Abstract
The overall goal of our study was to compare the proteins found in the saliva proteomes of three mammals: human, mouse and rat. Our first objective was to compare two human proteomes with very different analysis depths. The 89 shared proteins in this comparison apparently represent a core of highly-expressed human salivary proteins. Of the proteins unique to each proteome, one-half to 2/3 lack signal peptides and probably are contaminants instead of less highly-represented salivary proteins. We recently published the first rodent saliva proteomes with saliva collected from the genome mouse (C57BL/6) and the genome rat (BN/SsNHsd/Mcwi). Our second objective was to compare the proteins in the human proteome with those we identified in the genome mouse and rat to determine those common to all three mammals, as well as the specialized rodent subset. We also identified proteins unique to each of the three mammals, because differences in the secreted protein constitutions can provide clues to differences in the evolutionary adaptation of the secretions in the three different mammals.
1. Introduction
The advent of genomic and proteomic sciences has provided a flood of new information about genes expressed to produce the array of proteins characteristic of a particular tissue. Determining which genes are expressed in a particular type of cell and/or in the fluid it secretes can be done by assaying either RNA transcripts, translated protein products or, ideally, both. Mammals, including primates and rodents, produce and secrete proteins into saliva from three major salivary glands: the parotid, sublingual and submandibular glands, as well as other minor sources (e.g., tongue).
Salivary glands produce the proteins necessary to initiate digestion, to lubricate the hard and soft tissues of the mouth and to protect against infection. Primary salivary gland malfunction can occur due to viral or bacterial infection, autoimmune disease (e.g., Sjögren’s syndrome [1]), calcium stone formation, which blocks secretion, or tumor development and/or invasion. Medications and radiation treatment can also inhibit salivary gland function. A decrease in saliva production leads to the breakdown of teeth and the other oral cavity structures, thus much attention is focused on maintaining appropriate salivary gland function.
We previously obtained saliva proteomes of the genome mouse (C57BL/6) and the genome rat (BN/SsNHsd/Mcwi) using multidimensional protein identification technology (MUDPIT) for the purpose of studying rapidly evolving proteins and their genes [2]. That publication focused on the independent expansions of the mouse and rat kallikrein subfamilies expressed in saliva and how selection influenced their evolution.
The overall goal of the project we report here was to compare the proteins found in the saliva proteomes of three mammals, human, mouse and rat, in order to identify proteins shared and unique to one or more taxa. We selected two different human saliva proteomes to compare and contrast with our rodent saliva proteomes [2]. One human saliva proteome [3] was produced from whole saliva and analyzed at a depth similar to the rat and mouse proteomes we produced, while the second [4] reported a far more extensive human saliva proteome from salivary gland duct secretions collected by three different groups participating in a consortium. Because these two human proteomes differ both in collection and analysis techniques, our first objective was to compare the identifications made by the two studies. Our questions are:
- Which proteins are shared between the two human saliva proteomes and which are not?
- Does a deeper proteome necessarily improve the protein representation of salivary gland secretions?
- Does using saliva collected from individual salivary gland ducts, rather than whole saliva, improve the representation of salivary gland secretions in the final analysis?
The major advantage of proteomes is that proteins identified at a high probability from two or more high quality peptides can be confidently believed to be present in the protein mixture analyzed. However, in secretions, such as saliva or tears, one cannot conclude that every identified protein was secreted by the gland(s) producing that fluid. Proteins found in saliva are primarily secreted by salivary glands, but can also result from contamination from other sources (e.g., tracheal, naso-pharyngeal) or from cellular breakdown. We used the presence of a signal peptide as a surrogate for extracellular secretion [5] in order to eliminate from further consideration the contaminating proteins most likely produced by cellular breakdown.
The mouse and rat are widely used as experimental organisms in studies of human pathological conditions, and so, it is important to understand the ways in which their physiologies are comparable to human physiology and the ways in which they are not. Moreover, differences in the secreted salivary proteins can provide clues to differences in the evolutionary adaptation of the secretions in the three different mammals. Thus, our second objective was to determine which salivary proteins are shared among the three mammal proteomes and which are unique to one of them or shared by only two of them. For this objective, our questions were:
- What proteins are shared by the human, mouse and rat saliva proteomes, and which are shared by two of the three proteomes?
