Quantitative Proteogenomic Characterization of Inflamed Murine Colon Tissue Using an Integrated Discovery, Verification, and Validation Proteogenomic Workflow
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
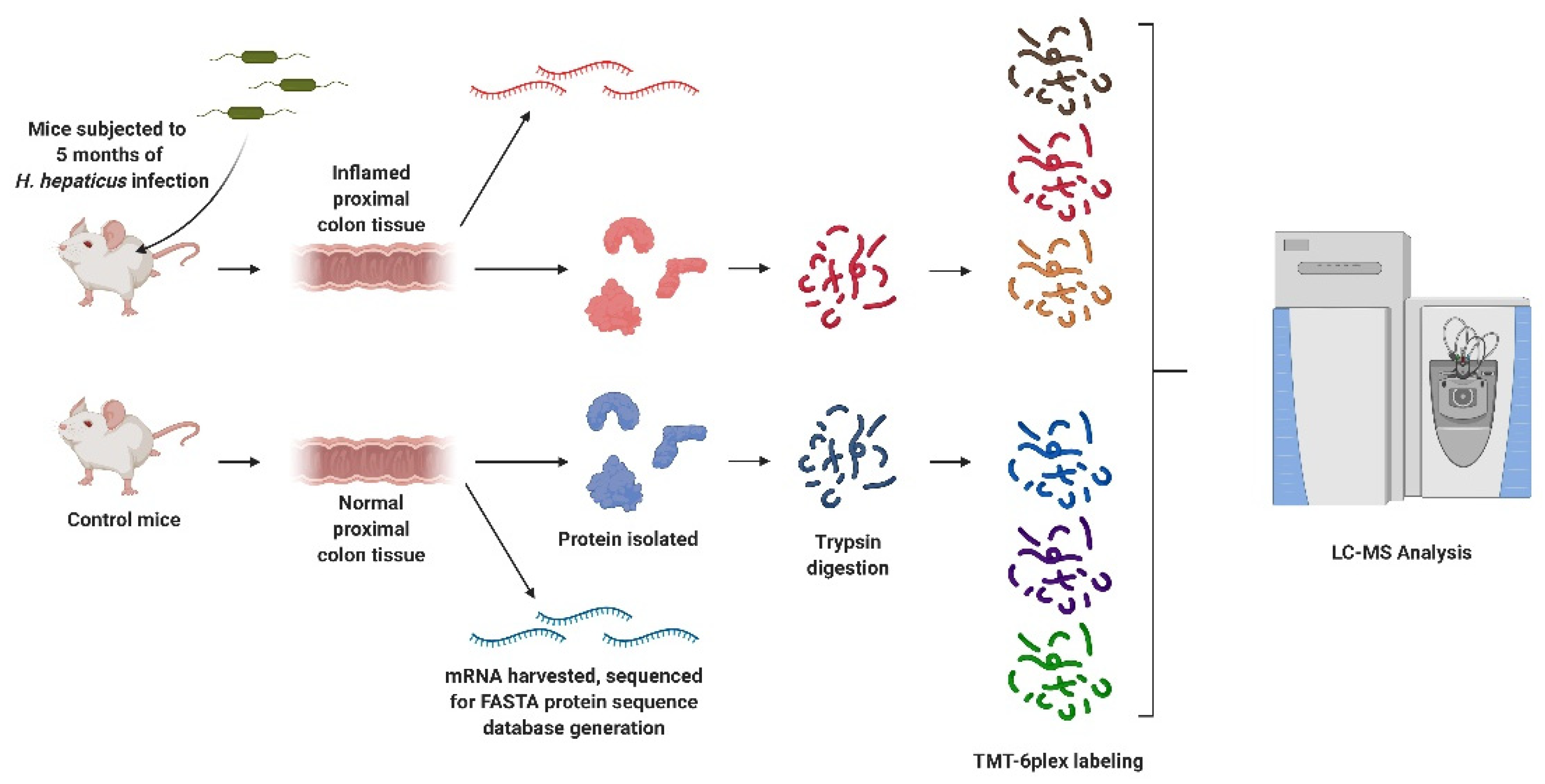
2.2. Treatment Conditions, Tissue and Protein Isolation and Proteolytic Digestion
2.3. Peptide Labeling, Fractionation, and LC–MS/MS Analysis
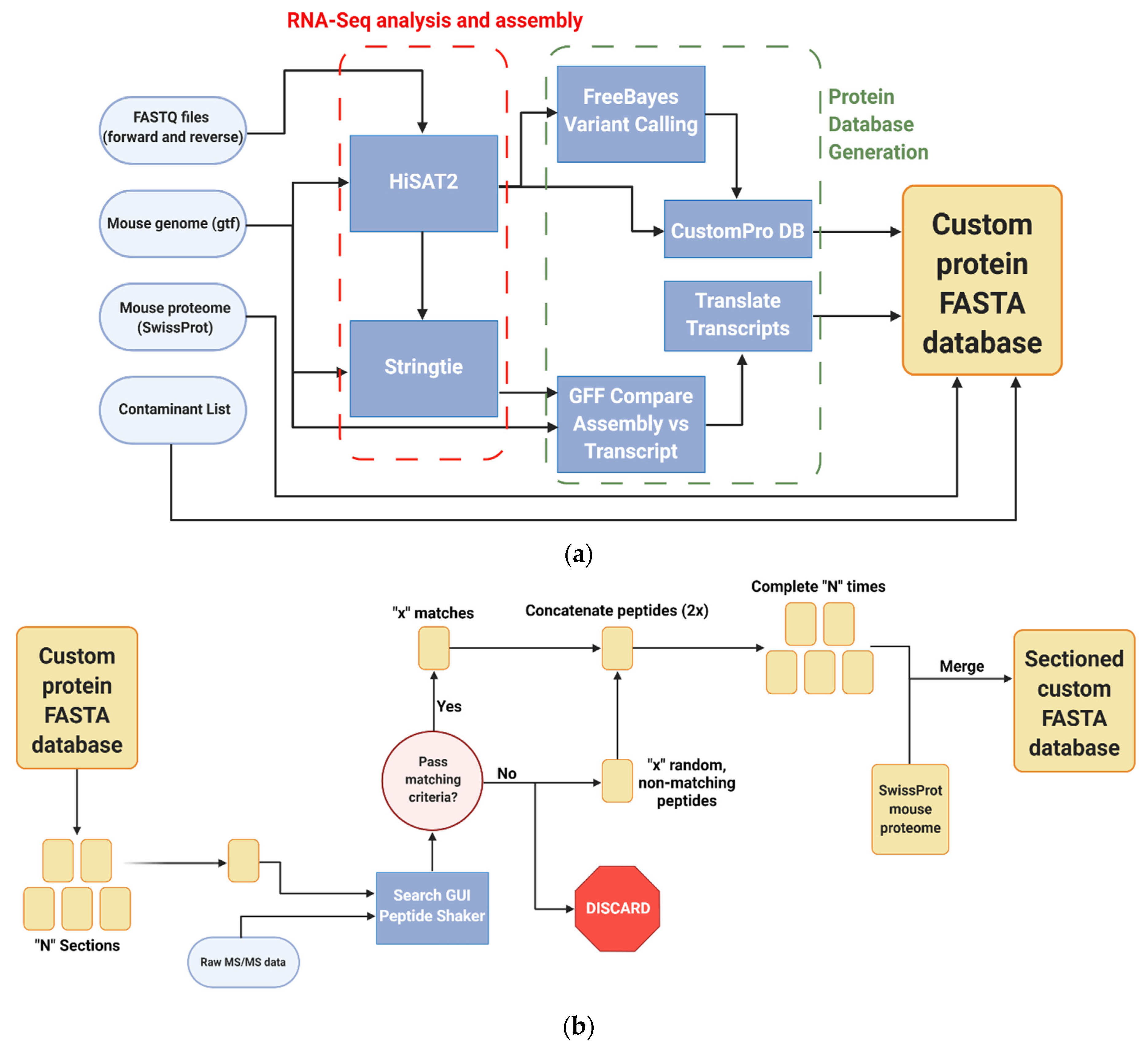
2.4. Database Construction
2.5. Database Sectioning
2.6. Differential Abundance Proteomic and Proteogenomic Analysis
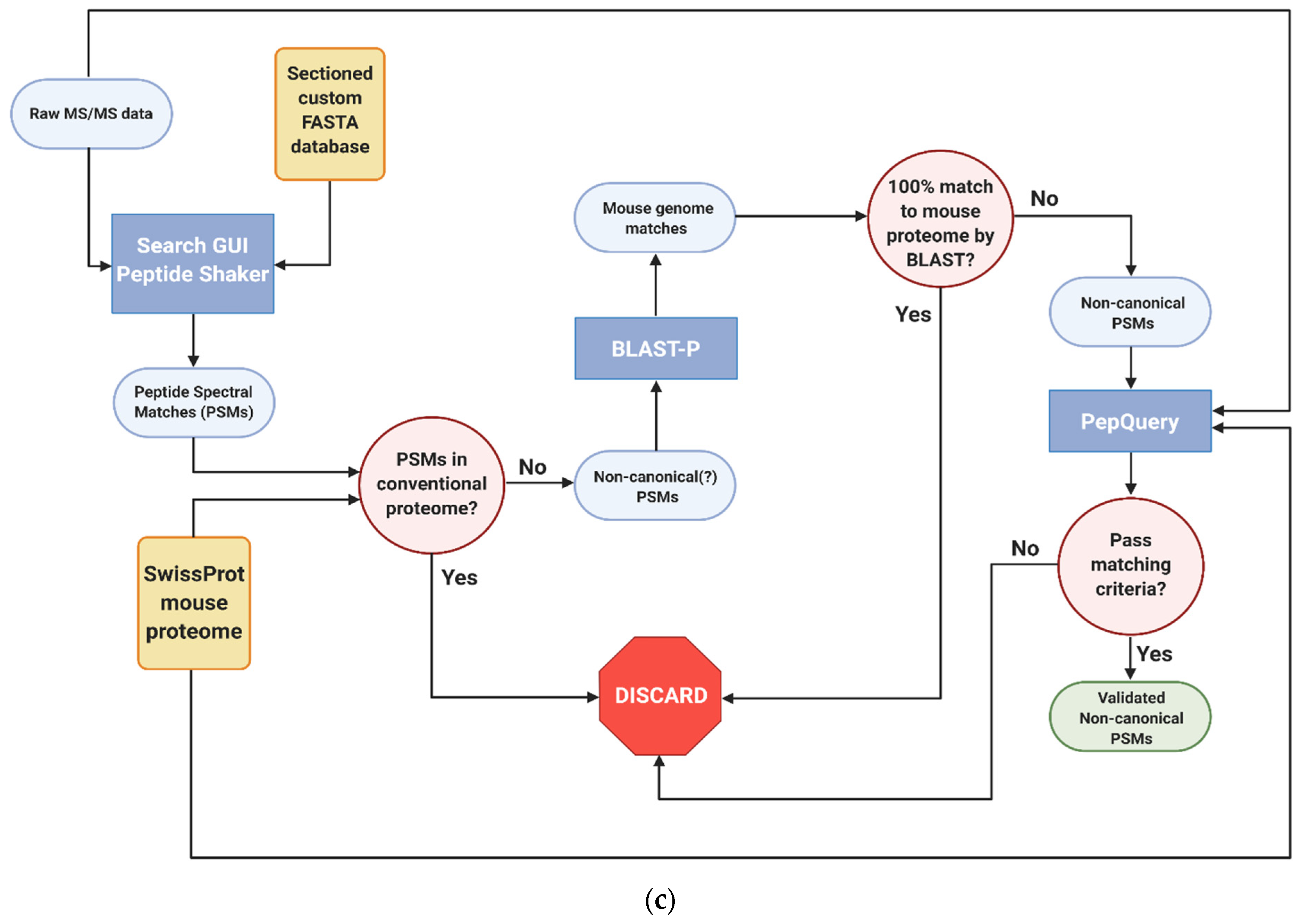
2.7. Identification, Verification and Validation of Non-Canonical Peptides
2.8. Validation and Quantitation of Non-Canonical Peptides
3. Results
3.1. Creation and Sectioning of a Custom RNA-Seq-Based FASTA Database
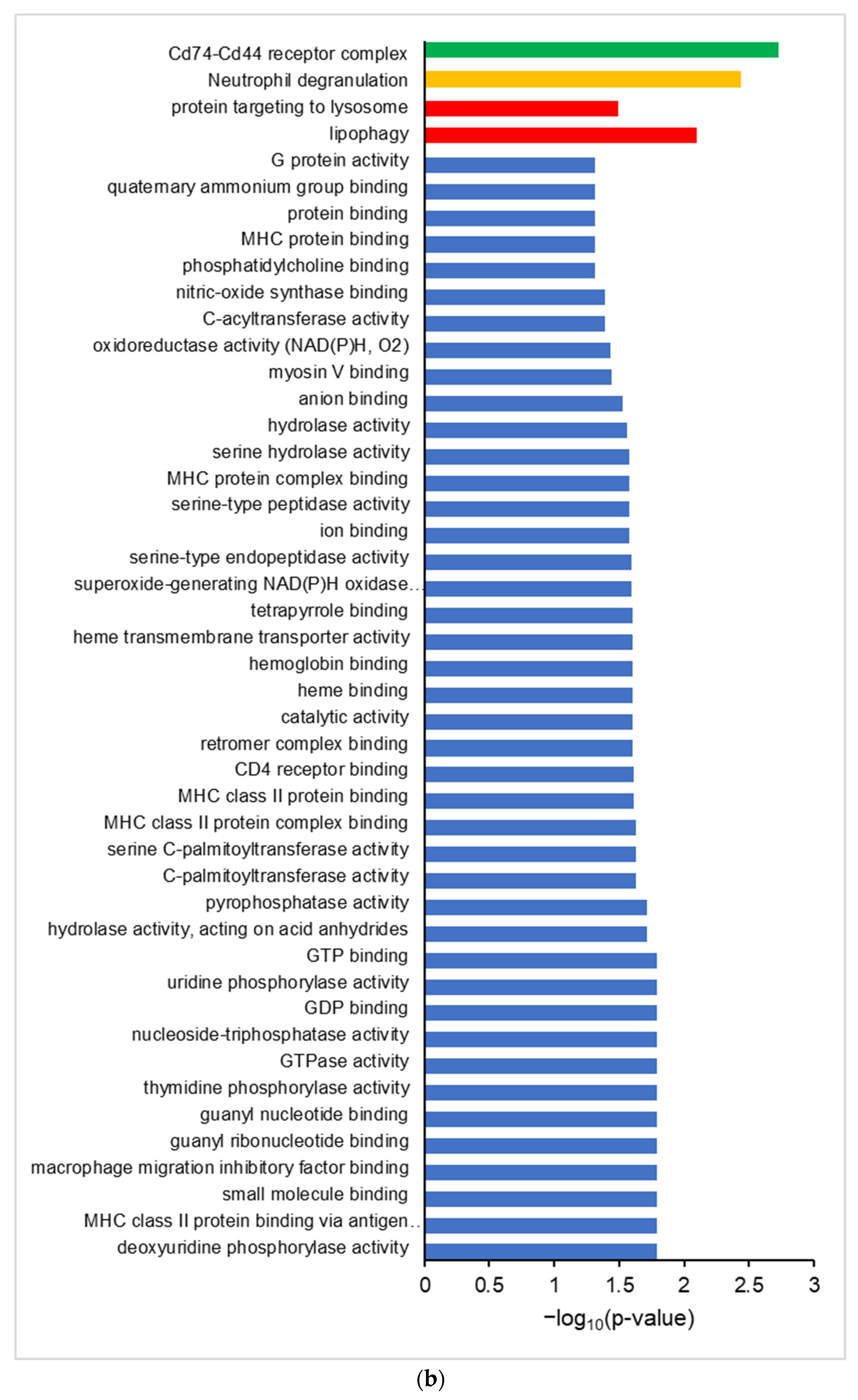
3.2. Global Proteogenomic Analysis Reveals Inflammation-Driven Changes in Protein Abundance
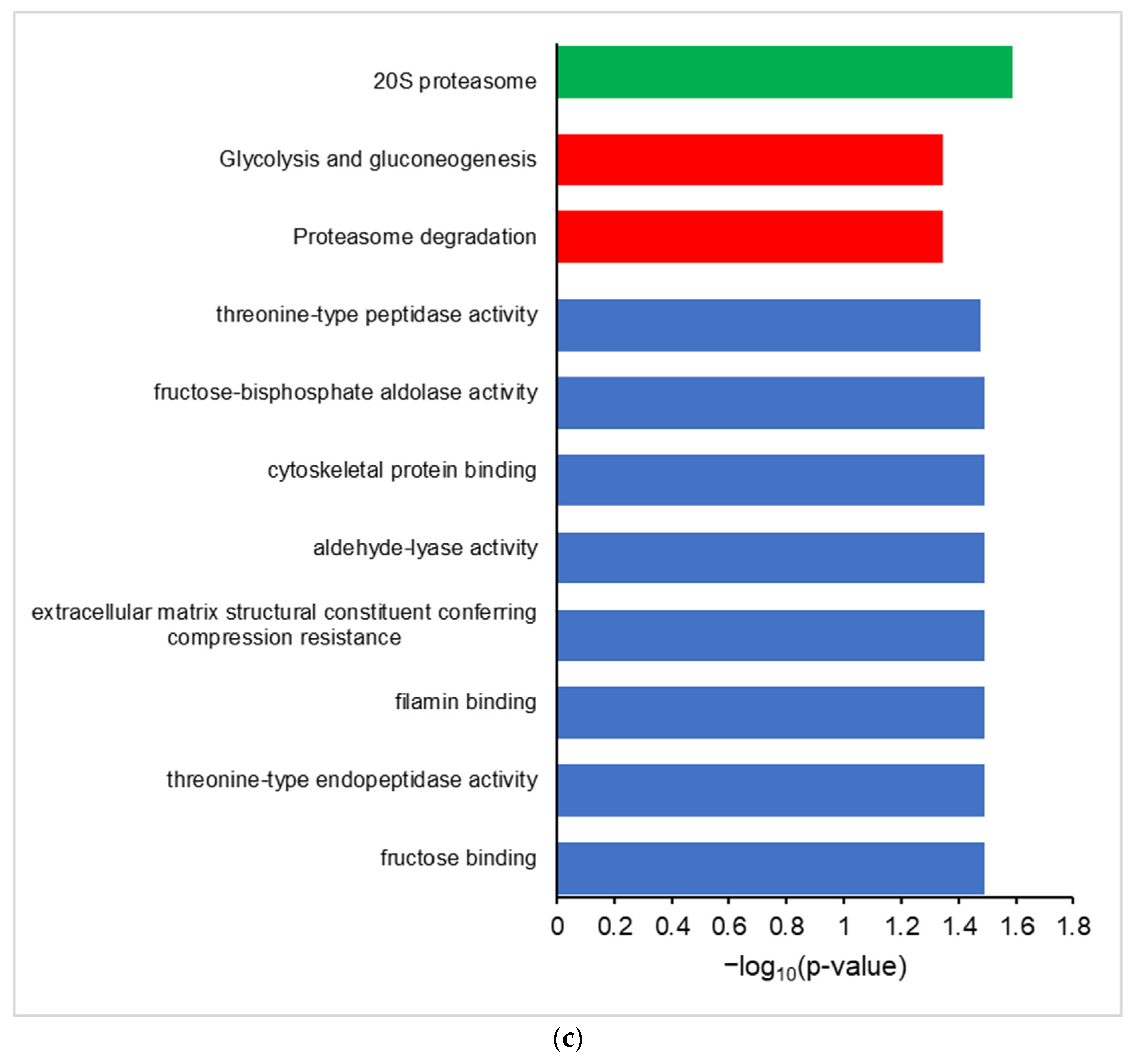
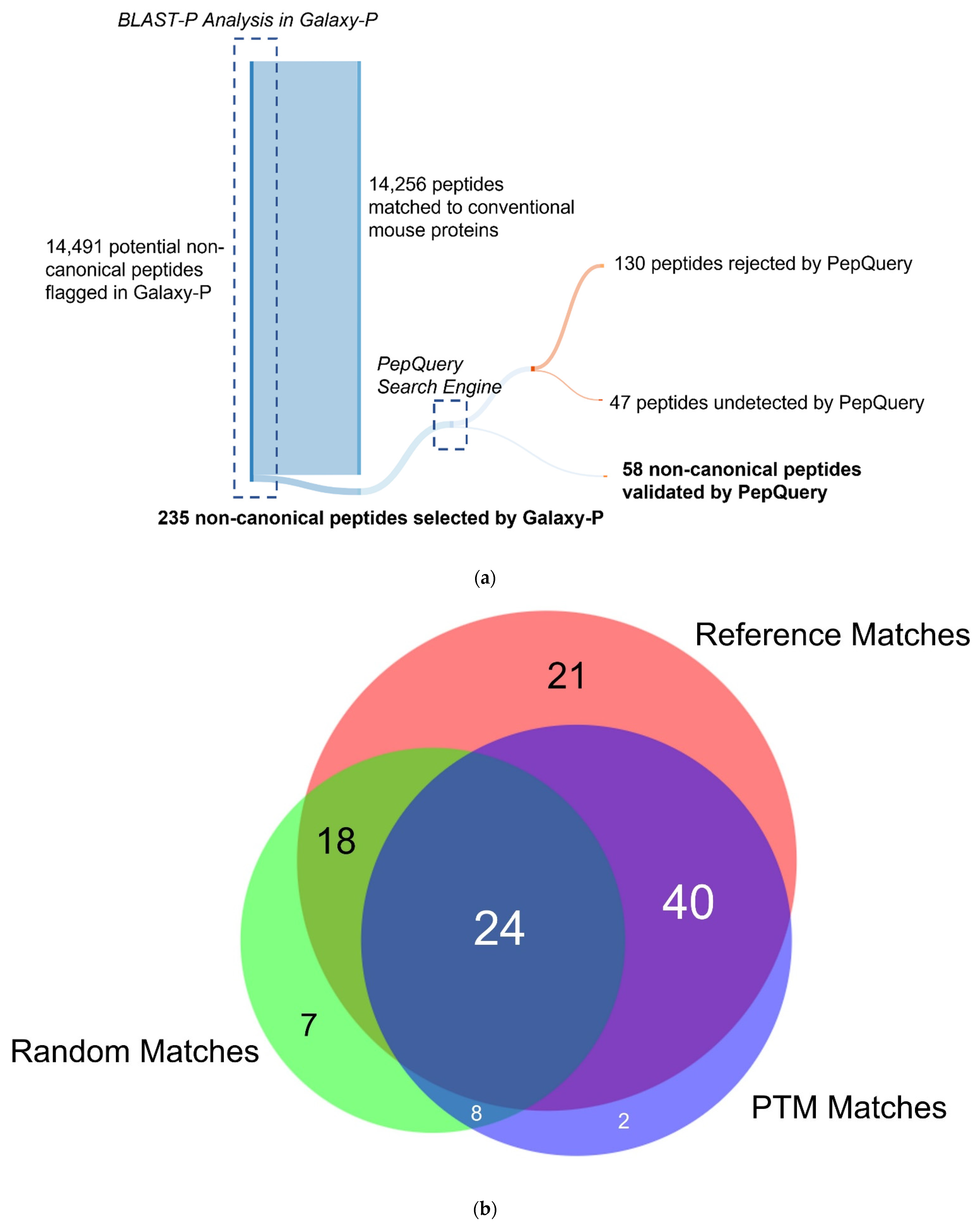
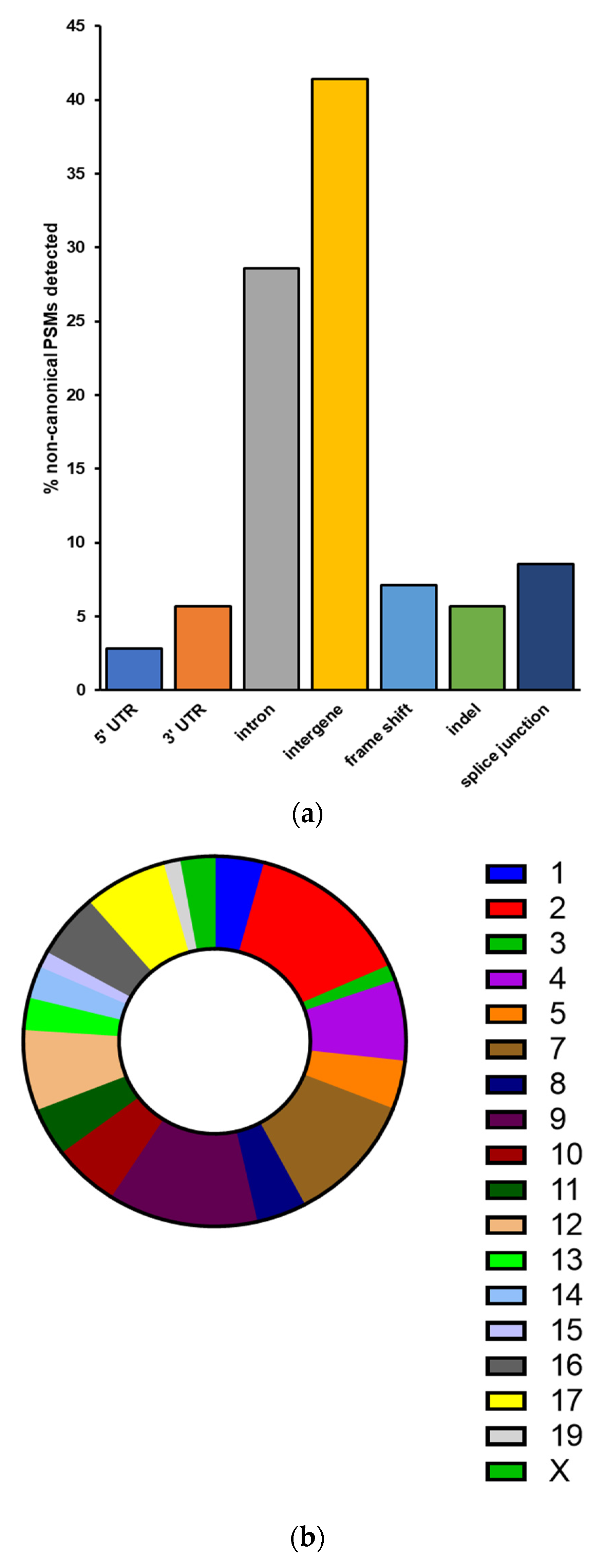
3.3. Galaxy-P Provides Peptide-Centric Discovery of Non-Canonical Sequences
3.4. PepQuery Verifies the Highest Confidence Non-Canonical Peptide Candidates
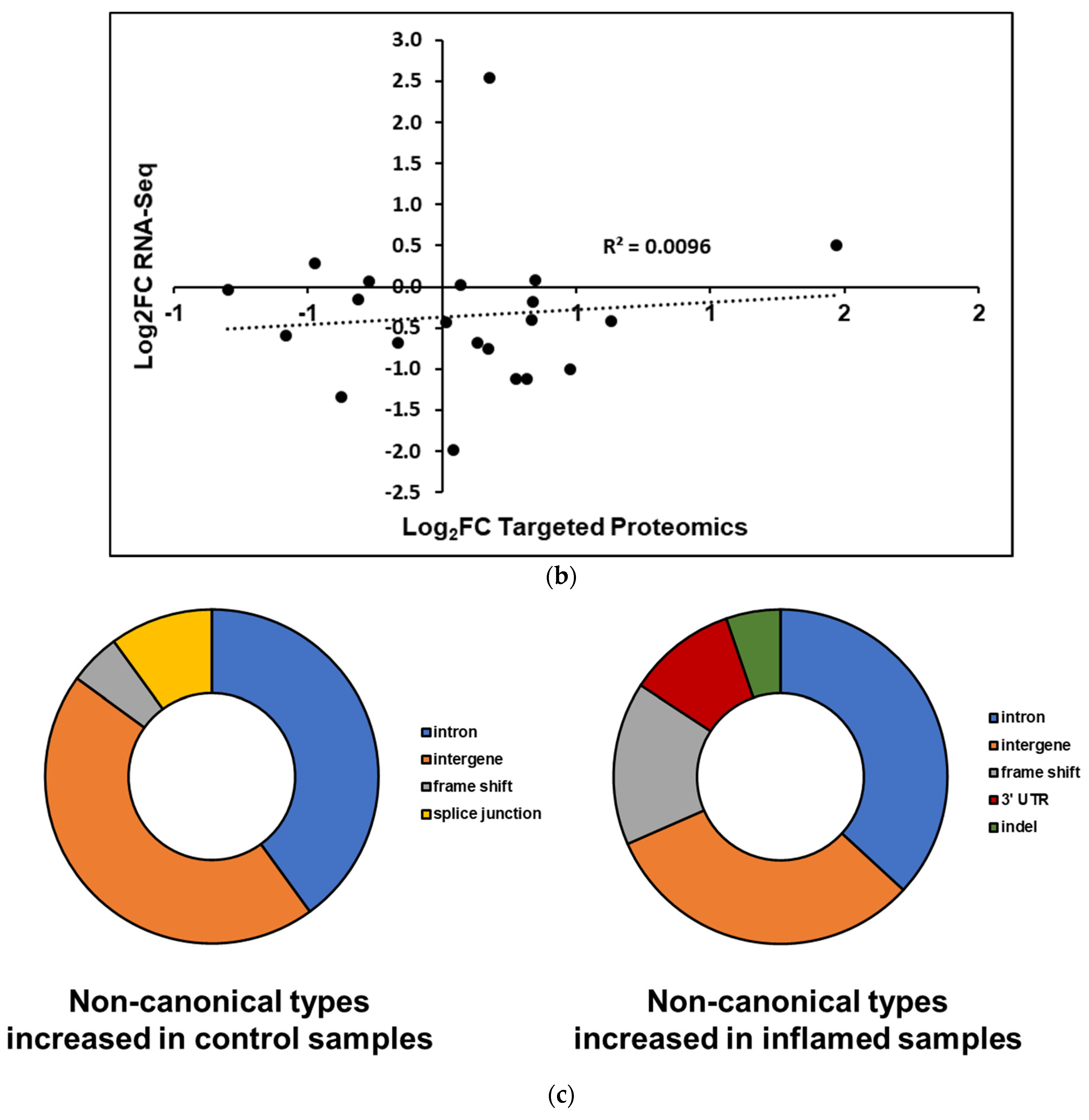
3.5. Targeted Proteomics Experiments Validate the Presence of Non-Canonical Peptides
