–1294. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fonslow, B.R.; Shan, B.; Baek, M.-C.; Yates, J.R. Protein Analysis by Shotgun/Bottom-up Proteomics. Chem. Rev. 2013, 113, 2343–2394. [Google Scholar] [CrossRef] [PubMed]
- Abolfathi, H.; Sheikhpour, M.; Shahraeini, S.S.; Khatami, S.; Nojoumi, S.A. Studies in Lung Cancer Cytokine Proteomics: A Review. Expert Rev. Proteom. 2021, 18, 49–64. [Google Scholar] [CrossRef]
- Alharbi, R.A. Proteomics Approach and Techniques in Identification of Reliable Biomarkers for Diseases. Saudi J. Biol. Sci. 2020, 27, 968–974. [Google Scholar] [CrossRef]
- Boys, E.L.; Liu, J.; Robinson, P.J.; Reddel, R.R. Clinical Applications of Mass Spectrometry-Based Proteomics in Cancer: Where Are We? Proteomics 2022, 23, 2200238. [Google Scholar] [CrossRef] [PubMed]
- Triple Negative Breast Cancer (Page 6): Pennmedicine.org. Available online: https://www.pennmedicine.org/cancer/types-of-cancer/breast-cancer/types-of-breast-cancer/triplenegative-breast-cancer#:~:text=Triple%2Dnegative%20breast%20cancers%20tend (accessed on 28 June 2023).
- Asleh, K.; Negri, G.L.; Spencer Miko, S.E.; Colborne, S.; Hughes, C.S.; Wang, X.Q.; Gao, D.; Gilks, C.B.; Chia, S.K.L.; Nielsen, T.O.; et al. Proteomic Analysis of Archival Breast Cancer Clinical Specimens Identifies Biological Subtypes with Distinct Survival Outcomes. Nat. Commun. 2022, 13, 896. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive Molecular Portraits of Human Breast Tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Hoadley, K.A.; Yau, C.; Wolf, D.M.; Cherniack, A.D.; Tamborero, D.; Ng, S.; Leiserson, M.D.M.; Niu, B.; McLellan, M.D.; Uzunangelov, V.; et al. Multiplatform Analysis of 12 Cancer Types Reveals Molecular Classification within and across Tissues of Origin. Cell 2014, 158, 929–944. [Google Scholar] [CrossRef]
- Shah, S.P.; Roth, A.; Goya, R.; Oloumi, A.; Ha, G.; Zhao, Y.; Turashvili, G.; Ding, J.; Tse, K.; Haffari, G.; et al. The Clonal and Mutational Evolution Spectrum of Primary Triple-Negative Breast Cancers. Nature 2012, 486, 395–399. [Google Scholar] [CrossRef]
- Curtis, C.; Shah, S.P.; Chin, S.-F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The Genomic and Transcriptomic Architecture of 2,000 Breast Tumours Reveals Novel Subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Stephens, P.J.; Tarpey, P.S.; Davies, H.; Van Loo, P.; Greenman, C.; Wedge, D.C.; Nik-Zainal, S.; Martin, S.; Varela, I.; Bignell, G.R.; et al. The Landscape of Cancer Genes and Mutational Processes in Breast Cancer. Nature 2012, 486, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Krug, K.; Jaehnig, E.J.; Satpathy, S.; Blumenberg, L.; Karpova, A.; Anurag, M.; Miles, G.; Mertins, P.; Geffen, Y.; Tang, L.C.; et al. Proteogenomic Landscape of Breast Cancer Tumorigenesis and Targeted Therapy. Cell 2020, 183, 1436–1456.e31. [Google Scholar] [CrossRef]
- Hughes, C.S.; McConechy, M.K.; Cochrane, D.R.; Nazeran, T.; Karnezis, A.N.; Huntsman, D.G.; Morin, G.B. Quantitative Profiling of Single Formalin Fixed Tumour Sections: Proteomics for Translational Research. Sci. Rep. 2016, 6, 34949. [Google Scholar] [CrossRef]
- Moggridge, S.; Sorensen, P.H.; Morin, G.B.; Hughes, C.S. Extending the Compatibility of the SP3 Paramagnetic Bead Processing Approach for Proteomics. J. Proteome Res. 2018, 17, 1730–1740. [Google Scholar] [CrossRef] [PubMed]
- Anurag, M.; Zhu, M.; Huang, C.; Vasaikar, S.; Wang, J.; Hoog, J.; Burugu, S.; Gao, D.; Suman, V.; Zhang, X.H.; et al. Immune Checkpoint Profiles in Luminal B Breast Cancer (Alliance). JNCI J. Natl. Cancer Inst. 2020, 112, 737–746. [Google Scholar] [CrossRef]
- Henle, A.M.; Nassar, A.; Puglisi-Knutson, D.; Youssef, B.; Knutson, K.L. Downregulation of TAP1 and TAP2 in Early Stage Breast Cancer. PLoS ONE 2017, 12, e0187323. [Google Scholar] [CrossRef]
- Liu, C.; Li, Y.; Xing, X.; Zhuang, J.; Wang, J.; Wang, C.; Zhang, L.; Liu, L.; Feng, F.; Li, H.; et al. Immunogenomic Landscape Analyses of Immune Molecule Signature-Based Risk Panel for Patients with Triple-Negative Breast Cancer. Mol. Ther. Nucleic. Acids 2022, 28, 670–684. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Li, J.; Zhang, X.Y.; Ge, X.; Ma, J. Differences in Clinical Features and Prognosis between Orbit Adenoid Cystic Carcinoma and Adenocarcinoma: A Study from the SEER 18 Database. Tumori 2023, 109, 61–70. [Google Scholar] [CrossRef]
- Yao, Q.; Hou, W.; Chen, J.; Bai, Y.; Long, M.; Huang, X.; Zhao, C.; Zhou, L.; Niu, D. Comparative Proteomic and Clinicopathological Analysis of Breast Adenoid Cystic Carcinoma and Basal-like Triple-Negative Breast Cancer. Front. Med. 2022, 9, 2087. [Google Scholar] [CrossRef]
- Gaur, P.; Bhattacharya, S.; Kant, S.; Kushwaha, R.A.; Garg, R.; Singh, G.; Pandey, S. Association of Inflammatory Biomarkers with Lung Cancer in North Indian Population. Afr. Health Sci. 2019, 19, 2147. [Google Scholar] [CrossRef] [PubMed]
- Enewold, L.; Mechanic, L.E.; Bowman, E.D.; Zheng, Y.-L.; Yu, Z.; Trivers, G.; Alberg, A.J.; Harris, C.C. Serum Concentrations of Cytokines and Lung Cancer Survival in African Americans and Caucasians. Cancer Epidemiol. Biomark. Prev. 2009, 18, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.Y.; Zhang, C.; Wang, X.; Zhai, L.; Ma, Y.; Mao, Y.; Qian, K.; Sun, C.; Liu, Z.; Jiang, S.; et al. Integrative Proteomic Characterization of Human Lung Adenocarcinoma. Cell 2020, 182, 245–261.e17. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The biology and management of non-small cell lung cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef]
- Hughes, P.E.; Caenepeel, S.; Wu, L.C. Targeted Therapy and Checkpoint Immunotherapy Combinations for the Treatment of Cancer. Trends Immunol. 2016, 37, 462–476. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.J.; Shaw, A.T. Resisting Resistance: Targeted Therapies in Lung Cancer. Trends Cancer 2016, 2, 350–364. [Google Scholar] [CrossRef]