- Are the proteins shared between two or all three mammal proteomes encoded by genes with known evolutionary relationships, that is to say that they are orthologous or paralogous; or is their apparent similarity an accident of naming that does not represent a true evolutionary relationship?
- What proteins are unique to the saliva proteomes of each of the three mammals?
It was our ultimate goal to determine whether the proteins that appeared to be similar in two or more mammal saliva proteomes actually shared an evolutionary history, i.e., they were orthologous/ paralogous, or whether the similarity was superficial and they do not share an evolutionary history. Superficial similarities can arise when characteristics that may have occurred as the result of convergent evolution (e.g., a high representation of an amino acid, such as proline) result in similar naming, but where a shared evolutionary history is lacking in the two taxa under scrutiny. In discussing potentially shared evolutionary histories, we tried to take into consideration the similarities and differences in rodent and human nutritional physiology and behavior.
2. Experimental
2.1. Protein Identification from Proteomic Data
The materials and LC-MS/MS methods were reported previously for the human [3,4], mouse (C57BL/6) and rat (BN/SsNHsd/Mcwi) [2] saliva proteomes. The information on rat Klk1 gene subfamily expression in the Sprague-Dawley strain can also be found in [2].
The spectra from the two human studies were identified by searching against two different databases, the human-only entries in Swiss-Prot (Swiss-Prot, Release 42.0, October 2003) [3] and the European Bioinformatics Institute (EBI) human International Protein Index (IPI) database (version 3.01; release date November 1, 2004) [4]. To compare these identifications, we first converted the two sets of data to the UniProt format, and this was especially important in view of the deactivation of the IPI database. We used the UniProt ID Mapping function to batch convert IPI numbers [6]. Some IPI numbers could not be converted to UniProt in that way; thus, we used the NCBI protein search function to convert the remaining IPI numbers. One hundred and eighty-eight proteins from [4] were not successfully converted from IPI to UniProt Accession numbers, and these were eliminated from further analysis. Furthermore, some proteins have several IPI numbers that convert to the same UniProt number, and there are also proteins with one IPI number that correspond to multiple UniProt numbers. In those cases, we evaluated each protein number and retained only the validated or most recently reviewed UniProt number. See Figure 1 for a summary of this and downstream processes.
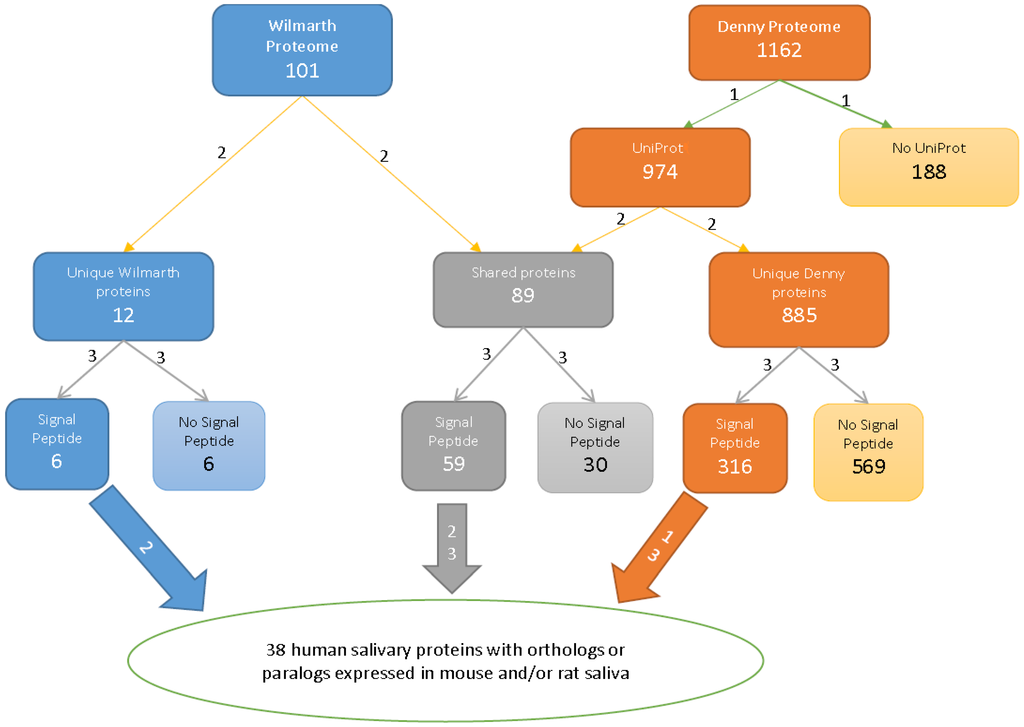
Figure 1.