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Chiba, T.; Marusawa, H.; Ushijima, T. Inflammation-Associated Cancer Development in Digestive Organs: Mechanisms and Roles for Genetic and Epigenetic Modulation. Gastroenterology 2012, 143, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.V.; Fernandes, T.A.A.D.M.; De Azevedo, J.C.V.; Cobucci, R.N.O.; De Carvalho, M.G.F.; Andrade, V.S.; De Araujo, J.M.G. Link between chronic inflammation and human papillomavirus-induced carcinogenesis. Oncol. Lett. 2015, 9, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Greten, F.R.; Eckmann, L.; Greten, T.F.; Park, J.M.; Li, Z.-W.; Egan, L.J.; Kagnoff, M.F.; Karin, M. IKKβ Links Inflammation and Tumorigenesis in a Mouse Model of Colitis-Associated Cancer. Cell 2004, 118, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Affara, N.I.; Coussens, L.M. IKKα at the Crossroads of Inflammation and Metastasis. Cell 2007, 129, 25–26. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mangerich, A.; Knutson, C.G.; Parry, N.M.; Muthupalani, S.; Ye, W.; Prestwich, E.; Cui, L.; McFaline, J.L.; Mobley, M.; Ge, Z.; et al. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc. Natl. Acad. Sci. USA 2012, 109, E1820–E1829. [Google Scholar] [CrossRef] [PubMed]
- Meira, L.B.; Bugni, J.M.; Green, S.L.; Lee, C.-W.; Pang, B.; Borenshtein, D.; Rickman, B.H.; Rogers, A.B.; Moroski-Erkul, C.A.; McFaline, J.L.; et al. DNA damage induced by chronic inflammation contributes to colon carcinogenesis in mice. J. Clin. Investig. 2008, 118, 2516–2525. [Google Scholar] [CrossRef]
- Huang, Z.; Huang, D.; Ni, S.; Peng, Z.; Sheng, W.; Du, X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int. J. Cancer 2010, 127, 118–126. [Google Scholar] [CrossRef]