- Peters, S.; Kerr, K.M.; Stahel, R. PD-1 blockade in advanced NSCLC: A focus on pembrolizumab. Cancer Treat. Rev. 2018, 62, 39–49. [Google Scholar] [CrossRef]
- Thomas, A.; Liu, S.V.; Subramaniam, D.S.; Giaccone, G. Refining the treatment of NSCLC according to histological and molecular subtypes. Nat. Rev. Clin. Oncol. 2015, 12, 511–526. [Google Scholar] [CrossRef]
- Hirsch, F.R.; Varella-Garcia, M.; Bunn, P.A., Jr.; Di Maria, M.V.; Veve, R.; Bremmes, R.M.; Barón, A.E.; Zeng, C.; Franklin, W.A. Epidermal growth factor receptor in non-small-cell lung carcinomas: Correlation between gene copy number and protein expression and impact on prognosis. J. Clin. Oncol. 2003, 21, 3798–3807. [Google Scholar] [CrossRef]
- Vargas, A.J.; Harris, C.C. Biomarker development in the precision medicine era: Lung cancer as a case study. Nat. Rev. Cancer 2016, 16, 525–537. [Google Scholar] [CrossRef]
- Wang, S.; Chen, A.; Zhu, W.; Feng, D.; Wei, J.; Li, Q.; Shi, X.; Lv, X.; Liu, M. Characterization of Fatty Acid Metabolism in Lung Adenocarcinoma. Front. Genet. 2022, 13, 905508. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V.; Reddy, E.P.; Shokat, K.M.; Soucek, L. Drugging the ‘undruggable’ cancer targets. Nat. Rev. Cancer 2017, 17, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Papke, B.; Der, C.J. Drugging RAS: Know the enemy. Science 2017, 355, 1158–1163. [Google Scholar] [CrossRef] [PubMed]
- Key Statistics for Prostate Cancer | Prostate Cancer Facts. Available online: https://www.cancer.org/cancer/types/prostate-cancer/about/key-statistics.html (accessed on 28 June 2023).
- Screening Tests for Prostate Cancer. Available online: https://www.cancer.org/cancer/types/prostate-cancer/detection-diagnosis-staging/tests.html (accessed on 28 June 2023).
- Khoo, A.; Liu, L.Y.; Nyalwidhe, J.O.; Semmes, O.J.; Vesprini, D.; Downes, M.R.; Boutros, P.C.; Liu, S.K.; Kislinger, T. Proteomic discovery of non-invasive biomarkers of localized prostate cancer using mass spectrometry. Nat. Rev. Urol. 2021, 18, 707–724. [Google Scholar] [CrossRef] [PubMed]
- Tonry, C.; Finn, S.; Armstrong, J.; Pennington, S.R. Clinical proteomics for prostate cancer: Understanding prostate cancer pathology and protein biomarkers for improved disease management. Clin. Proteom. 2020, 17, 41. [Google Scholar] [CrossRef]
- Ummanni, R.; Duscharla, D.; Barett, C.; Venz, S.; Schlomm, T.; Heinzer, H.; Walther, R.; Bokemeyer, C.; Brümmendorf, T.H.; Murthy, P.V.; et al. Prostate cancer-associated autoantibodies in serum against tumor-associated antigens as potential new biomarkers. J. Proteom. 2015, 119, 218–229. [Google Scholar] [CrossRef]
- Leng, S.X.; McElhaney, J.E.; Walston, J.D.; Xie, D.; Fedarko, N.S.; Kuchel, G.A. ELISA and Multiplex Technologies for Cytokine Measurement in Inflammation and Aging Research. J. Gerontol. A Biol. Sci. Med. Sci. 2008, 63, 879–884. [Google Scholar] [CrossRef]
- Addona, T.A.; Shi, X.; Keshishian, H.; Mani, D.R.; Burgess, M.; Gillette, M.A.; Clauser, K.R.; Shen, D.; Lewis, G.D.; Farrell, L.A.; et al. A Pipeline That Integrates the Discovery and Verification of Plasma Protein Biomarkers Reveals Candidate Markers for Cardiovascular Disease. Nat. Biotechnol. 2011, 29, 635–643. [Google Scholar] [CrossRef]
- Reis, B.S.; Jungbluth, A.A.; Frosina, D.; Holz, M.; Ritter, E.; Nakayama, E.; Ishida, T.; Obata, Y.; Carver, B.; Scher, H.; et al. Prostate Cancer Progression Correlates with Increased Humoral Immune Response to a Human Endogenous Retrovirus GAG Protein. Clin. Cancer Res. 2013, 19, 6112–6125. [Google Scholar] [CrossRef]
- Kragstrup, T.W.; Vorup-Jensen, T.; Deleuran, B.; Hvid, M. A Simple Set of Validation Steps Identifies and Removes False Results in a Sandwich Enzyme-Linked Immunosorbent Assay Caused by Anti-Animal IgG Antibodies in Plasma from Arthritis Patients. Springerplus 2013, 2, 263. [Google Scholar] [CrossRef]
- Sun, R.; Hunter, C.; Chen, C.; Ge, W.; Morrice, N.; Liang, S.; Zhu, T.; Yuan, C.; Ruan, G.; Zhang, Q.; et al. Accelerated Protein Biomarker Discovery from FFPE Tissue Samples Using Single-Shot, Short Gradient Microflow SWATH MS. J. Proteome Res. 2020, 19, 2732–2741. [Google Scholar] [CrossRef] [PubMed]
- Randall, E.C.; Zadra, G.; Chetta, P.; Lopez, B.G.C.; Syamala, S.; Basu, S.S.; Agar, J.N.; Loda, M.; Tempany, C.M.; Fennessy, F.M.; et al. Molecular Characterization of Prostate Cancer with Associated Gleason Score Using Mass Spectrometry Imaging. Mol. Cancer Res. 2019, 17, 1155–1165. [Google Scholar] [CrossRef] [PubMed]
- Clendinen, C.S.; Gaul, D.A.; Monge, M.E.; Arnold, R.S.; Edison, A.S.; Petros, J.A.; Fernández, F.M. Preoperative Metabolic Signatures of Prostate Cancer Recurrence Following Radical Prostatectomy. J. Proteome Res. 2019, 18, 1316–1327. [Google Scholar] [CrossRef]
- Sadeesh, N.; Scaravilli, M.; Latonen, L. Proteomic Landscape of Prostate Cancer: The View Provided by Quantitative Proteomics, Integrative Analyses, and Protein Interactomes. Cancers 2021, 13, 4829. [Google Scholar] [CrossRef]
- Sinha, A.; Huang, V.; Livingstone, J.; Wang, J.; Fox, N.S.; Kurganovs, N.; Ignatchenko, V.; Fritsch, K.; Donmez, N.; Heisler, L.E.; et al. The Proteogenomic Landscape of Curable Prostate Cancer. Cancer Cell 2019, 35, 414–427.e6. [Google Scholar] [CrossRef]
- Latonen, L.; Afyounian, E.; Jylhä, A.; Nättinen, J.; Aapola, U.; Annala, M.; Kivinummi, K.K.; Tammela, T.T.L.; Beuerman, R.W.; Uusitalo, H.; et al. Integrative Proteomics in Prostate Cancer Uncovers Robustness against Genomic and Transcriptomic Aberrations during Disease Progression. Nat. Commun. 2018, 9, 1176. [Google Scholar] [CrossRef]
- Customized Drug to Kill Brain Cancer Cells. National Institutes of Health (NIH). Available online: https://www.nih.gov/news-events/nih-research-matters/customized-drug-kill-brain-cancer-cells#:~:text=A%20type%20of%20tumor%20called (accessed on 5 June 2023).