Flow chart for comparing the two human proteomes (Steps 1, 2 and 3) and the human with rodent saliva proteomes. Step 1: the IPI accession numbers of the proteome [4] were converted to UniProt accession numbers; Step 2: proteins in the two proteomes were sorted by their UniProt numbers; Step 3: proteins were grouped by signal peptide status.
2.3. Identifying Secreted and Non-Secreted Proteins in the Saliva Proteomes
SignalP [7,8] was used to predict the presence or absence of a signal-peptide cleavage site for each protein to help determine whether or not that protein will be processed for secretion (Figure 1). Proteins with a D score greater than 0.45 were predicted to have a signal peptide and signal-peptide cleavage site, designating them as putative secreted proteins. Proteins with a D score below 0.45 were categorized as lacking a signal peptide.
2.4. Identifying Similar Proteins
We grouped the shared human proteins with the most similar rodent proteins by UniProt ID and then tested for the orthology and paralogy of their genes. Orthology between human, mouse and rat were computed using the “orthology” feature on [9], which identifies the best Basic Local Alignment Search Tool for Protein sequences (BLASTP) match and filters out non-syntenic hits [10]. For unclear protein identities, the Genome Browser Convert utility was used to locate the position of a gene in the genome assembly of other species [10]. During the conversion process, portions of the genome in the coordinate range of the original assembly were aligned to the new assembly, while preserving their order and orientation. We double-checked all proteins found only in two of three taxa against the other taxon by identifying the ortholog’s UniProt number with BLASTP and manually searching the appropriate proteome for that protein.
3. Results and Discussion
3.1. Comparing and Contrasting the Proteins Identified in Two Human Saliva Proteomes
We chose two human saliva proteomes of very different depths to compare and contrast. One study collected whole saliva from a single adult male and separated peptides with two-dimensional chromatography linked to mass spectrometry [3]. The second study was far more extensive, involving three different institutions in a consortium that produced a deeper proteome [4]. In that study, salivas were collected from subjects of both sexes using collection devices designed for each duct. The peptides were separated by a number of different methods before LC-MS/MS analysis of the peptide mixtures was performed. We wished to determine how the results from these two very different human saliva proteome studies compared and contrasted.
3.4. Proteins Unique to Rodent Saliva
Clearly, the three mammals share a core of proteins that play important roles in the early stages of digestion, in protecting and lubricating hard and soft surfaces and in immunological protection and maintenance of the oral cavity generally. Given the many decades of research on individual proteins playing these roles, this is hardly surprising. Perhaps more intriguing are the proteins shared by mouse and rat, but absent from human saliva, especially since the mouse and rat are widely used as experimental organisms in studies of human pathological conditions, and rodent-specific proteins may limit the applicability of these models. The rodent-shared protein group (Table 2) is 25% as large (seven) as the core shared between human and one or both rodents (29; Table 1). Four of the seven rodent-unique proteins are clearly orthologous, while half of the proteins shared between humans and rodents include complex paralog/ortholog sets, reflecting more complex evolutionary histories.
The mouse and rat secrete chitinase, common salivary protein, deoxyribonuclease, odorant binding protein, ovostatin, proline-rich lacrimal 1 protein and submandibular gland protein into their saliva that humans do not. Other studies have shown that both rodents are capable of expressing an impressive array of kallikreins from subfamilies that are unique to each genome [14] (see below).
