Exosomal Proteome Profiling: A Potential Multi-Marker Cellular Phenotyping Tool to Characterize Hypoxia-Induced Radiation Resistance in Breast Cancer
Abstract
:1. Introduction
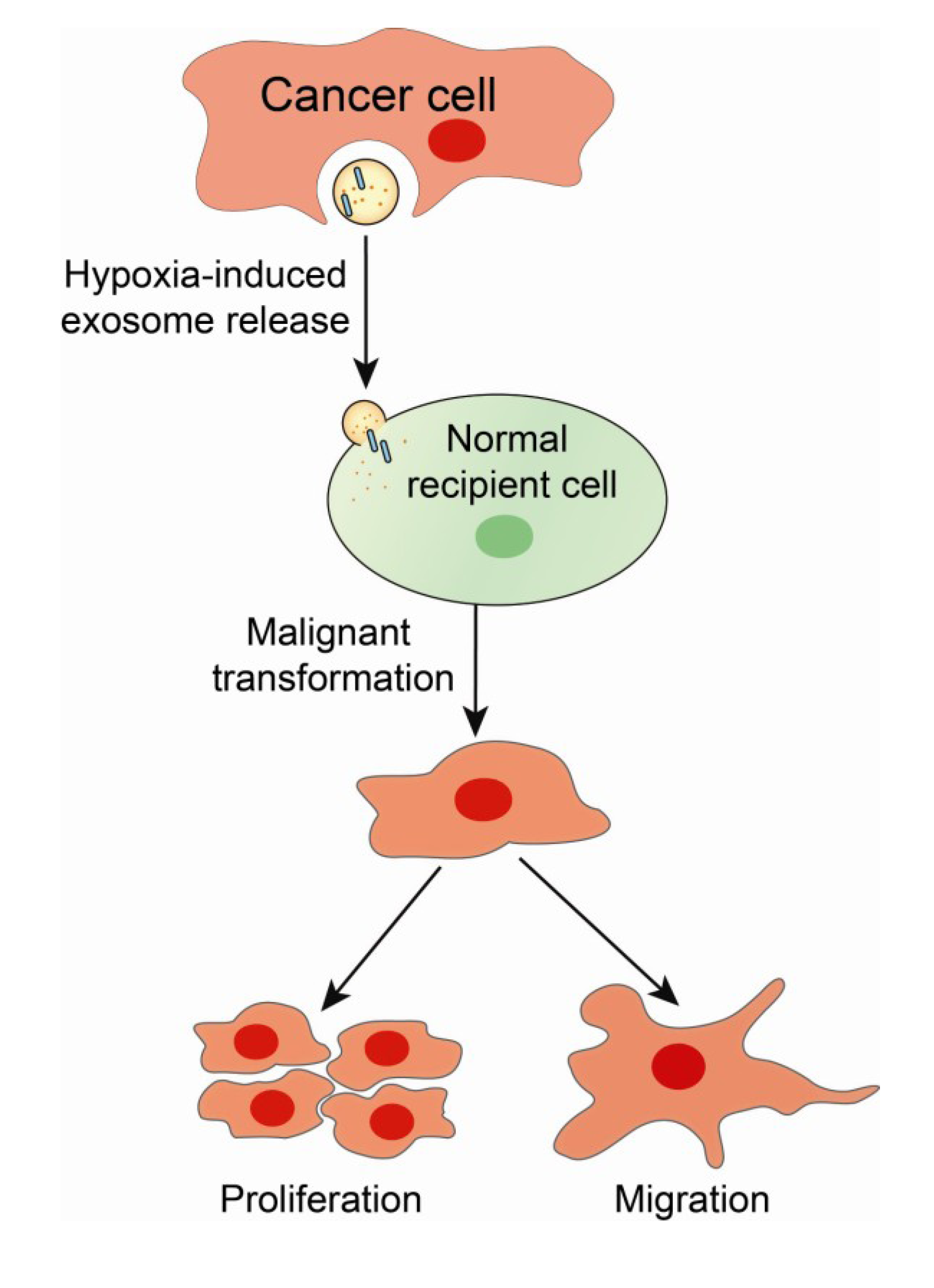
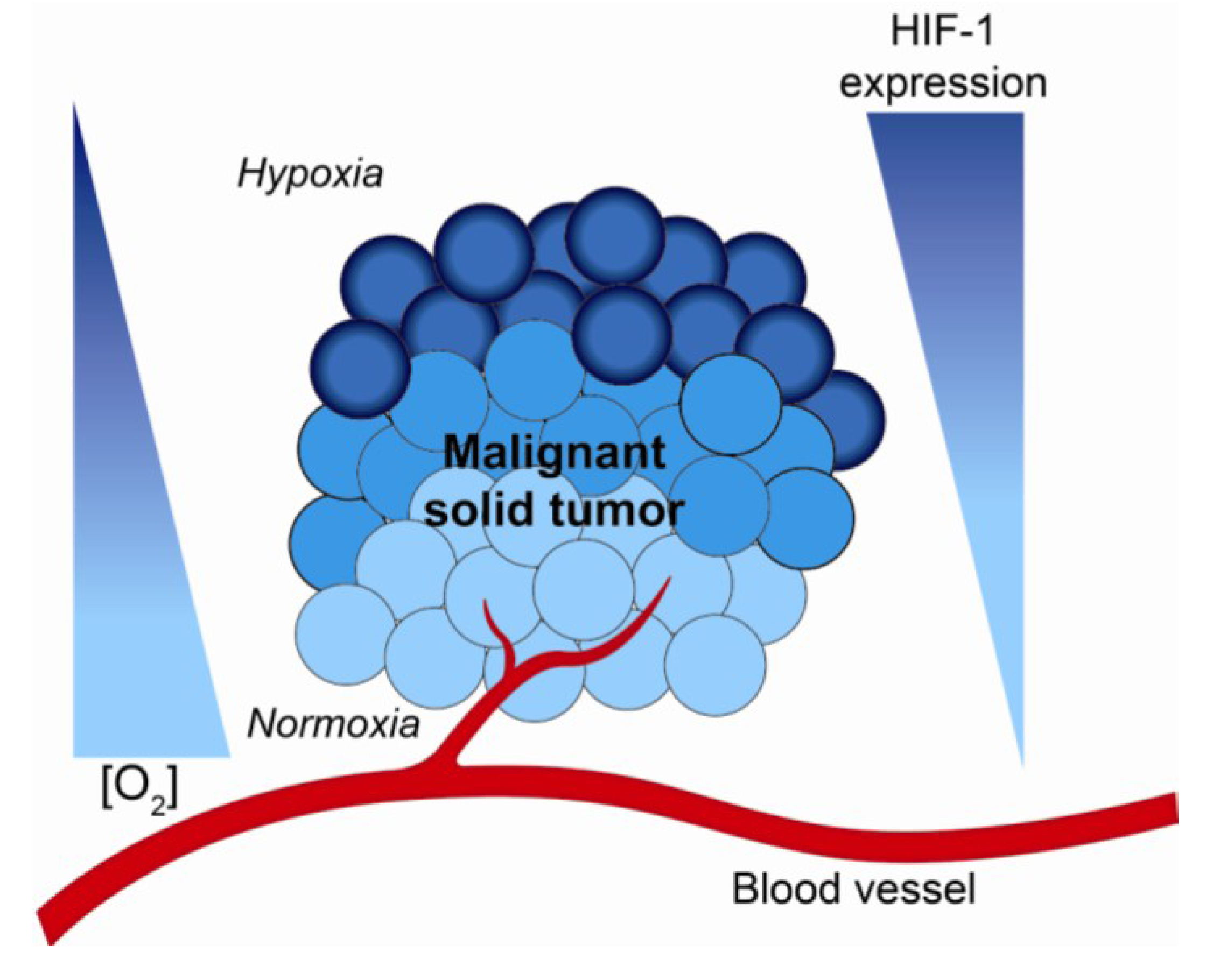
2. Hypoxia and the Tumor Microenvironment—Mechanisms of Radiation Resistance
3. Morphological Characteristics of Exosomes
4. Isolation of Exosomes

5. Exosomes as Biological Effectors and Carriers of Oncogenic Signatures in Cancer
6. Proteomic Profiling of Exosomes
7. Proteomic Analysis of the Exosome Proteome for the Development of Biomarkers
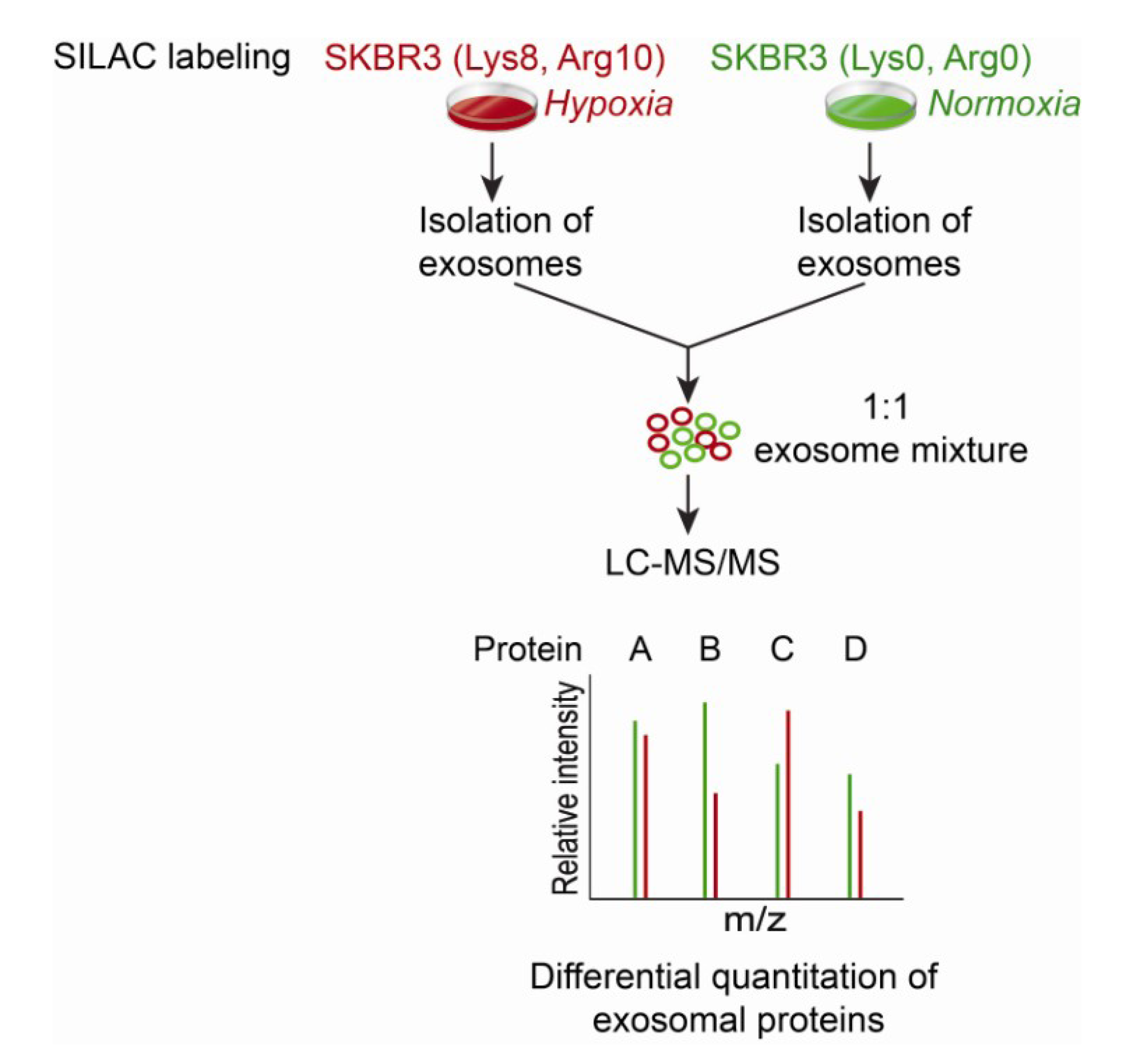
8. Targeted Proteomic Analysis of the Exosome Proteome
9. Proteomic Data Analysis
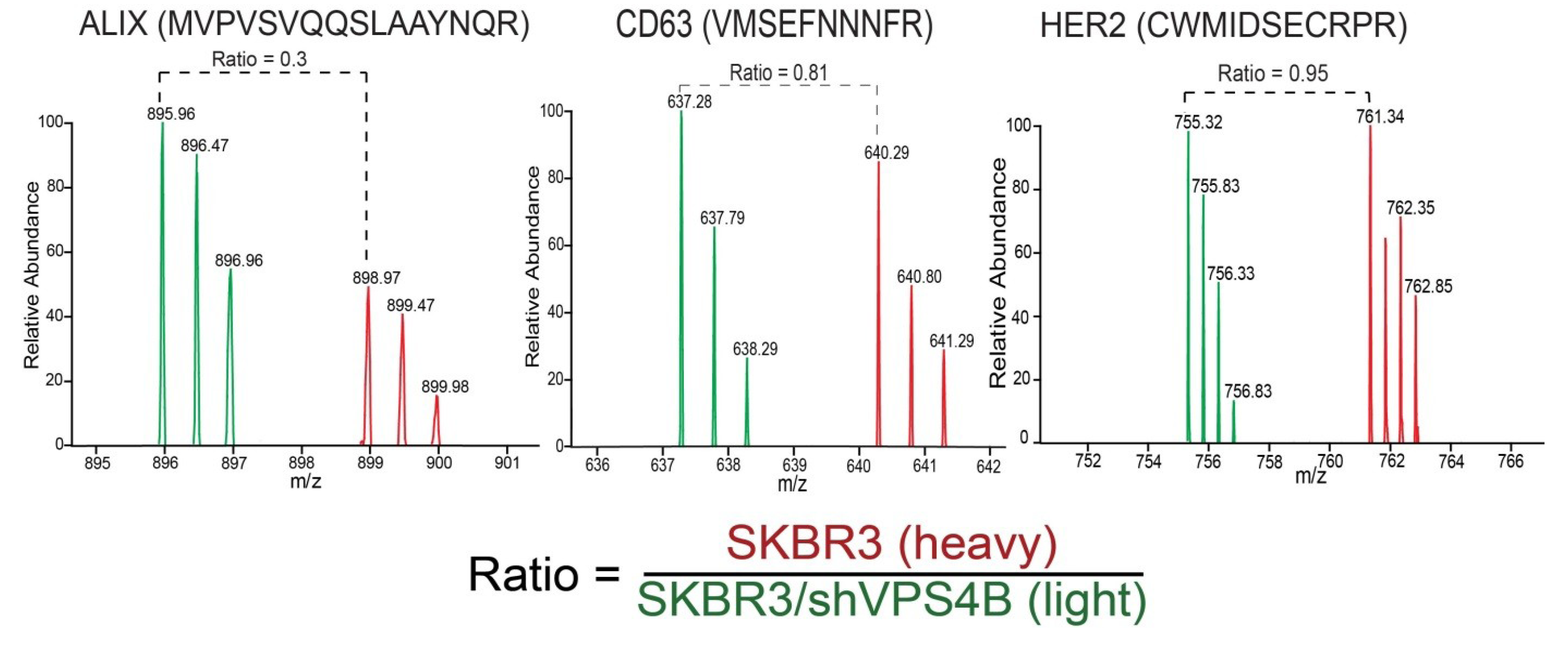


10. Conclusions
Acknowledgments
Conflicts of Interest
References
- Slamon, D.; Eiermann, W.; Robert, N.; Pienkowski, T.; Martin, M.; Press, M.; Mackey, J.; Glaspy, J.; Chan, A.; Pawlicki, M.; et al. Adjuvant trastuzumab in HER2-positive breast cancer. N. Engl. J. Med. 2011, 365, 1273–1283. [Google Scholar] [CrossRef]
- Vaupel, P.; Briest, S.; Hockel, M. Hypoxia in breast cancer: Pathogenesis, characterization and biological/therapeutic implications. Wien. Med. Wochenschr. 2002, 152, 334–342. [Google Scholar]
- Vaupel, P.; Mayer, A. Hypoxia in cancer: Significance and impact on clinical outcome. Cancer Metastasis Rev. 2007, 26, 225–239. [Google Scholar] [CrossRef]
- Kucharzewska, P.; Christianson, H.C.; Welch, J.E.; Svensson, K.J.; Fredlund, E.; Ringner, M.; Morgelin, M.; Bourseau-Guilmain, E.; Bengzon, J.; Belting, M. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc. Natl. Acad. Sci.USA 2013, 110, 7312–7317. [Google Scholar] [CrossRef]
- Garnier, D.; Jabado, N.; Rak, J. Extracellular vesicles as prospective carriers of oncogenic protein signatures in adult and paediatric brain tumours. Proteomics 2012, 13, 1595–1607. [Google Scholar] [CrossRef]
- Bellingham, S.A.; Guo, B.B.; Coleman, B.M.; Hill, A.F. Exosomes: Vehicles for the transfer of toxic proteins associated with neurodegenerative diseases? Front. Physiol. 2012, 3, e124. [Google Scholar]
- Yang, M.; Chen, J.; Su, F.; Yu, B.; Lin, L.; Liu, Y.; Huang, J.D.; Song, E. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol. Cancer 2011, 10, e117. [Google Scholar]
- Park, J.E.; Tan, H.S.; Datta, A.; Lai, R.C.; Zhang, H.; Meng, W.; Lim, S.K.; Sze, S.K. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol. Cell. Proteomics 2011, 9, 1085–1099. [Google Scholar]