- Chen, L.; Qin, D.; Guo, X.; Wang, Q.; Li, J. Putting Proteomics into Immunotherapy for Glioblastoma. Front. Immunol. 2021, 12, 593255. [Google Scholar] [CrossRef] [PubMed]
- Lam, K.H.B.; Leon, A.J.; Hui, W.; Lee, S.C.-E.; Batruch, I.; Faust, K.; Klekner, A.; Hutóczki, G.; Koritzinsky, M.; Richer, M.; et al. Topographic Mapping of the Glioblastoma Proteome Reveals a Triple-Axis Model of Intra-Tumoral Heterogeneity. Nat. Commun. 2022, 13, 116. [Google Scholar] [CrossRef]
- Dai, X.; Shen, L. Advances and Trends in Omics Technology Development. Front. Med. 2022, 9, 911861. [Google Scholar] [CrossRef]
- Jaoude, D.A.; Moore, J.A.; Moore, M.B.; Twumasi-Ankrah, P.; Ablah, E.; Moore, D.F. Glioblastoma and Increased Survival with Longer Chemotherapy Duration. Kans. J. Med. 2019, 12, 65–69. [Google Scholar] [CrossRef]
- Yi, H.-G.; Jeong, Y.H.; Kim, Y.; Choi, Y.-J.; Moon, H.E.; Park, S.H.; Kang, K.S.; Bae, M.; Jang, J.; Youn, H.; et al. A Bioprinted Human-Glioblastoma-On-a-Chip for the Identification of Patient-Specific Responses to Chemoradiotherapy. Nat. Biomed. Eng. 2019, 3, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Truong, D.; Fiorelli, R.; Barrientos, E.S.; Melendez, E.L.; Sanai, N.; Mehta, S.; Nikkhah, M. A Three-Dimensional (3D) Organotypic Microfluidic Model for Glioma Stem Cells—Vascular Interactions. Biomaterials 2019, 198, 63–77. [Google Scholar] [CrossRef]
- El Hage, S.; Wakim, E.; Daou, L.; El Masri, J.; Salameh, P. Epidemiology and Incidence of Retinoblastoma in the Middle East: A Nationwide Study in Lebanon. Cureus 2021, 13, e18696. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Zhang, W.; Wang, Y.; Huang, D.; Shi, J.; Li, B.; Zhang, Y.; Zhou, Y. Characterization, Treatment and Prognosis of Retinoblastoma with Central Nervous System Metastasis. BMC Ophthalmol. 2018, 18, 207. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Yang, J.; Xiao, W.; Jiang, Z.; Chen, S.; Guo, D.; Zhang, P.; Liu, C.; Yang, H.; Xie, Z. Single-Cell Characterization of Malignant Phenotypes and Microenvironment Alteration in Retinoblastoma. Cell Death Dis. 2022, 13, 458. [Google Scholar] [CrossRef] [PubMed]
- Mao, P.; Shen, Y.; Xu, X.; Zhong, J. Comprehensive Analysis of the Immune Cell Infiltration Landscape and Immune-Related Methylation in Retinoblastoma. Front. Genet. 2022, 13, 864473. [Google Scholar] [CrossRef]
- Galardi, A.; Colletti, M.; Lavarello, C.; Di Paolo, V.; Mascio, P.; Russo, I.; Cozza, R.; Romanzo, A.; Valente, P.; De Vito, R.; et al. Proteomic Profiling of Retinoblastoma-Derived Exosomes Reveals Potential Biomarkers of Vitreous Seeding. Cancers 2020, 12, 1555. [Google Scholar] [CrossRef]
- Munier, F.L. Classification and management of seeds in retinoblastoma. Ellsworth Lecture Ghent August 24th 2013. Ophthalmic Genet. 2014, 35, 193–207. [Google Scholar] [CrossRef]
- Munier, F.L.; Gaillard, M.-C.; Balmer, A.; Soliman, S.; Podilsky, G.; Moulin, A.P.; Beck-Popovic, M. Intravitreal Chemotherapy for Vitreous Disease in Retinoblastoma Revisited: From Prohibition to Conditional Indications. Br. J. Ophthalmol. 2012, 96, 1078–1083. [Google Scholar] [CrossRef]
- “AIDS Related Malignancies”. Johns Hopkins Medicine. Available online: www.hopkinsmedicine.org/health/conditions-and-diseases/hiv-and-aids/aidsrelated-malignancies (accessed on 23 January 2023).