- Alves Martins, B.A.; De Bulhões, G.F.; Cavalcanti, I.N.; Martins, M.M.; de Oliveira, P.G.; Martins, A.M.A. Biomarkers in colorectal cancer: The role of translational proteomics research. Front. Oncol. 2019, 9, 1284. [Google Scholar] [CrossRef]
- Petralia, F.; Tignor, N.; Reva, B.; Koptyra, M.; Chowdhury, S.; Rykunov, D.; Krek, A.; Ma, W.; Zhu, Y.; Ji, J.; et al. Integrated Proteogenomic Characterization across Major Histological Types of Pediatric Brain Cancer. Cell 2020, 183, 1962–1985. [Google Scholar] [CrossRef]
- Jardim-Perassi, B.V.; Alexandre, P.A.; Sonehara, N.M.; De Paula-Junior, R.; Reis Júnior, O.; Fukumasu, H.; Chammas, R.; Coutinho, L.L.; Zuccari, D.A.P.D.C. RNA-Seq transcriptome analysis shows anti-tumor actions of melatonin in a breast cancer xenograft model. Sci. Rep. 2019, 9, 966. [Google Scholar] [CrossRef]
- Jia, J.; Liu, X.; Li, L.; Lei, C.; Dong, Y.; Wu, G.; Hu, G. Transcriptional and Translational Relationship in Environmental Stress: RNAseq and ITRAQ Proteomic Analysis Between Sexually Reproducing and Parthenogenetic Females in Moina micrura. Front. Physiol. 2018, 9, 812. [Google Scholar] [CrossRef] [PubMed]
- Kisluk, J.; Ciborowski, M.; Niemira, M.; Kretowski, A.; Niklinski, J. Proteomics biomarkers for non-small cell lung cancer. J. Pharm. Biomed. Anal. 2014, 101, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Hegde, P.S.; White, I.R.; Debouck, C. Interplay of transcriptomics and proteomics. Curr. Opin. Biotechnol. 2003, 14, 647–651. [Google Scholar] [CrossRef] [PubMed]