- Subra, C.; Grand, D.; Laulagnier, K.; Stella, A.; Lambeau, G.; Paillasse, M.; de Medina, P.; Monsarrat, B.; Perret, B.; Silvente-Poirot, S.; et al. Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J. Lipid Res. 2010, 51, 2105–2120. [Google Scholar] [CrossRef]
- Taylor, D.D.; Gercel-Taylor, C. Tumour-derived exosomes and their role in cancer-associated T-cell signalling defects. Br. J. Cancer 2005, 92, 305–311. [Google Scholar]
- Azmi, A.S.; Bao, B.; Sarkar, F.H. Exosomes in cancer development, metastasis, and drug resistance: A comprehensive review. Cancer Metastasis Rev. 2013. [Google Scholar] [CrossRef]
- EL Andaloussi, S.; Mager, I.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef]
- Simona, F.; Laura, S.; Simona, T.; Riccardo, A. Contribution of proteomics to understanding the role of tumor-derived exosomes in cancer progression: State of the art and new perspectives. Proteomics 2013, 13, 1581–1594. [Google Scholar] [CrossRef]
- Martins, V.R.; Dias, M.S.; Hainaut, P. Tumor-cell-derived microvesicles as carriers of molecular information in cancer. Curr. Opin. Oncol. 2013, 25, 66–75. [Google Scholar] [CrossRef]
- Yu, X.; Harris, S.L.; Levine, A.J. The regulation of exosome secretion: A novel function of the p53 protein. Cancer Res. 2006, 66, 4795–4801. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Paine, M.S.; Brooks, A.M.; McCubrey, J.A.; Renegar, R.H.; Wang, R.; Terrian, D.M. Senescence-associated exosome release from human prostate cancer cells. Cancer Res. 2008, 68, 7864–7871. [Google Scholar] [CrossRef]
- Lespagnol, A.; Duflaut, D.; Beekman, C.; Blanc, L.; Fiucci, G.; Marine, J.C.; Vidal, M.; Amson, R.; Telerman, A. Exosome secretion, including the DNA damage-induced p53-dependent secretory pathway, is severely compromised in TSAP6/Steap3-null mice. Cell Death Differ. 2008, 15, 1723–1733. [Google Scholar] [CrossRef]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brugger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Savina, A.; Fader, C.M.; Damiani, M.T.; Colombo, M.I. Rab11 promotes docking and fusion of multivesicular bodies in a calcium-dependent manner. Traffic 2005, 6, 131–143. [Google Scholar] [CrossRef]
- Parolini, I.; Federici, C.; Raggi, C.; Lugini, L.; Palleschi, S.; de Milito, A.; Coscia, C.; Iessi, E.; Logozzi, M.; Molinari, A.; et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J. Biol. Chem. 2009, 284, 34211–34222. [Google Scholar] [CrossRef]