These important differences in secreted salivary proteins may provide clues to differences in the evolutionary adaptation of the secretions in the three different mammals. For example, it is possible that chitinase and deoxyribonuclease in rodent saliva provide the potential for digesting food sources more available to rodents than to humans. We also note that some of the proteins unique to rodent saliva proteomes may play a primary or secondary role in grooming and pelage maintenance. Humans are one of the few mammals without a pelage of fur or wool covering nearly the entire body, and thus, the potential roles of proteins involved in grooming and pelage maintenance are not included in most human-centric discussions of saliva constitution. For example, we have previously shown that mice coat their pelts with salivary androgen-binding protein (ABP; [15]), and we suggested that this was a means of advertising the subspecies of the animal, since ABP has been implicated in mediating subspecies identification [15,16,17,18]. A general role in coating surfaces was later proposed for secretoglobins, such as ABP, by Dominguez [19] following the first report of substantial identities among rabbit uteroglobin, cat Fel dI and mouse ABP by Karn [20]. One can envision that a surface coating might include a chitinase that could defend against ectoparasites by attacking their exoskeletons.
The presence of the unique array of salivary kallikreins in rodent saliva is a knotty problem, given that, at least in mouse saliva, they show extensive sex-limited expression. Rodent species, including the house mouse (Mus musculus) and some strains of rats (Rattus norvegicus), show impressive elaboration of a specific tissue of the submandibular gland, the granulated convoluted tubular (GCT) tissue, often only in males following puberty [21]. This sex-limited tissue differentiation causes the submandibular glands with elaborated GCT to produce kallikrein serine proteases encoded in Klk1 gene subfamilies that have recently expanded independently in house mice and rats [14]. This results in a clear sex-limited expression of all, but a few, of these Klk1b subfamily kallikrein genes in male mice, but the picture is not so clear in rats [2]. The two strains of rat that have been studied to date show a very different expression of their Klk1c subfamily kallikrein genes, with the genome rat not expressing any of them, while the Sprague-Dawley rat expresses the Klk1c kallikrein genes in both sexes. Unfortunately, neither human saliva proteome project [3,4] addressed the issue of differential expression of proteins in males and females. Thus, we cannot currently assess the contribution of sex-limited expression to the complement of proteins found by [4] that were not found by [3,4].
3.5. Proteins Unique to Each Saliva Proteome
Removing the salivary proteins shared by two or three of the mammal proteomes allowed the identification of the proteins unique to each of them (SF4, SF5). The human saliva proteome contains a number of salivary proteins that distinguish it from the rodent proteomes, including the statherin-like PRPs, the histatins, zinc alpha glycoprotein and the Ig saliva secretory complex. Statherin prevents calcium phosphate precipitation in saliva, thus allowing calcium to be maintained at a supersaturated level in saliva to prevent deterioration of the teeth [22]. In addition to the physical shielding properties of the epithelial layer and mucin, components of innate immunity including lysozyme, lactoferrin and cystatins likely cooperate with adaptive humoral immunity mediated by antibodies in the Ig secretory complex to fight infection in the human oral cavity [23]. The presence of lysozyme and the Ig secretory complex in human, but not in rodent, saliva suggests that humans have more need of such weapons against infection. The remaining proteins appear to have an assortment of unrelated functions. Strikingly, the addition of the proteins unique to [3] and to [4] that have signal peptides brought the human list to 381. A brief survey of these proteins produced descriptions, such as: uncharacterized protein, protein existence uncertain and tissue specificity = epidermis, protein existence inferred from homology, and subcellular location = lysosome. In other words, the majority of these protein identifications seem to make up a highly heterogeneous collection of proteins, and we suspect that many of them are contaminants in spite of having signal peptides.
Of the 22 unique mouse salivary proteins, 2/3 consist of eleven Klk1b-encoded subfamily kallikreins and three androgen-binding protein (ABP) subunits (total of 14), none of which have human equivalents. The Klk1b subfamily kallikreins are expressed almost exclusively in males, and we have suggested, on the basis of new data, that the previous speculative function of the species-specific rodent kallikreins as important solely in wound healing in males be investigated further. In addition to or instead of that function, we proposed that their sex-limited expression, coupled with their rapid evolution, may be clues to an as-yet-undetermined interaction between the sexes [2]. The three ABP subunit proteins, which form dimers to produce mouse pheromones (reviewed in [24]), are found in both sexes of mice and have been proposed to be involved in incipient reinforcement, where subspecies of mice make secondary contact [17]. Mice also secrete trypsinogen, a peptidase inhibitor, MUP5, EGF binding protein, vomeromodulin, a glycoprotein and two poorly characterized proteins.