- Alfaro, J.A.; Sinha, A.; Kislinger, T.; Boutros, P.C. Onco-proteogenomics: Cancer proteomics joins forces with genomics. Nat. Methods 2014, 11, 1107–1113. [Google Scholar] [CrossRef]
- Vasaikar, S.; Huang, C.; Wang, X.; Petyuk, V.A.; Savage, S.R.; Wen, B.; Dou, Y.; Zhang, Y.; Shi, Z.; Arshad, O.A.; et al. Proteogenomic Analysis of Human Colon Cancer Reveals New Therapeutic Opportunities. Cell 2019, 177, 1035–1049. [Google Scholar] [CrossRef]
- Smith, L.M.; Kelleher, N.L.; The Consortium for Top Down Proteomics. Proteoform: A single term describing protein complexity. Nat. Methods 2013, 10, 186–187. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, T.; Zhang, Z.; Payne, S.H.; Zhang, B.; McDermott, J.E.; Zhou, J.-Y.; Petyuk, V.A.; Chen, L.; Ray, D.; et al. Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer. Cell 2016, 166, 755–765. [Google Scholar] [CrossRef]
- Nesvizhskii, A. Proteogenomics: Concepts, applications and computational strategies. Nat. Methods 2014, 11, 1114–1125. [Google Scholar] [CrossRef]
- Tariq, M.U.; Haseeb, M.; Aledhari, M.; Razzak, R.; Parizi, R.M.; Saeed, F. Methods for Proteogenomics Data Analysis, Challenges, and Scalability Bottlenecks: A Survey. IEEE Access 2021, 9, 5497–5516. [Google Scholar] [CrossRef]
- Erdman, S.E.; Rao, V.P.; Poutahidis, T.; Rogers, A.B.; Taylor, C.L.; Jackson, E.A.; Ge, Z.; Lee, C.W.; Schauer, D.B.; Wogan, G.N.; et al. Nitric oxide and TNF-α trigger colonic inflammation and carcinogenesis in Helicobacter hepaticus-infected, Rag2-deficient mice. Proc. Natl. Acad. Sci. USA 2009, 106, 1027–1032. [Google Scholar] [CrossRef]