- Isaguliants, M.; Bayurova, E.; Avdoshina, D.; Kondrashova, A.; Chiodi, F.; Palefsky, J.M. Oncogenic Effects of HIV-1 Proteins, Mechanisms Behind. Cancers 2021, 13, 305. [Google Scholar] [CrossRef]
- Gao, Q.; Zhu, H.; Dong, L.; Shi, W.; Chen, R.; Song, Z.; Huang, C.; Li, J.; Dong, X.; Zhou, Y.; et al. Integrated Proteogenomic Characterization of HBV-Related Hepatocellular Carcinoma. Cell 2019, 179, 561–577.e22. [Google Scholar] [CrossRef]
- Mertins, P.; Mani, D.R.; Ruggles, K.V.; Gillette, M.A.; Clauser, K.R.; Wang, P.; Wang, X.; Qiao, J.W.; Cao, S.; Petralia, F.; et al. Proteogenomics Connects Somatic Mutations to Signalling in Breast Cancer. Nature 2016, 534, 55–62. [Google Scholar] [CrossRef]
- Tang, L.; Zeng, J.; Geng, P.; Fang, C.; Wang, Y.; Sun, M.; Wang, C.; Wang, J.; Yin, P.; Hu, C.; et al. Global Metabolic Profiling Identifies a Pivotal Role of Proline and Hydroxyproline Metabolism in Supporting Hypoxic Response in Hepatocellular Carcinoma. Clin. Cancer Res. 2018, 24, 474–485. [Google Scholar] [CrossRef] [PubMed]
- Eckert, M.A.; Coscia, F.; Chryplewicz, A.; Chang, J.W.; Hernandez, K.M.; Pan, S.; Tienda, S.M.; Nahotko, D.A.; Li, G.; Blaženović, I.; et al. Proteomics Reveals NNMT as a Master Metabolic Regulator of Cancer-Associated Fibroblasts. Nature 2019, 569, 723–728. [Google Scholar] [CrossRef] [PubMed]
- Bienvenu, E.; Mukanyangezi, M.F.; Rulisa, S.; Martner, A.; Hasséus, B.; Vorontsov, E.; Tobin, G.; Giglio, D. Changes in the Proteome in the Development of Chronic Human Papillomavirus Infection—A Prospective Study in HIV Positive and HIV Negative Rwandan Women. Cancers 2021, 13, 5983. [Google Scholar] [CrossRef] [PubMed]
- Pai, S.-Y.; Lurain, K.; Yarchoan, R. How Immunodeficiency Can Lead to Malignancy. Hematol. Am. Soc. Hematol. Educ. Program 2021, 2021, 287–295. [Google Scholar] [CrossRef]
- Kadiu, I.; Ricardo-Dukelow, M.; Ciborowski, P.; Gendelman, H.E. Cytoskeletal Protein Transformation in HIV-1-Infected Macrophage Giant Cells. J. Immunol. 2007, 178, 6404–6415. [Google Scholar] [CrossRef]
- Haverland, N.A.; Fox, H.S.; Ciborowski, P. Quantitative Proteomics by SWATH-MS Reveals Altered Expression of Nucleic Acid Binding and Regulatory Proteins in HIV-1-Infected Macrophages. J. Proteome Res. 2014, 13, 2109–2119. [Google Scholar] [CrossRef]
- Valentín-Guillama, G.; López, S.; Kucheryavykh, Y.; Chorna, N.; Pérez, J.; Ortiz-Rivera, J.; Inyushin, M.; Makarov, V.; Valentín-Acevedo, A.; Quinones-Hinojosa, A.; et al. HIV-1 Envelope Protein Gp120 Promotes Proliferation and the Activation of Glycolysis in Glioma Cell. Cancers 2018, 10, 301. [Google Scholar] [CrossRef]
- López, S.N.; Rodríguez-Valentín, M.; Rivera, M.; Rodríguez, M.; Babu, M.; Cubano, L.A.; Xiong, H.; Wang, G.; Kucheryavykh, L.; Boukli, N.M. HIV-1 Gp120 Clade B/c Induces a GRP78 Driven Cytoprotective Mechanism in Astrocytoma. Oncotarget 2017, 8, 68415–68438. [Google Scholar] [CrossRef]
- Yarchoan, R.; Uldrick, T.S. HIV-Associated Cancers and Related Diseases. N. Engl. J. Med. 2018, 378, 1029–1041. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute. HIV Infection and Cancer Risk. Available online: https://www.cancer.gov/about-cancer/causes-prevention/risk/infectious-agents/hiv-fact-sheet (accessed on 28 June 2023).
- Hernández-Ramírez, R.U.; Shiels, M.S.; Dubrow, R.; Engels, E.A. Cancer Risk in HIV-Infected People in the USA from 1996 to 2012: A Population-Based, Registry-Linkage Study. Lancet HIV 2017, 4, e495–e504. [Google Scholar] [CrossRef] [PubMed]
- Choy, W.; Lagman, C.; Lee, S.J.; Bui, T.T.; Safaee, M.; Yang, I. Impact of Human Immunodeficiency Virus in the Pathogenesis and Outcome of Patients with Glioblastoma Multiforme. Brain Tumor Res. Treat. 2016, 4, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Jokonya, L.; Musara, A.; Esene, I.N.; Kabulo, K.D.M.; Kabeya, C.M.; Kalangu, K.K.N. Prevalence of Human Immunodeficiency Virus Infection in Brain Glioma Patients: Is the Virus Protective from Gliomas? Surg. Neurol. Int. 2018, 9, 103. [Google Scholar] [CrossRef]
- Oliveira, V.C.M.D.; Gomes, T.; Ferreira, L.C.L.; Damian, M.M.; Silva, V.M.F.Q.; Araújo, J.R.; Safe, I.P.; Ramasawmy, R. Glioblastoma Multiforme in an HIV-Infected Patient: An Unexpected Diagnosis. J. Int. Assoc. Provid. AIDS Care (JIAPAC) 2014, 13, 411–413. [Google Scholar] [CrossRef]
- Grande, F.; Occhiuzzi, M.; Rizzuti, B.; Ioele, G.; De Luca, M.; Tucci, P.; Svicher, V.; Aquaro, S.; Garofalo, A. CCR5/CXCR4 Dual Antagonism for the Improvement of HIV Infection Therapy. Molecules 2019, 24, 550. [Google Scholar] [CrossRef]
- Alkhatib, G. The Biology of CCR5 and CXCR4. Curr. Opin. HIV AIDS 2009, 4, 96–103. [Google Scholar] [CrossRef]
- Ullah, T.R. The Role of CXCR4 in Multiple Myeloma: Cells’ Journey from Bone Marrow to Beyond. J. Bone Oncol. 2019, 17, 100253. [Google Scholar] [CrossRef]
- Anitha, A.K.; Narayanan, P.; Ajayakumar, N.; Sivakumar, K.C.; Kumar, K.S. Novel Small Synthetic HIV-1 v3 Crown Variants: CCR5 Targeting Ligands. J. Biochem. 2022, 172, 149–164. [Google Scholar] [CrossRef]
- Burger, M.; Glodek, A.; Hartmann, T.; Schmitt-Gräff, A.; Silberstein, L.E.; Fujii, N.; Kipps, T.J.; Burger, J.A. Functional Expression of CXCR4 (CD184) on Small-Cell Lung Cancer Cells Mediates Migration, Integrin Activation, and Adhesion to Stromal Cells. Oncogene 2003, 22, 8093–8101. [Google Scholar] [CrossRef]
- Kijima, T.; Maulik, G.; Ma, P.C.; Tibaldi, E.V.; Turner, R.E.; Rollins, B.; Sattler, M.; Johnson, B.E.; Salgia, R. Regulation of Cellular Proliferation, Cytoskeletal Function, and Signal Transduction through CXCR4 and C-Kit in Small Cell Lung Cancer Cells. Cancer Res. 2002, 62, 6304–6311. [Google Scholar] [PubMed]
- Choi, W.-T.; Yang, Y.; Xu, Y.; An, J. Targeting Chemokine Receptor CXCR4 for Treatment of HIV-1 Infection, Tumor Progression, and Metastasis. Curr. Top. Med. Chem. 2014, 14, 1574–1589. [Google Scholar] [CrossRef] [PubMed]
- Li, R.-J.; Zhao, L.-J.; Zhan, Z.-L.; Lü, X.; Gong, L.-L.; Wang, P. Significance of Expression of Chemokine Receptor and Matrix Metalloproteinase in Small Cell Lung Cancer. Zhonghua Yi Xue Za Zhi 2012, 92, 532–535. [Google Scholar]