- Svensson, K.J.; Kucharzewska, P.; Christianson, H.C.; Skold, S.; Lofstedt, T.; Johansson, M.C.; Morgelin, M.; Bengzon, J.; Ruf, W.; Belting, M. Hypoxia triggers a proangiogenic pathway involving cancer cell microvesicles and PAR-2-mediated heparin-binding EGF signaling in endothelial cells. Proc. Natl. Acad. Sci. USA 2011, 108, 13147–13152. [Google Scholar] [CrossRef]
- Orriss, I.R.; Knight, G.E.; Utting, J.C.; Taylor, S.E.; Burnstock, G.; Arnett, T.R. Hypoxia stimulates vesicular ATP release from rat osteoblasts. J. Cell. Physiol. 2009, 220, 155–162. [Google Scholar] [CrossRef]
- Wysoczynski, M.; Ratajczak, M.Z. Lung cancer secreted microvesicles: Underappreciated modulators of microenvironment in expanding tumors. Int. J. Cancer 2009, 125, 1595–1603. [Google Scholar] [CrossRef]
- King, H.W.; Michael, M.Z.; Gleadle, J.M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 2012, 12, e421. [Google Scholar] [CrossRef]
- Simpson, R.J.; Jensen, S.S.; Lim, J.W. Proteomic profiling of exosomes: Current perspectives. Proteomics 2008, 8, 4083–4099. [Google Scholar] [CrossRef]
- Simpson, R.J.; Lim, J.W.; Moritz, R.L.; Mathivanan, S. Exosomes: Proteomic insights and diagnostic potential. Expert Rev. Proteomics 2009, 6, 267–283. [Google Scholar] [CrossRef]
- Ward, C.; Langdon, S.P.; Mullen, P.; Harris, A.L.; Harrison, D.J.; Supuran, C.T.; Kunkler, I.H. New strategies for targeting the hypoxic tumour microenvironment in breast cancer. Cancer Treat. Rev. 2013, 39, 171–179. [Google Scholar] [CrossRef]
- Gatenby, R.A.; Smallbone, K.; Maini, P.K.; Rose, F.; Averill, J.; Nagle, R.B.; Worrall, L.; Gillies, R.J. Cellular adaptations to hypoxia and acidosis during somatic evolution of breast cancer. Br. J. Cancer 2007, 97, 646–653. [Google Scholar] [CrossRef]
- Bristow, R.G.; Hill, R.P. Hypoxia and metabolism: Hypoxia, DNA repair and genetic instability. Nat. Rev. Cancer 2008, 8, 180–192. [Google Scholar] [CrossRef]
- Thomlinson, R.H.; Gray, L.H. The histological structure of some human lung cancers and the possible implications for radiotherapy. Br. J. Cancer 1955, 9, 539–549. [Google Scholar] [CrossRef]
- Bindra, R.S.; Crosby, M.E.; Glazer, P.M. Regulation of DNA repair in hypoxic cancer cells. Cancer Metastasis Rev. 2007, 26, 249–260. [Google Scholar] [CrossRef]
- Yoshimura, M.; Itasaka, S.; Harada, H.; Hiraoka, M. Microenvironment and radiation therapy. Biomed. Res. Int. 2013, 2013, e685308. [Google Scholar]
- Harada, H. How can we overcome tumor hypoxia in radiation therapy? J. Radiat. Res. 2011, 52, 545–556. [Google Scholar] [CrossRef]
- Brown, J.M. Exploiting the hypoxic cancer cell: Mechanisms and therapeutic strategies. Mol. Med. Today 2000, 6, 157–162. [Google Scholar] [CrossRef]