- Han, Q.; Kono, T.J.Y.; Knutson, C.G.; Parry, N.M.; Seiler, C.L.; Fox, J.G.; Tannenbaum, S.R.; Tretyakova, N.Y. Multi-Omics Characterization of Inflammatory Bowel Disease-Induced Hyperplasia/Dysplasia in the Rag2−/−/Il10−/− Mouse Model. Int. J. Mol. Sci. 2021, 22, 364. [Google Scholar] [CrossRef] [PubMed]
- Erdman, S.E.; Poutahidis, T.; Tomczak, M.; Rogers, A.B.; Cormier, K.; Plank, B.; Horwitz, B.H.; Fox, J.G. CD4+ CD25+ Regulatory T Lymphocytes Inhibit Microbially Induced Colon Cancer in Rag2-Deficient Mice. Am. J. Pathol. 2003, 162, 691–702. [Google Scholar] [CrossRef]
- Brennan, M.L.; Wu, W.; Fu, X.; Shen, Z.; Song, W.; Frost, H.; Vadseth, C.; Narine, L.; Lenkiewicz, E.; Borchers, M.T.; et al. A tale of two controversies: Defining both the role of peroxidases in nitrotyrosine formation in vivo using eosinophil peroxidase and myeloperoxidase-deficient mice, and the nature of peroxidase-generated reactive nitrogen species. J. Biol. Chem. 2002, 277, 17415–17427. [Google Scholar] [CrossRef]
- Boekel, J.; Chilton, J.M.; Cooke, I.R.; Horvatovich, P.L.; Jagtap, P.D.; Käll, L.; Lehtiö, J.; Lukasse, P.; Moerland, P.D.; Griffin, T. Multi-omic data analysis using Galaxy. Nat. Biotechnol. 2015, 33, 137–139. [Google Scholar] [CrossRef]
- Kumar, P.; Johnson, J.E.; Easterly, C.; Mehta, S.; Sajulga, R.; Nunn, B.; Jagtap, P.D.; Griffin, T.J. A Sectioning and Database Enrichment Approach for Improved Peptide Spectrum Matching in Large, Genome-Guided Protein Sequence Databases. J. Proteome Res. 2020, 19, 2772–2785. [Google Scholar] [CrossRef]