- Akashi, T.; Koizumi, K.; Tsuneyama, K.; Saiki, I.; Takano, Y.; Fuse, H. Chemokine Receptor CXCR4 Expression and Prognosis in Patients with Metastatic Prostate Cancer. Cancer Sci. 2008, 99, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Debnath, B.; Xu, S.; Grande, F.; Garofalo, A.; Neamati, N. Small Molecule Inhibitors of CXCR4. Theranostics 2013, 3, 47–75. [Google Scholar] [CrossRef]
- Walenkamp, A.M.E.; Lapa, C.; Herrmann, K.; Wester, H.-J. CXCR4 Ligands: The next Big Hit? J. Nucl. Med. 2017, 58, 77S–82S. [Google Scholar] [CrossRef] [PubMed]
- Sicoli, D.; Jiao, X.; Ju, X.; Velasco-Velazquez, M.; Ertel, A.; Addya, S.; Li, Z.; Ando, S.; Fatatis, A.; Paudyal, B.; et al. CCR5 Receptor Antagonists Block Metastasis to Bone of V-Src-Oncogene-Transformed Metastatic Prostate Cancer Cell Lines. Cancer Res. 2014, 74, 7103–7114. [Google Scholar] [CrossRef]
- Jiao, X.; Nawab, O.; Patel, T.; Kossenkov, A.V.; Halama, N.; Jaeger, D.; Pestell, R.G. Recent Advances Targeting CCR5 for Cancer and Its Role in Immuno-Oncology. Cancer Res. 2019, 79, 4801–4807. [Google Scholar] [CrossRef]
- Hsu, S.-K.; Chiu, C.-C.; Dahms, H.-U.; Chou, C.-K.; Cheng, C.-M.; Chang, W.-T.; Cheng, K.-C.; Wang, H.-M.D.; Lin, I.L. Unfolded Protein Response (UPR) in Survival, Dormancy, Immunosuppression, Metastasis, and Treatments of Cancer Cells. Int. J. Mol. Sci. 2019, 20, 2518. [Google Scholar] [CrossRef]
- Chonghaile, T.N.; Gupta, S.; John, M.; Szegezdi, E.; Logue, S.E.; Samali, A. BCL-2 Modulates the Unfolded Protein Response by Enhancing Splicing of X-Box Binding Protein-1. Biochem. Biophys. Res. Commun. 2015, 466, 40–45. [Google Scholar] [CrossRef]
- Hatok, J.; Racay, P. Bcl-2 Family Proteins: Master Regulators of Cell Survival. Biomol. Concepts 2016, 7, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Jin, Y.; Fan, Z. The Mechanism of Warburg Effect-Induced Chemoresistance in Cancer. Front. Oncol. 2021, 11, 698023. [Google Scholar] [CrossRef] [PubMed]
- Papadopoli, D.; Pollak, M.; Topisirovic, I. The Role of GSK3 in Metabolic Pathway Perturbations in Cancer. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 119059. [Google Scholar] [CrossRef]
- Shah, A.; Kumar, S.; Simon, S.D.; Singh, D.P.; Kumar, A. HIV Gp120- and Methamphetamine-Mediated Oxidative Stress Induces Astrocyte Apoptosis via Cytochrome P450 2E1. Cell Death Dis. 2013, 4, e850. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.V.; Valuev-Elliston, V.T.; Ivanova, O.N.; Kochetkov, S.N.; Starodubova, E.S.; Bartosch, B.; Isaguliants, M.G. Oxidative Stress during HIV Infection: Mechanisms and Consequences. Oxid. Med. Cell. Longev. 2016, 2016, 8910396. [Google Scholar] [CrossRef]
- Bayurova, E.; Jansons, J.; Skrastina, D.; Smirnova, O.; Mezale, D.; Kostyusheva, A.; Kostyushev, D.; Petkov, S.; Podschwadt, P.; Valuev-Elliston, V.; et al. HIV-1 Reverse Transcriptase Promotes Tumor Growth and Metastasis Formation via ROS-Dependent Upregulation of Twist. Oxid. Med. Cell. Longev. 2019, 2019, 6016278. [Google Scholar] [CrossRef] [PubMed]
- Miller, I.P.; Pavlović, I.; Poljšak, B.; Šuput, D.; Milisav, I. Beneficial Role of ROS in Cell Survival: Moderate Increases in H2O2 Production Induced by Hepatocyte Isolation Mediate Stress Adaptation and Enhanced Survival. Antioxidants 2019, 8, 434. [Google Scholar] [CrossRef]
- Yip, H.Y.K.; Papa, A. Signaling Pathways in Cancer: Therapeutic Targets, Combinatorial Treatments, and New Developments. Cells 2021, 10, 659. [Google Scholar] [CrossRef]
- Huang, J.; Pan, H.; Wang, J.; Wang, T.; Huo, X.; Ma, Y.; Lu, Z.; Sun, B.; Jiang, H. Unfolded Protein Response in Colorectal Cancer. Cell Biosci. 2021, 11, 26. [Google Scholar] [CrossRef]
- Khaled, J.; Kopsida, M.; Lennernäs, H.; Heindryckx, F. Drug Resistance and Endoplasmic Reticulum Stress in Hepatocellular Carcinoma. Cells 2022, 11, 632. [Google Scholar] [CrossRef]
- Guan, R.; Zhang, X.; Guo, M. Glioblastoma Stem Cells and Wnt Signaling Pathway: Molecular Mechanisms and Therapeutic Targets. Chin. Neurosurg. J. 2020, 6, 25. [Google Scholar] [CrossRef] [PubMed]
- Latour, M.; Her, N.-G.; Kesari, S.; Nurmemmedov, E. WNT Signaling as a Therapeutic Target for Glioblastoma. Int. J. Mol. Sci. 2021, 22, 8428. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, M.; Xu, F.; Jiang, S. Wnt Signaling in Breast Cancer: Biological Mechanisms, Challenges and Opportunities. Mol. Cancer 2020, 19, 165. [Google Scholar] [CrossRef] [PubMed]
- Abreu de Oliveira, W.A.; El Laithy, Y.; Bruna, A.; Annibali, D.; Lluis, F. Wnt Signaling in the Breast: From Development to Disease. Front. Cell Dev. Biol. 2022, 10, 884467. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Qin, L.; Li, X. Role of STAT3 Signaling Pathway in Breast Cancer. Cell Commun. Signal. 2020, 18, 33. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Nag, S.; Zhang, R. Targeting the NFκB Signaling Pathways for Breast Cancer Prevention and Therapy. Curr. Med. Chem. 2015, 22, 264–289. [Google Scholar] [CrossRef]
- BeLow, M.; Osipo, C. Notch Signaling in Breast Cancer: A Role in Drug Resistance. Cells 2020, 9, 2204. [Google Scholar] [CrossRef]
- Shorning, B.Y.; Dass, M.S.; Smalley, M.J.; Pearson, H.B. The PI3K-AKT-MTOR Pathway and Prostate Cancer: At the Crossroads of AR, MAPK, and WNT Signaling. Int. J. Mol. Sci. 2020, 21, 4507. [Google Scholar] [CrossRef]
- Wang, C.; Chen, Q.; Xu, H. Wnt/β-Catenin Signal Transduction Pathway in Prostate Cancer and Associated Drug Resistance. Discov. Oncol. 2021, 12, 1–11. [Google Scholar] [CrossRef]
- Brighi, N.; Conteduca, V.; Lolli, C.; Gurioli, G.; Schepisi, G.; Palleschi, M.; Mariotti, M.; Casadei, C.; De Giorgi, U. The Cyclin-Dependent Kinases Pathway as a Target for Prostate Cancer Treatment: Rationale and Future Perspectives. Crit. Rev. Oncol. Hematol. 2021, 157, 103199. [Google Scholar] [CrossRef]
- Siddiqui, F.; Vaqar, S.; Siddiqui, A.H. Lung Cancer; StatPearls Publishing: Treasure Island, FL, USA, 2022. Available online: https://www.ncbi.nlm.nih.gov/books/NBK482357/ (accessed on 28 June 2023).