- Brown, J.M.; Wilson, W.R. Exploiting tumour hypoxia in cancer treatment. Nat. Rev. Cancer 2004, 4, 437–447. [Google Scholar] [CrossRef]
- Ren, Y.; Hao, P.; Dutta, B.; Cheow, E.S.; Sim, K.H.; Gan, C.S.; Lim, S.K.; Sze, S.K. Hypoxia modulates A431 cellular pathways association to tumor radioresistance and enhanced migration revealed by comprehensive proteomic and functional studies. Mol. Cell. Proteomics 2013, 12, 485–498. [Google Scholar] [CrossRef]
- Sirbu, B.M.; Cortez, D. DNA damage response: Three levels of DNA repair regulation. Cold Spring Harb. Perspect. Biol. 2013. [Google Scholar] [CrossRef]
- Lin, H.H.; Li, X.; Chen, J.L.; Sun, X.; Cooper, F.N.; Chen, Y.R.; Zhang, W.; Chung, Y.; Li, A.; Cheng, C.T.; et al. Identification of an AAA ATPase VPS4B-dependent pathway that modulates epidermal growth factor receptor abundance and signaling during hypoxia. Mol. Cell. Biol. 2012, 32, 1124–1138. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, J.; Wang, X.; Li, Y.; Chen, Y.; Li, K.; Zhang, J.; Yao, L.; Guo, G. HIF-1 and NDRG2 contribute to hypoxia-induced radioresistance of cervical cancer Hela cells. Exp. Cell. Res. 2010, 316, 1985–1993. [Google Scholar] [CrossRef]
- Moeller, B.J.; Dewhirst, M.W. HIF-1 and tumour radiosensitivity. Br. J. Cancer 2006, 95, 1–5. [Google Scholar] [CrossRef]
- Kaidar-Person, O.; Lai, C.; Kuten, A.; Belkacemi, Y. “The Infinite Maze” of breast cancer, signaling pathways and radioresistance. Breast 2013, 22, 411–418. [Google Scholar] [CrossRef]
- Grosso, S.; Doyen, J.; Parks, S.K.; Bertero, T.; Paye, A.; Cardinaud, B.; Gounon, P.; Lacas-Gervais, S.; Noel, A.; Pouyssegur, J.; et al. MiR-210 promotes a hypoxic phenotype and increases radioresistance in human lung cancer cell lines. Cell Death Dis. 2013, 4, e544. [Google Scholar] [CrossRef]
- Potiron, V.A.; Abderrhamani, R.; Giang, E.; Chiavassa, S.; di Tomaso, E.; Maira, S.M.; Paris, F.; Supiot, S. Radiosensitization of prostate cancer cells by the dual PI3K/mTOR inhibitor BEZ235 under normoxic and hypoxic conditions. Radiother. Oncol. 2013, 106, 138–146. [Google Scholar] [CrossRef]
- Tamara Marie-Egyptienne, D.; Lohse, I.; Hill, R.P. Cancer stem cells, the epithelial to mesenchymal transition (EMT) and radioresistance: Potential role of hypoxia. Cancer Lett. 2012, in press. [Google Scholar]