The genome rat saliva proteome has only three unique proteins: contiguous repeat polypeptide, an alpha-2 microglobulin distinct from the shared version, and an uncharacterized protein with similarity to GRPCB. Although, as we noted above, the saliva of another rat strain also contains numerous rat-specific kallikreins. Thus, the question of whether the expression of species-specific kallikrein family genes is shared between the two rodents or unique to mice depends on the strain of rat in the comparison.
4. Conclusions
Much work has been done on individual salivary proteins in humans and other animals over the past five decades, and there are relatively recent research papers and reviews that have focused on the human salivary proteins (e.g., PRPs [25]; a human saliva glycoprotein proteome [26]; a human proteomic study from a consortium of institutions [4]; and a 2013 review of human salivary proteins [27]). Less has been done with rodent salivary proteins. We published the results of the application of multidimensional protein identification technology (MUDPIT), an LC-LC-MS/MS analysis, to stimulated mouse and rat saliva for the purpose of studying rapidly evolving proteins and their genes [2].
It is possible that the comparison and contrast of the salivary protein components of human and rodent saliva that we have presented here has raised more questions than it has provided insights. Given that there has been no previous such study, we hope that at least we have framed some important questions, especially evolutionary ones, for us and for others to pursue. Our one conclusion that we feel will be useful for future studies involving one or the other rodent as a model for human oral physiology is that there are significant differences in the protein constituents between the salivas of humans and rodents, which could be misleading if not taken into consideration.
Supplementary Materials
Supplementary File 1Acknowledgments
C.M.L. received salary support from the University of Arizona’s Specialized Program of Research Excellence in Gastrointestinal Cancer (P50 CA95060) and the partnership for Native American Cancer Prevention (U54 CA143924). Amanda Chung was supported in part by a grant to the University of Arizona by the Howard Hughes Medical Institute (52006942). Laboratory space was supported by a Cancer Center Support Grant (P30 CA23074). The authors thank Vaclav Janoušek for an orientation to the Access Program.
Conflicts of Interest
The authors declare no conflict of interest.
Supplementary Files
- SF1: Human salivary proteins shared by [3,4] with SignalP and shared expression status;
- SF2: Human salivary proteins unique to [4] with signal peptides, listing shared expression;
- SF3: Human salivary proteins unique to [3] with SignalP and shared expression status;
- SF4: Mouse salivary proteins [2] with SignalP and shared expression status;
- SF5: Rat salivary proteins [2] with SignalP and shared expression status.
References and Notes
- Peria, Y.; Agmon-Levina, N.; Theodora, E.; Shoenfeld, Y. Sjögren’s syndrome, the old and the new. Best Pract. Res. Clin. Rheumatol. 2012, 26, 105–117. [Google Scholar] [CrossRef]
- Karn, R.C.; Laukaitis, C.M. Positive selection shaped the convergent evolution of independently expanded kallikrein subfamilies expressed in mouse and rat saliva proteomes. PLoS One 2011, 6, e20979. [Google Scholar] [CrossRef]
- Wilmarth, P.A.; Riviere, M.A.; Rustvold, D.L.; Lauten, J.D.; Madden, T.E.; David, L.L. Two-dimensional liquid chromatography study of the human whole saliva proteome. J. Proteome Res. 2004, 3, 1017–1023. [Google Scholar] [CrossRef]
- Denny, P.; Hagen, F.K.; Hardt, M.; Liao, L.; Yan, W.; Arellanno, M.; Bassilian, S.; Bedi, G.S.; Boontheung, P.; Cociorva, D.; et al. The proteomes of human parotid and submandibular/ sublingual gland salivas collected as the ductal secretions. J. Proteome Res. 2008, 7, 1994–2006. [Google Scholar] [CrossRef]
- Castle, D.; Castle, A. Intracellular transport and secretion of salivary proteins. Crit. Rev. Oral Biol. Med. 1998, 9, 4–22. [Google Scholar] [CrossRef]
- UniProt. Available online: http://www.uniprot.org/ (accessed on 1 October 2013).
- SignalP 4.1 server. Available online: http://www.cbs.dtu.dk/services/SignalP/ (accessed on 1 October 2013).