- Sudhesh Dev, S.; Zainal Abidin, S.A.; Farghadani, R.; Othman, I.; Naidu, R. Receptor Tyrosine Kinases and Their Signaling Pathways as Therapeutic Targets of Curcumin in Cancer. Front. Pharmacol. 2021, 12, 772510. [Google Scholar] [CrossRef] [PubMed]
- Saigí, M.; Carcereny, E.; Morán, T.; Cucurull, M.; Domènech, M.; Hernandez, A.; Martinez-Cardús, A.; Pros, E.; Sanchez-Cespedes, M. Biological and Clinical Perspectives of the Actionable Gene Fusions and Amplifications Involving Tyrosine Kinase Receptors in Lung Cancer. Cancer Treat. Rev. 2022, 109, 102430. [Google Scholar] [CrossRef] [PubMed]
- Makino, Y.; Arakawa, Y.; Yoshioka, E.; Shofuda, T.; Minamiguchi, S.; Kawauchi, T.; Tanji, M.; Kanematsu, D.; Nonaka, M.; Okita, Y.; et al. Infrequent RAS Mutation Is Not Associated with Specific Histological Phenotype in Gliomas. BMC Cancer 2021, 21, 1025. [Google Scholar] [CrossRef] [PubMed]
- Kciuk, M.; Gielecińska, A.; Budzinska, A.; Mojzych, M.; Kontek, R. Metastasis and MAPK Pathways. Int. J. Mol. Sci. 2022, 23, 3847. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Nie, J.; Ma, X.; Wei, Y.; Peng, Y.; Wei, X. Targeting PI3K in Cancer: Mechanisms and Advances in Clinical Trials. Mol. Cancer 2019, 18, 26. [Google Scholar] [CrossRef]
- Colardo, M.; Segatto, M.; Di Bartolomeo, S. Targeting RTK-PI3K-MTOR Axis in Gliomas: An Update. Int. J. Mol. Sci. 2021, 22, 4899. [Google Scholar] [CrossRef]
- Read, A.; Schröder, M. The Unfolded Protein Response: An Overview. Biology 2021, 10, 384. [Google Scholar] [CrossRef]
- Bhattarai, K.R.; Chaudhary, M.; Kim, H.-R.; Chae, H.-J. Endoplasmic Reticulum (ER) Stress Response Failure in Diseases. Trends Cell Biol. 2020, 30, 672–675. [Google Scholar] [CrossRef]
- Choi, J.-A.; Song, C.-H. Insights into the Role of Endoplasmic Reticulum Stress in Infectious Diseases. Front. Immunol. 2020, 10, 3147. [Google Scholar] [CrossRef]
- Goda, J.; Pachpor, T.; Basu, T.; Chopra, S.; Gota, V. Targeting the AKT Pathway: Repositioning HIV Protease Inhibitors as Radiosensitizers. Indian J. Med. Res. 2016, 143, 145. [Google Scholar] [CrossRef]
- Gupta, A.K.; Cerniglia, G.J.; Mick, R.; McKenna, W.G.; Muschel, R.J. HIV Protease Inhibitors Block Akt Signaling and Radiosensitize Tumor Cells Both in Vitro and in Vivo. Cancer Res. 2005, 65, 8256–8265. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y. The Co-Receptor Signaling Model of HIV-1 Pathogenesis in Peripheral CD4 T Cells. Retrovirology 2009, 6, 41. [Google Scholar] [CrossRef] [PubMed]
- Borsa, M.; Ferreira, P.L.C.; Petry, A.; Ferreira, L.G.E.; Camargo, M.M.; Bou-Habib, D.C.; Pinto, A.R. HIV Infection and Antiretroviral Therapy Lead to Unfolded Protein Response Activation. Virol. J. 2015, 12, 77. [Google Scholar] [CrossRef]
- Liu, H.; Yang, J.; Li, L.; Shi, W.; Yuan, X.; Wu, L. The Natural Occurring Compounds Targeting Endoplasmic Reticulum Stress. Evid. Based Complement. Alternat. Med. 2016, 2016, 7831282. [Google Scholar] [CrossRef] [PubMed]
- Martucciello, S.; Masullo, M.; Cerulli, A.; Piacente, S. Natural Products Targeting ER Stress, and the Functional Link to Mitochondria. Int. J. Mol. Sci. 2020, 21, 1905. [Google Scholar] [CrossRef]
- Kim, C.; Kim, B. Anti-Cancer Natural Products and Their Bioactive Compounds Inducing ER Stress-Mediated Apoptosis: A Review. Nutrients 2018, 10, 1021. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Ou, S.; Huang, H. Green Tea Polyphenols Induce Cell Death in Breast Cancer MCF-7 Cells through Induction of Cell Cycle Arrest and Mitochondrial-Mediated Apoptosis. J. Zhejiang Univ. Sci. B 2017, 18, 89–98. [Google Scholar] [CrossRef]
- Fatima, I.; Kanwal, S.; Mahmood, T. Natural Products Mediated Targeting of Virally Infected Cancer. Dose Response 2019, 17, 155932581881322. [Google Scholar] [CrossRef]
- Rivera, M.; Ramos, Y.; Rodríguez-Valentín, M.; López-Acevedo, S.; Cubano, L.A.; Zou, J.; Zhang, Q.; Wang, G.; Boukli, N.M. Targeting Multiple Pro-Apoptotic Signaling Pathways with Curcumin in Prostate Cancer Cells. PLoS ONE 2017, 12, e0179587. [Google Scholar] [CrossRef]
- Burton, L.J.; Rivera, M.; Hawsawi, O.; Zou, J.; Hudson, T.; Wang, G.; Zhang, Q.; Cubano, L.; Boukli, N.; Odero-Marah, V. Muscadine Grape Skin Extract Induces an Unfolded Protein Response-Mediated Autophagy in Prostate Cancer Cells: A TMT-Based Quantitative Proteomic Analysis. PLoS ONE 2016, 11, e0164115. [Google Scholar] [CrossRef]
- Lin, C.-L.; Lee, C.-H.; Chen, C.-M.; Cheng, C.-W.; Chen, P.-N.; Ying, T.-H.; Hsieh, Y.-H. Protodioscin Induces Apoptosis through ROS-Mediated Endoplasmic Reticulum Stress via the JNK/P38 Activation Pathways in Human Cervical Cancer Cells. Cell. Physiol. Biochem. 2018, 46, 322–334. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.-L.; Lee, H.-L.; Yang, S.-F.; Wang, S.-W.; Lin, C.-P.; Hsieh, Y.-H.; Chiou, H.-L. Protodioscin Induces Mitochondrial Apoptosis of Human Hepatocellular Carcinoma Cells through Eliciting ER Stress-Mediated IP3R Targeting Mfn1/Bak Expression. J. Hepatocell. Carcinoma 2022, 9, 327–341. [Google Scholar] [CrossRef] [PubMed]
- Lewinska, A.; Adamczyk-Grochala, J.; Deregowska, A.; Wnuk, M. Sulforaphane-Induced Cell Cycle Arrest and Senescence Are Accompanied by DNA Hypomethylation and Changes in MicroRNA Profile in Breast Cancer Cells. Theranostics 2017, 7, 3461–3477. [Google Scholar] [CrossRef]