- Chambers, M.C.; Jagtap, P.D.; Johnson, J.E.; McGowan, T.; Kumar, P.; Onsongo, G.; Guerrero, C.R.; Barsnes, H.; Vaudel, M.; Martens, L.; et al. An Accessible Proteogenomics Informatics Resource for Cancer Researchers. Cancer Res. 2017, 77, e43–e46. [Google Scholar] [CrossRef] [PubMed]
- Jagtap, P.D.; Johnson, J.E.; Onsongo, G.; Sadler, F.W.; Murray, K.; Wang, Y.; Shenykman, G.M.; Bandhakavi, S.; Smith, L.M.; Griffin, T.J. Flexible and Accessible Workflows for Improved Proteogenomic Analysis Using the Galaxy Framework. J. Proteome Res. 2014, 13, 5898–5908. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Wang, X.; Zhang, B. customProDB: An R package to generate customized protein databases from RNA-Seq data for proteomics search. Bioinformatics 2013, 29, 3235–3237. [Google Scholar] [CrossRef]
- Mellacheruvu, D.; Wright, Z.; Couzens, A.L.; Lambert, J.-P.; St-Denis, N.A.; Li, T.; Miteva, Y.V.; Hauri, S.; Sardiu, M.E.; Low, T.Y.; et al. The CRAPome: A contaminant repository for affinity purification–mass spectrometry data. Nat. Methods 2013, 10, 730–736. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.; Griffin, T.J.; Jagtap, P.; Sajulga, R.; Johnson, J.; Kumar, P. Proteogenomics 1: Database Creation (Galaxy Training Materials). 2021. Available online: http:///training-material/topics/proteomics/tutorials/proteogenomics-dbcreation/tutorial.html (accessed on 29 April 2021).