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. Signalp 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- UCSC genome bioinformatics. Available online: http://www.genome.ucsc.edu/ (accessed on 1 October 2013).
- Kent, W.J. Blat—The blast-like alignment tool. Genome Res. 2002, 12, 656–664. [Google Scholar]
- Fitch, W.M. Distinguishing homologous from analogous proteins. Syst. Zool. 1970, 19, 99–113. [Google Scholar] [CrossRef]
- Fitch, W.M. Homology a personal view on some of the problems. Trends Genet. 2000, 16, 227–231. [Google Scholar] [CrossRef]
- Gabaldon, T.; Koonin, E.V. Functional and evolutionary implications of gene orthology. Nat. Rev. Genet. 2013, 14, 360–366. [Google Scholar] [CrossRef]
- Lawrence, M.G.; Lai, J.; Clements, J.A. Kallikreins on steroids: Structure, function, and hormonal regulation of prostate-specific antigen and the extended kallikrein locus. Endocr. Rev. 2010, 31, 407–446. [Google Scholar] [CrossRef]
- Laukaitis, C.M.; Critser, E.S.; Karn, R.C. Salivary androgen-binding protein (abp) mediates sexual isolation in mus musculus. Evolution 1997, 51, 2000–2005. [Google Scholar] [CrossRef]
- Talley, H.M.; Laukaitis, C.M.; Karn, R.C. Female preference for male saliva: Implications for sexual isolation of Mus musculus subspecies. Evolution 2001, 55, 631–634. [Google Scholar] [CrossRef]
- Vošlajerová Bímová, B.; Macholán, M.; Baird, S.E.B.; Munclinger, P.; Laukaitis, C.M.; Karn, R.C.; Luzynski, K.; Tucker, P.; Piálek, J. Reinforcement selection acting on the European house mouse hybrid zone. Mol. Ecol. 2011, 20, 2403–2424. [Google Scholar] [CrossRef]
- Bímová, B.; Karn, R.C.; Pialek, J. The role of salivary androgen-binding protein in reproductive isolation between two subspecies of house mouse: Mus musculus musculus and Mus musculus domesticus. Biol. J. Linn. Soc. Lond. 2005, 84, 349–361. [Google Scholar] [CrossRef]
- Dominguez, P. Cloning of a syrian hamster cdna related to sexual dimorphism: Establishment of a new family of proteins. FEBS Lett. 1995, 376, 257–261. [Google Scholar] [CrossRef]
- Karn, R.C. The mouse salivary androgen-binding protein (ABP) alpha subunit closely resembles chain 1 of the cat allergen Fel di. Biochem. Genet. 1994, 32, 271–277. [Google Scholar] [CrossRef]
- Gresik, E.W. The granular convoluted tubule (GCT) cell of rodent submandibular glands. Microsc. Res. Tech. 1994, 27, 1–24. [Google Scholar] [CrossRef]
- Hay, D.I.; Smith, D.J.; Schluckebier, S.K.; Moreno, E.C. Relationship between concentration of human salivary statherin and inhibition of calcium-phosphate precipitation in stimulated human-parotid saliva. J. Dent. Res. 1984, 63, 857–863. [Google Scholar] [CrossRef]
- Brandtzaeg, P. Secretory immunity with special reference to the oral cavity. J. Oral Microbiol. 2013, 5, 1–24. [Google Scholar]
- Laukaitis, C.; Karn, R.C. Recognition of subspecies status mediated by androgen-binding protein (ABP) in the evolution of incipient reinforcement on the european house mouse hybrid zone. In Evolution of the House Mouse; Macholan, M., Munclinger, P., Baird, S.J., Pialek, J., Eds.; Cambridge University Press: Cambridge, UK, 2012. [Google Scholar]
- Williamson, M.P. The structure and function of proline-rich regions in proteins. Biochem. J. 1994, 297, 249–260. [Google Scholar]
- Ramachandran, P.; Boontheung, P.; Xie, Y.; Sondej, M.; Wong, D.T.; Loo, J.A. Identification of n-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J. Proteome Res. 2006, 5, 1493–1503. [Google Scholar] [CrossRef]
- Carpenter, G.H. The secretion, components, and properties of saliva. Annu. Rev. Food Sci. Technol. 2013, 4, 267–276. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