- Zou, X.; Qu, Z.; Fang, Y.; Shi, X.; Ji, Y. Endoplasmic Reticulum Stress Mediates Sulforaphane-Induced Apoptosis of HepG2 Human Hepatocellular Carcinoma Cells. Mol. Med. Rep. 2017, 15, 331–338. [Google Scholar] [CrossRef]
- Piotrowski, J.; Jędrzejewski, T.; Kozak, W. Immunomodulatory and Antitumor Properties of Polysaccharide Peptide (PSP). Postepy. Hig. Med. Dosw. 2015, 69, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Saleh, M.H.; Rashedi, I.; Keating, A. Immunomodulatory Properties of Coriolus Versicolor: The Role of Polysaccharopeptide. Front. Immunol. 2017, 8, 1087. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Valentín, M.; López, S.; Rivera, M.; Ríos-Olivares, E.; Cubano, L.; Boukli, N.M. Naturally Derived Anti-HIV Polysaccharide Peptide (PSP) Triggers a Toll-like Receptor 4-Dependent Antiviral Immune Response. J. Immunol. Res. 2018, 2018, 8741698. [Google Scholar] [CrossRef]
- Alvarez-Rivera, E.; Rodríguez-Valentín, M.; Boukli, N.M. The Antiviral Compound PSP Inhibits HIV-1 Entry via PKR-Dependent Activation in Monocytic Cells. Viruses 2023, 15, 804. [Google Scholar] [CrossRef]
- Tang, M.; Bolderson, E.; O’Byrne, K.J.; Richard, D.J. Tumor Hypoxia Drives Genomic Instability. Front. Cell Dev. Biol. 2021, 9, 626229. [Google Scholar] [CrossRef]
- Vadakekolathu, J.; Boocock, D.J.; Pandey, K.; Guinn, B.; Legrand, A.; Miles, A.K.; Coveney, C.; Ayala, R.; Purcell, A.W.; McArdle, S.E. Multi-Omic Analysis of Two Common P53 Mutations: Proteins Regulated by Mutated P53 as Potential Targets for Immunotherapy. Cancers 2022, 14, 3975. [Google Scholar] [CrossRef]
- Alvarado-Ortiz, E.; de la Cruz-López, K.G.; Becerril-Rico, J.; Sarabia-Sánchez, M.A.; Ortiz-Sánchez, E.; García-Carrancá, A. Mutant P53 Gain-of-Function: Role in Cancer Development, Progression, and Therapeutic Approaches. Front. Cell Dev. Biol. 2021, 8, 607670. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, J.; Xu, D.; Zhang, T.; Hu, W.; Feng, Z. Gain-of-Function Mutant P53 in Cancer Progression and Therapy. J. Mol. Cell Biol. 2020, 12, 674–687. [Google Scholar] [CrossRef] [PubMed]
- Doll, S.; Gnad, F.; Mann, M. The Case for Proteomics and Phospho-Proteomics in Personalized Cancer Medicine. Proteom. Clin. Appl. 2019, 13, 1800113. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, H.; Zenklusen, J.C.; Staudt, L.M.; Doroshow, J.H.; Lowy, D.R. The next Horizon in Precision Oncology—Proteogenomics to Inform Cancer Diagnosis and Treatment. Cell 2021, 184, 1661–1670. [Google Scholar] [CrossRef] [PubMed]
- Kumbale, C.M.; Voit, E.O. Toward Personalized Medicine for HIV/AIDS. J. AIDS HIV Treat. 2021, 3, 37–41. [Google Scholar] [CrossRef]
- Mu, Y.; Kodidela, S.; Wang, Y.; Kumar, S.; Cory, T.J. The Dawn of Precision Medicine in HIV: State of the Art of Pharmacotherapy. Expert Opin. Pharmacother. 2018, 19, 1581–1595. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, H.; Yu, Y.; Zhang, Y. Application of Nanomaterials in Proteomics-Driven Precision Medicine. Theranostics 2022, 12, 2674–2686. [Google Scholar] [CrossRef]
- Turriziani, B.; Garcia-Munoz, A.; Pilkington, R.; Raso, C.; Kolch, W.; von Kriegsheim, A. On-Beads Digestion in Conjunction with Data-Dependent Mass Spectrometry: A Shortcut to Quantitative and Dynamic Interaction Proteomics. Biology 2014, 3, 320–332. [Google Scholar] [CrossRef]
- Hughes, C.S.; Moggridge, S.; Müller, T.; Sorensen, P.H.; Morin, G.B.; Krijgsveld, J. Single-Pot, Solid-Phase-Enhanced Sample Preparation for Proteomics Experiments. Nat. Protoc. 2019, 14, 68–85. [Google Scholar] [CrossRef]
- Serrels, A.; Lund, T.; Serrels, B.; Byron, A.; McPherson, R.C.; von Kriegsheim, A.; Gómez-Cuadrado, L.; Canel, M.; Muir, M.; Ring, J.E.; et al. Nuclear FAK Controls Chemokine Transcription, Tregs, and Evasion of Anti-Tumor Immunity. Cell 2015, 163, 160–173. [Google Scholar] [CrossRef]
- Zhou, H.; Wang, F.; Wang, Y.; Ning, Z.; Hou, W.; Wright, T.G.; Sundaram, M.; Zhong, S.; Yao, Z.; Figeys, D. Improved Recovery and Identification of Membrane Proteins from Rat Hepatic Cells Using a Centrifugal Proteomic Reactor. Mol. Cell. Proteom. 2011, 10, O111.008425. [Google Scholar] [CrossRef] [PubMed]
- Müller, T.; Kalxdorf, M.; Longuespée, R.; Kazdal, D.N.; Stenzinger, A.; Krijgsveld, J. Automated Sample Preparation with SP 3 for Low-Input Clinical Proteomics. Mol. Syst. Biol. 2020, 16, e9111. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, C.; Schallenberg, S.; Kirchner, M.; Ziehm, M.; Niquet, S.; Haji, M.; Beier, C.; Neudecker, J.; Klauschen, F.; Mertins, P. Comprehensive Micro-Scaled Proteome and Phosphoproteome Characterization of Archived Retrospective Cancer Repositories. Nat. Commun 2021, 12, 3576. [Google Scholar] [CrossRef]
- Ruprecht, B.; Di Bernardo, J.; Wang, Z.; Mo, X.; Ursu, O.; Christopher, M.; Fernandez, R.B.; Zheng, L.; Dill, B.D.; Wang, H.; et al. A Mass Spectrometry-Based Proteome Map of Drug Action in Lung Cancer Cell Lines. Nat. Chem. Biol. 2020, 16, 1111–1119. [Google Scholar] [CrossRef] [PubMed]
- Atallah-Yunes, S.A.; Murphy, D.J.; Noy, A. HIV-Associated Burkitt Lymphoma. Lancet Haematol. 2020, 7, e594–e600. [Google Scholar] [CrossRef]