- Vaudel, M.; Barsnes, H.; Berven, F.S.; Sickmann, A.; Martens, L. SearchGUI: An open-source graphical user interface for simultaneous OMSSA and X!Tandem searches. Proteomics 2011, 11, 996–999. [Google Scholar] [CrossRef] [PubMed]
- Vaudel, M.; Burkhart, J.M.; Zahedi, R.P.; Oveland, E.; Berven, F.S.; Sickmann, A.; Martens, L.; Barsnes, H. PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat. Biotechnol. 2015, 33, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Choi, M.; Tzouros, M.; Golling, S.; Pandya, N.J.; Banfai, B.; Dunkley, T.; Vitek, O. MSstatsTMT: Statistical Detection of Differentially Abundant Proteins in Experiments with Isobaric Labeling and Multiple Mixtures. Mol. Cell. Proteom. 2020, 19, 1706–1723. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Gish, W.; States, D.J. Identification of protein coding regions by database similarity search. Nat. Genet. 1993, 3, 266–272. [Google Scholar] [CrossRef]
- Available online: https://github.com/galaxyproteomics/tools-galaxyp/tree/master/tools/pep_pointer (accessed on 6 April 2022).
- Wen, B.; Wang, X.; Zhang, B. PepQuery enables fast, accurate, and convenient proteomic validation of novel genomic alterations. Genome Res. 2019, 29, 485–493. [Google Scholar] [CrossRef]
- Peterson, A.C.; Russell, J.D.; Bailey, D.J.; Westphall, M.S.; Coon, J.J. Parallel Reaction Monitoring for High Resolution and High Mass Accuracy Quantitative, Targeted Proteomics. Mol. Cell. Proteom. 2012, 11, 1475–1488. [Google Scholar] [CrossRef]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef]
- Gessulat, S.; Schmidt, T.; Zolg, D.P.; Samaras, P.; Schnatbaum, K.; Zerweck, J.; Knaute, T.; Rechenberger, J.; Delanghe, B.; Huhmer, A.; et al. Prosit: Proteome-wide prediction of peptide tandem mass spectra by deep learning. Nat. Methods 2019, 16, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Yadav, A.K.; Dash, D. Choosing an Optimal Database for Protein Identification from Tandem Mass Spectrometry Data; Springer: New York, NY, USA, 2017; Volume 1549, pp. 17–29. [Google Scholar]
- Li, K.; Vaudel, M.; Zhang, B.; Ren, Y.; Wen, B. PDV: An integrative proteomics data viewer. Bioinformatics 2019, 35, 1249–1251. [Google Scholar] [CrossRef] [PubMed]
- Farinati, F.; Cardin, R.; Degan, P.; Rugge, M.; Di Mario, F.; Bonvicini, P.; Naccarato, R. Oxidative DNA damage accumulation in gastric carcinogenesis. Gut 1998, 42, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Hatziapostolou, M.; Iliopoulos, D. Epigenetic aberrations during oncogenesis. Cell. Mol. Life Sci. 2011, 68, 1681–1702. [Google Scholar] [CrossRef] [PubMed]
- Knutson, C.G.; Mangerich, A.; Zeng, Y.; Raczynski, A.R.; Liberman, R.G.; Kang, P.; Ye, W.; Prestwich, E.G.; Lu, K.; Wishnok, J.S.; et al. Chemical and cytokine features of innate immunity characterize serum and tissue profiles in inflammatory bowel disease. Proc. Natl. Acad. Sci. USA 2013, 110, E2332–E2341. [Google Scholar] [CrossRef]
- Janz, D.R.; Bastarache, J.A.; Sills, G.; Wickersham, N.; May, A.K.; Bernard, G.R.; Ware, L.B. Association between haptoglobin, hemopexin and mortality in adults with sepsis. Crit. Care 2013, 17, R272. [Google Scholar] [CrossRef]
- Winter, N.A.; Gibson, P.G.; Fricker, M.; Simpson, J.L.; Wark, P.A.; McDonald, V.M. Hemopexin: A Novel Anti-inflammatory Marker for Distinguishing COPD From Asthma. Allergy, Asthma Immunol. Res. 2021, 13, 450–467. [Google Scholar] [CrossRef]
- Zhang, X.; Xiao, Z.; Liu, X.; Du, L.; Wang, L.; Wang, S.; Zheng, N.; Zheng, G.; Li, W.; Zhang, X.; et al. The Potential Role of ORM2 in the Development of Colorectal Cancer. PLoS ONE 2012, 7, e31868. [Google Scholar] [CrossRef]
- Hayashi, K.-G.; Hosoe, M.; Kizaki, K.; Fujii, S.; Kanahara, H.; Takahashi, T.; Sakumoto, R. Differential gene expression profiling of endometrium during the mid-luteal phase of the estrous cycle between a repeat breeder (RB) and non-RB cows. Reprod. Biol. Endocrinol. 2017, 15, 20. [Google Scholar] [CrossRef] [PubMed]
- Wynn, T.A. Cellular and molecular mechanisms of fibrosis. J. Pathol. 2008, 214, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wu, L.; Wu, X.; Li, K.; Li, T.; Xu, B.; Liu, W. Combined analysis of serum SAP and PRSS2 for the differential diagnosis of CD and UC. Clin. Chim. Acta 2021, 514, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Waithman, J.; Moffat, J.M.; Patterson, N.L.; van Beek, A.E.; Mintern, J.D. Antigen Presentation. In Reference Module in Biomedical Sciences; Elsevier: Amsterdam, The Netherlands, 2014. [Google Scholar]
- Keller, C.W.; Kotur, M.B.; Mundt, S.; Dokalis, N.; Ligeon, L.-A.; Shah, A.M.; Prinz, M.; Becher, B.; Münz, C.; Lünemann, J.D. CYBB/NOX2 in conventional DCs controls T cell encephalitogenicity during neuroinflammation. Autophagy 2021, 17, 1244–1258. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Kabir, I.; Tietelman, G.; Huan, C.; Fan, J.; Worgall, T.; Jiang, X.-C. Sphingolipid de novo biosynthesis is essential for intestine cell survival and barrier function. Cell Death Dis. 2018, 9, 173. [Google Scholar] [CrossRef]
- Nardella, C.; Chen, Z.; Salmena, L.; Carracedo, A.; Alimonti, A.; Egia, A.; Carver, B.; Gerald, W.; Cordon-Cardo, C.; Pandolfi, P.P. Aberrant Rheb-mediated mTORC1 activation and Pten haploinsufficiency are cooperative oncogenic events. Genes Dev. 2008, 22, 2172–2177. [Google Scholar] [CrossRef]