- Eldh, M.; Ekstrom, K.; Valadi, H.; Sjostrand, M.; Olsson, B.; Jernas, M.; Lotvall, J. Exosomes communicate protective messages during oxidative stress; possible role of exosomal shuttle RNA. PLoS One 2010, 5, e15353. [Google Scholar]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta 2012, 1820, 940–948. [Google Scholar]
- Thery, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar]
- Conde-Vancells, J.; Rodriguez-Suarez, E.; Embade, N.; Gil, D.; Matthiesen, R.; Valle, M.; Elortza, F.; Lu, S.C.; Mato, J.M.; Falcon-Perez, J.M. Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. J. Proteome Res. 2008, 7, 5157–5166. [Google Scholar] [CrossRef]
- Wubbolts, R.; Leckie, R.S.; Veenhuizen, P.T.; Schwarzmann, G.; Mobius, W.; Hoernschemeyer, J.; Slot, J.W.; Geuze, H.J.; Stoorvogel, W. Proteomic and biochemical analyses of human B cell-derived exosomes. J. Biol. Chem. 2003, 278, 10963–10972. [Google Scholar] [CrossRef]
- Balaj, L.; Lessard, R.; Dai, L.; Cho, Y.J.; Pomeroy, S.L.; Breakefield, X.O.; Skog, J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011, 2, e180. [Google Scholar] [CrossRef]
- Gibbings, D.J.; Ciaudo, C.; Erhardt, M.; Voinnet, O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009, 11, 1143–1149. [Google Scholar] [CrossRef]
- Lee, Y.S.; Shibata, Y.; Malhotra, A.; Dutta, A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009, 23, 2639–2649. [Google Scholar] [CrossRef]
- Haussecker, D.; Huang, Y.; Lau, A.; Parameswaran, P.; Fire, A.Z.; Kay, M.A. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 2010, 16, 673–695. [Google Scholar] [CrossRef]
- Thomas, S.N.; Wan, Y.; Liao, Z.; Hanson, P.I.; Yang, A.J. Stable isotope labeling with amino acids in cell culture based mass spectrometry approach to detect transient protein interactions using substrate trapping. Anal. Chem. 2011, 83, 5511–5518. [Google Scholar] [CrossRef]
- Liao, Z.; Thomas, S.N.; Wan, Y.; Lin, H.H.; Ann, D.K.; Yang, A.J. An internal standard-assisted synthesis and degradation proteomic approach reveals the potential linkage between VPS4B depletion and activation of fatty acid beta-oxidation in breast cancer cells. Int. J. Proteomics 2013, 2013, e291415. [Google Scholar]
- Battke, C.; Ruiss, R.; Welsch, U.; Wimberger, P.; Lang, S.; Jochum, S.; Zeidler, R. Tumour exosomes inhibit binding of tumour-reactive antibodies to tumour cells and reduce ADCC. Cancer Immunol. Immunother. 2011, 60, 639–648. [Google Scholar] [CrossRef]
- Ciravolo, V.; Huber, V.; Ghedini, G.C.; Venturelli, E.; Bianchi, F.; Campiglio, M.; Morelli, D.; Villa, A.; Della Mina, P.; Menard, S.; et al. Potential role of HER2-overexpressing exosomes in countering trastuzumab-based therapy. J. Cell. Physiol. 2012, 227, 658–667. [Google Scholar] [CrossRef]