- Clifford, D.B.; Ances, B.M. HIV-Associated Neurocognitive Disorder. Lancet Infect. Dis. 2013, 13, 976–986. [Google Scholar] [CrossRef]
- Eggers, C.; Arendt, G.; Hahn, K.; Husstedt, I.W.; Maschke, M.; Neuen-Jacob, E.; Obermann, M.; Rosenkranz, T.; Schielke, E.; Straube, E. HIV-1-Associated Neurocognitive Disorder: Epidemiology, Pathogenesis, Diagnosis, and Treatment. J. Neurol. 2017, 264, 1715–1727. [Google Scholar] [CrossRef]
- Tsimberidou, A.M.; Fountzilas, E.; Nikanjam, M.; Kurzrock, R. Review of Precision Cancer Medicine: Evolution of the Treatment Paradigm. Cancer Treat. Rev. 2020, 86, 102019. [Google Scholar] [CrossRef]
- Sicklick, J.K.; Kato, S.; Okamura, R.; Schwaederle, M.; Hahn, M.E.; Williams, C.B.; De, P.; Krie, A.; Piccioni, D.E.; Miller, V.A.; et al. Molecular Profiling of Cancer Patients Enables Personalized Combination Therapy: The I-PREDICT Study. Nat. Med. 2019, 25, 744–750. [Google Scholar] [CrossRef]
- Dale, B.; Cheng, M.; Park, K.-S.; Kaniskan, H.Ü.; Xiong, Y.; Jin, J. Advancing Targeted Protein Degradation for Cancer Therapy. Nat. Rev. Cancer 2021, 21, 638–654. [Google Scholar] [CrossRef]
- Davis-Marcisak, E.F.; Deshpande, A.; Stein-O’Brien, G.L.; Ho, W.J.; Laheru, D.; Jaffee, E.M.; Fertig, E.J.; Kagohara, L.T. From Bench to Bedside: Single-Cell Analysis for Cancer Immunotherapy. Cancer Cell 2021, 39, 1062–1080. [Google Scholar] [CrossRef] [PubMed]
- Schwaederle, M.; Zhao, M.; Lee, J.J.; Lazar, V.; Leyland-Jones, B.; Schilsky, R.L.; Mendelsohn, J.; Kurzrock, R. Association of Biomarker-Based Treatment Strategies with Response Rates and Progression-Free Survival in Refractory Malignant Neoplasms. JAMA Oncol. 2016, 2, 1452. [Google Scholar] [CrossRef] [PubMed]
- Schapira, M.; Calabrese, M.F.; Bullock, A.N.; Crews, C.M. Targeted Protein Degradation: Expanding the Toolbox. Nat. Rev. Drug Discov. 2019, 18, 949–963. [Google Scholar] [CrossRef] [PubMed]
- Kasztura, M.; Richard, A.; Bempong, N.-E.; Loncar, D.; Flahault, A. Cost-Effectiveness of Precision Medicine: A Scoping Review. Int. J. Public Health 2019, 64, 1261–1271. [Google Scholar] [CrossRef] [PubMed]
- Naithani, N.; Atal, A.T.; Tilak, T.V.S.V.G.K.; Vasudevan, B.; Misra, P.; Sinha, S. Precision Medicine: Uses and Challenges. Med. J. Armed Forces India 2021, 77, 258–265. [Google Scholar] [CrossRef]
- Goetz, L.H.; Schork, N.J. Personalized Medicine: Motivation, Challenges, and Progress. Fertil. Steril. 2018, 109, 952–963. [Google Scholar] [CrossRef]
- Fu, Y.; Ling, Z.; Arabnia, H.; Deng, Y. Current Trend and Development in Bioinformatics Research. BMC Bioinform. 2020, 21, 538. [Google Scholar] [CrossRef]
- Behl, T.; Kaur, I.; Sehgal, A.; Singh, S.; Bhatia, S.; Al-Harrasi, A.; Zengin, G.; Babes, E.E.; Brisc, C.; Stoicescu, M.; et al. Bioinformatics Accelerates the Major Tetrad: A Real Boost for the Pharmaceutical Industry. Int. J. Mol. Sci. 2021, 22, 6184. [Google Scholar] [CrossRef]
- Guo, H.; Zhang, Z.; Wang, Y.; Xue, S. Identification of Crucial Genes and Pathways Associated with Prostate Cancer in Multiple Databases. J. Int. Med. Res. 2021, 49, 030006052110166. [Google Scholar] [CrossRef]
- Ren, Y.; Deng, R.; Zhang, Q.; Li, J.; Han, B.; Ye, P. Bioinformatics Analysis of Key Genes in Triple Negative Breast Cancer and Validation of Oncogene PLK1. Ann. Transl. Med. 2020, 8, 1637. [Google Scholar] [CrossRef]
- Sun, B.; Zhao, H. The Bioinformatics Analysis of RIOX2 Gene in Lung Adenocarcinoma and Squamous Cell Carcinoma. PLoS ONE 2021, 16, e0259447. [Google Scholar] [CrossRef] [PubMed]
- Harel, M.; Lahav, C.; Jacob, E.; Dahan, N.; Sela, I.; Elon, Y.; Raveh Shoval, S.; Yahalom, G.; Kamer, I.; Zer, A.; et al. Longitudinal Plasma Proteomic Profiling of Patients with Non-Small Cell Lung Cancer Undergoing Immune Checkpoint Blockade. J. Immunother. Cancer 2022, 10, e004582. [Google Scholar] [CrossRef] [PubMed]
- Moresi, F.; Rossetti, D.V.; Vincenzoni, F.; Simboli, G.A.; La Rocca, G.; Olivi, A.; Urbani, A.; Sabatino, G.; Desiderio, C. Investigating Glioblastoma Multiforme Sub-Proteomes: A Computational Study of CUSA Fluid Proteomic Data. Int. J. Mol. Sci. 2022, 23, 2058. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alvarez-Rivera, E.; Ortiz-Hernández, E.J.; Lugo, E.; Lozada-Reyes, L.M.; Boukli, N.M. Oncogenic Proteomics Approaches for Translational Research and HIV-Associated Malignancy Mechanisms. Proteomes 2023, 11, 22. https://doi.org/10.3390/proteomes11030022
Alvarez-Rivera E, Ortiz-Hernández EJ, Lugo E, Lozada-Reyes LM, Boukli NM. Oncogenic Proteomics Approaches for Translational Research and HIV-Associated Malignancy Mechanisms. Proteomes. 2023; 11(3):22. https://doi.org/10.3390/proteomes11030022
Chicago/Turabian StyleAlvarez-Rivera, Eduardo, Emanuel J. Ortiz-Hernández, Elyette Lugo, Lorraine M. Lozada-Reyes, and Nawal M. Boukli. 2023. "Oncogenic Proteomics Approaches for Translational Research and HIV-Associated Malignancy Mechanisms" Proteomes 11, no. 3: 22. https://doi.org/10.3390/proteomes11030022
APA StyleAlvarez-Rivera, E., Ortiz-Hernández, E. J., Lugo, E., Lozada-Reyes, L. M., & Boukli, N. M. (2023). Oncogenic Proteomics Approaches for Translational Research and HIV-Associated Malignancy Mechanisms. Proteomes, 11(3), 22. https://doi.org/10.3390/proteomes11030022