- Wang, J.; Xu, S.; Lv, W.; Shi, F.; Mei, S.; Shan, A.; Xu, J.; Yang, Y. Uridine phosphorylase 1 is a novel immune-related target and predicts worse survival in brain glioma. Cancer Med. 2020, 9, 5940–5947. [Google Scholar] [CrossRef]
- Miyashita, H.; Takebayashi, Y.; Eliason, J.F.; Fujimori, F.; Nitta, Y.; Sato, A.; Morikawa, H.; Ohashi, A.; Motegi, K.; Fukumoto, M.; et al. Uridine phosphorylase is a potential prognostic factor in patients with oral squamous cell carcinoma. Cancer 2002, 94, 2959–2966. [Google Scholar] [CrossRef]
- Chen, W.; Lu, C.; Hirota, C.; Iacucci, M.; Ghosh, S.; Gui, X. Smooth Muscle Hyperplasia/Hypertrophy is the Most Prominent Histological Change in Crohn’s Fibrostenosing Bowel Strictures: A Semiquantitative Analysis by Using a Novel Histological Grading Scheme. J. Crohn’s Colitis 2017, 11, 92–104. [Google Scholar] [CrossRef]
- Hartnett, L.; Egan, L.J. Inflammation, DNA methylation and colitis-associated cancer. Carcinogenesis 2012, 33, 723–731. [Google Scholar] [CrossRef]
- Janssen, W.J.; Danhorn, T.; Harris, C.; Mould, K.; Lee, F.F.-Y.; Hedin, B.R.; D’Alessandro, A.; Leach, S.M.; Alper, S. Inflammation-Induced Alternative Pre-mRNA Splicing in Mouse Alveolar Macrophages. G3 2020, 10, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The Human Genome Browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.; Easterly, C.W.; Sajulga, R.; Millikin, R.J.; Argentini, A.; Eguinoa, I.; Martens, L.; Shortreed, M.R.; Smith, L.M.; McGowan, T.; et al. Precursor Intensity-Based Label-Free Quantification Software Tools for Proteomic and Multi-Omic Analysis within the Galaxy Platform. Proteomes 2020, 8, 15. [Google Scholar] [CrossRef] [PubMed]



| Accession | Description | Gene | Coverage (%) | No. Peptides | log2FC | p-Value | q-Value |
|---|---|---|---|---|---|---|---|
| Q61646 | Haptoglobin | Hp | 37 | 12 | 2.60 | 1.27 × 10−7 | 1.79 × 10−4 |
| P07361 | Alpha-1-acid glycoprotein 2 | Orm2 | 11 | 3 | 2.08 | 2.90 × 10−5 | 8.13 × 10−3 |
| P07146 | Anionic trypsin 2 | Prss2 | 17 | 3 | 1.94 | 1.41 × 10−7 | 1.79 × 10−4 |
| P52624 | Uridine phosphorylase 1 | Upp1 | 37 | 9 | 1.60 | 7.86 × 10−6 | 2.85 × 10−3 |
| Q61093 | Cytochrome b-245 heavy chain | Cybb | 1 | 1 | 1.46 | 1.50 × 10−4 | 2.37 × 10−2 |
| P04441 | H-2 class II histocompatibility antigen gamma chain | Cd74 | 30 | 8 | 1.23 | 5.77 × 10−6 | 2.66 × 10−3 |
| STRG.18707.1_i_2_260 | chr8: 73261429–73261687+ | - | 7 | 1 | 1.09 | 5.45 × 10−5 | 1.15 × 10−2 |
| Q91X72 | Hemopexin | Hpx | 43 | 18 | 0.98 | 6.29 × 10−6 | 2.66 × 10−3 |
| O35704 | Serine palmitoyltransferase 1 | Sptlc1 | 15 | 6 | 0.39 | 2.25 × 10−6 | 1.43 × 10−3 |
| Q9CPW4 | Actin-related protein 2/3 complex subunit 5 | Arpc5 | 48 | 7 | 0.38 | 3.94 × 10−7 | 3.33 × 10−4 |
| O35114 | Lysosome membrane protein 2 | Scarb2 | 14 | 6 | 0.36 | 4.75 × 10−5 | 1.10 × 10−2 |
| P51150 | Ras-related protein Rab-7a | Rab7a | 64 | 12 | 0.31 | 1.79 × 10−4 | 2.67 × 10−2 |
| Q9WTL2 | Ras-related protein Rab-25 | Rab25 | 44 | 8 | 0.28 | 1.23 × 10−4 | 2.24 × 10−2 |
| Q921J2 | GTP-binding protein Rheb | Rheb | 28 | 6 | 0.24 | 2.40 × 10−4 | 3.20 × 10−2 |
| A6ZI44 | Fructose-bisphosphate aldolase | Aldoa | 63 | 23 | −0.47 | 3.20 × 10−5 | 8.13 × 10−3 |
| P57016 | Ladinin-1 | Lad1 | 17 | 8 | −0.60 | 3.74 × 10−4 | 4.31 × 10−2 |
| Q62000 | Mimecan | Ogn | 37 | 9 | −0.70 | 3.28 × 10−4 | 3.96 × 10−2 |
| P35385 | Heat shock protein beta-7 | Hspb7 | 33 | 4 | −0.78 | 3.25 × 10−4 | 3.96 × 10−2 |
| Q7TQD2 | Tubulin polymerization-promoting protein | Tppp | 17 | 3 | −0.91 | 1.10 × 10−4 | 2.16 × 10−2 |
| O55234 | Proteasome subunit beta type-5 | Psmb5 | 24 | 6 | −1.28 | 2.36 × 10−5 | 7.50 × 10−3 |
| Q99JI1 | Musculoskeletal embryonic nuclear protein 1 | Mustn1 | 18 | 1 | −1.39 | 2.29 × 10−4 | 3.20 × 10−2 |
| Q19LI2 | Alpha-1B-glycoprotein | A1bg | 2 | 1 | −1.68 | 1.38 × 10−4 | 2.33 × 10−2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajczewski, A.T.; Han, Q.; Mehta, S.; Kumar, P.; Jagtap, P.D.; Knutson, C.G.; Fox, J.G.; Tretyakova, N.Y.; Griffin, T.J. Quantitative Proteogenomic Characterization of Inflamed Murine Colon Tissue Using an Integrated Discovery, Verification, and Validation Proteogenomic Workflow. Proteomes 2022, 10, 11. https://doi.org/10.3390/proteomes10020011
Rajczewski AT, Han Q, Mehta S, Kumar P, Jagtap PD, Knutson CG, Fox JG, Tretyakova NY, Griffin TJ. Quantitative Proteogenomic Characterization of Inflamed Murine Colon Tissue Using an Integrated Discovery, Verification, and Validation Proteogenomic Workflow. Proteomes. 2022; 10(2):11. https://doi.org/10.3390/proteomes10020011
Chicago/Turabian StyleRajczewski, Andrew T., Qiyuan Han, Subina Mehta, Praveen Kumar, Pratik D. Jagtap, Charles G. Knutson, James G. Fox, Natalia Y. Tretyakova, and Timothy J. Griffin. 2022. "Quantitative Proteogenomic Characterization of Inflamed Murine Colon Tissue Using an Integrated Discovery, Verification, and Validation Proteogenomic Workflow" Proteomes 10, no. 2: 11. https://doi.org/10.3390/proteomes10020011
APA StyleRajczewski, A. T., Han, Q., Mehta, S., Kumar, P., Jagtap, P. D., Knutson, C. G., Fox, J. G., Tretyakova, N. Y., & Griffin, T. J. (2022). Quantitative Proteogenomic Characterization of Inflamed Murine Colon Tissue Using an Integrated Discovery, Verification, and Validation Proteogenomic Workflow. Proteomes, 10(2), 11. https://doi.org/10.3390/proteomes10020011