- Thery, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006. [Google Scholar] [CrossRef]
- Thery, C.; Boussac, M.; Veron, P.; Ricciardi-Castagnoli, P.; Raposo, G.; Garin, J.; Amigorena, S. Proteomic analysis of dendritic cell-derived exosomes: A secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 2001, 166, 7309–7318. [Google Scholar]
- Thery, C.; Regnault, A.; Garin, J.; Wolfers, J.; Zitvogel, L.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Molecular characterization of dendritic cell-derived exosomes. J. Cell Biol. 1999, 147, 599–610. [Google Scholar] [CrossRef]
- Cheruvanky, A.; Zhou, H.; Pisitkun, T.; Kopp, J.B.; Knepper, M.A.; Yuen, P.S.; Star, R.A. Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. Am. J. Physiol. Renal Physiol. 2007, 292, F1657–F1661. [Google Scholar] [CrossRef]
- de Bock, K.; Mazzone, M.; Carmeliet, P. Antiangiogenic therapy, hypoxia, and metastasis: Risky liaisons, or not? Nat. Rev. Clin. Oncol. 2011, 8, 393–404. [Google Scholar] [CrossRef]
- Marleau, A.M.; Chen, C.S.; Joyce, J.A.; Tullis, R.H. Exosome removal as a therapeutic adjuvant in cancer. J. Transl. Med. 2012, 10, e134. [Google Scholar] [CrossRef]
- Koga, K.; Matsumoto, K.; Akiyoshi, T.; Kubo, M.; Yamanaka, N.; Tasaki, A.; Nakashima, H.; Nakamura, M.; Kuroki, S.; Tanaka, M.; et al. Purification, characterization and biological significance of tumor-derived exosomes. Anticancer Res. 2005, 25, 3703–3707. [Google Scholar]
- Nahta, R.; Yu, D.; Hung, M.C.; Hortobagyi, G.N.; Esteva, F.J. Mechanisms of disease: Understanding resistance to HER2-targeted therapy in human breast cancer. Nat. Clin. Pract. Oncol. 2006, 3, 269–280. [Google Scholar] [CrossRef]
- Abdel-Razeq, H.; Marei, L. Current neoadjuvant treatment options for HER2-positive breast cancer. Biologics 2011, 5, 87–94. [Google Scholar]
- von Minckwitz, G.; Loibl, S.; Untch, M. What is the current standard of care for anti-HER2 neoadjuvant therapy in breast cancer? Oncology 2012, 26, 20–26. [Google Scholar]
- Aung, T.; Chapuy, B.; Vogel, D.; Wenzel, D.; Oppermann, M.; Lahmann, M.; Weinhage, T.; Menck, K.; Hupfeld, T.; Koch, R.; et al. Exosomal evasion of humoral immunotherapy in aggressive B-cell lymphoma modulated by ATP-binding cassette transporter A3. Proc. Natl. Acad. Sci. USA 2011, 108, 15336–15341. [Google Scholar] [CrossRef]
- Safaei, R.; Larson, B.J.; Cheng, T.C.; Gibson, M.A.; Otani, S.; Naerdemann, W.; Howell, S.B. Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells. Mol. Cancer Ther. 2005, 4, 1595–1604. [Google Scholar] [CrossRef]
- Chen, K.G.; Valencia, J.C.; Lai, B.; Zhang, G.; Paterson, J.K.; Rouzaud, F.; Berens, W.; Wincovitch, S.M.; Garfield, S.H.; Leapman, R.D.; et al. Melanosomal sequestration of cytotoxic drugs contributes to the intractability of malignant melanomas. Proc. Natl. Acad. Sci. USA 2006, 103, 9903–9907. [Google Scholar] [CrossRef]
- Shedden, K.; Xie, X.T.; Chandaroy, P.; Chang, Y.T.; Rosania, G.R. Expulsion of small molecules in vesicles shed by cancer cells: Association with gene expression and chemosensitivity profiles. Cancer Res. 2003, 63, 4331–4337. [Google Scholar]
- Choi, D.S.; Lee, J.M.; Park, G.W.; Lim, H.W.; Bang, J.Y.; Kim, Y.K.; Kwon, K.H.; Kwon, H.J.; Kim, K.P.; Gho, Y.S. Proteomic analysis of microvesicles derived from human colorectal cancer cells. J. Proteome Res. 2007, 6, 4646–4655. [Google Scholar] [CrossRef]
- Ji, H.; Greening, D.W.; Barnes, T.W.; Lim, J.W.; Tauro, B.J.; Rai, A.; Xu, R.; Adda, C.; Mathivanan, S.; Zhao, W.; et al. Proteome profiling of exosomes derived from human primary and metastatic colorectal cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics 2013, 13, 1672–1686. [Google Scholar] [CrossRef]
- Wolfers, J.; Lozier, A.; Raposo, G.; Regnault, A.; Thery, C.; Masurier, C.; Flament, C.; Pouzieux, S.; Faure, F.; Tursz, T.; et al. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat. Med. 2001, 7, 297–303. [Google Scholar] [CrossRef]
- Craven, R.A.; Totty, N.; Harnden, P.; Selby, P.J.; Banks, R.E. Laser capture microdissection and two-dimensional polyacrylamide gel electrophoresis: Evaluation of tissue preparation and sample limitations. Am. J. Pathol. 2002, 160, 815–822. [Google Scholar] [CrossRef]
- Marton, A.; Vizler, C.; Kusz, E.; Temesfoi, V.; Szathmary, Z.; Nagy, K.; Szegletes, Z.; Varo, G.; Siklos, L.; Katona, R.L.; et al. Melanoma cell-derived exosomes alter macrophage and dendritic cell functions in vitro. Immunol. Lett. 2012, 148, 34–38. [Google Scholar] [CrossRef]
- Hegmans, J.P.; Bard, M.P.; Hemmes, A.; Luider, T.M.; Kleijmeer, M.J.; Prins, J.B.; Zitvogel, L.; Burgers, S.A.; Hoogsteden, H.C.; Lambrecht, B.N. Proteomic analysis of exosomes secreted by human mesothelioma cells. Am. J. Pathol. 2004, 164, 1807–1815. [Google Scholar] [CrossRef]
- Graner, M.W.; Alzate, O.; Dechkovskaia, A.M.; Keene, J.D.; Sampson, J.H.; Mitchell, D.A.; Bigner, D.D. Proteomic and immunologic analyses of brain tumor exosomes. FASEB J. 2009, 23, 1541–1557. [Google Scholar] [CrossRef]
- Caby, M.P.; Lankar, D.; Vincendeau-Scherrer, C.; Raposo, G.; Bonnerot, C. Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 2005, 17, 879–887. [Google Scholar] [CrossRef]
- Looze, C.; Yui, D.; Leung, L.; Ingham, M.; Kaler, M.; Yao, X.; Wu, W.W.; Shen, R.F.; Daniels, M.P.; Levine, S.J. Proteomic profiling of human plasma exosomes identifies PPARgamma as an exosome-associated protein. Biochem. Biophys. Res. Commun. 2009, 378, 433–438. [Google Scholar] [CrossRef]
- Sabapatha, A.; Gercel-Taylor, C.; Taylor, D.D. Specific isolation of placenta-derived exosomes from the circulation of pregnant women and their immunoregulatory consequences. Am. J. Reprod. Immunol. 2006, 56, 345–355. [Google Scholar] [CrossRef]
- Gonzales, P.A.; Pisitkun, T.; Hoffert, J.D.; Tchapyjnikov, D.; Star, R.A.; Kleta, R.; Wang, N.S.; Knepper, M.A. Large-scale proteomics and phosphoproteomics of urinary exosomes. J. Am. Soc. Nephrol. 2009, 20, 363–379. [Google Scholar] [CrossRef]
- Nilsson, J.; Skog, J.; Nordstrand, A.; Baranov, V.; Mincheva-Nilsson, L.; Breakefield, X.O.; Widmark, A. Prostate cancer-derived urine exosomes: A novel approach to biomarkers for prostate cancer. Br. J. Cancer 2009, 100, 1603–1607. [Google Scholar] [CrossRef]
- Gonzalez-Begne, M.; Lu, B.; Han, X.; Hagen, F.K.; Hand, A.R.; Melvin, J.E.; Yates, J.R. Proteomic analysis of human parotid gland exosomes by multidimensional protein identification technology (MudPIT). J. Proteome Res. 2009, 8, 1304–1314. [Google Scholar] [CrossRef]
- Andre, F.; Schartz, N.E.; Movassagh, M.; Flament, C.; Pautier, P.; Morice, P.; Pomel, C.; Lhomme, C.; Escudier, B.; Le Chevalier, T.; et al. Malignant effusions and immunogenic tumour-derived exosomes. Lancet 2002, 360, 295–305. [Google Scholar] [CrossRef]
- Admyre, C.; Johansson, S.M.; Qazi, K.R.; Filen, J.J.; Lahesmaa, R.; Norman, M.; Neve, E.P.; Scheynius, A.; Gabrielsson, S. Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 2007, 179, 1969–1978. [Google Scholar]
- Ong, S.E.; Blagoev, B.; Kratchmarova, I.; Kristensen, D.B.; Steen, H.; Pandey, A.; Mann, M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 2002, 1, 376–386. [Google Scholar] [CrossRef]
- Liao, Z.; Wan, Y.; Thomas, S.N.; Yang, A.J. IsoQuant: A software tool for stable isotope labeling by amino acids in cell culture-based mass spectrometry quantitation. Anal. Chem. 2012, 84, 4535–4543. [Google Scholar] [CrossRef]
- Aebersold, R.; Mann, M. Mass spectrometry-based proteomics. Nature 2003, 422, 198–207. [Google Scholar] [CrossRef]
- Picotti, P.; Aebersold, R. Selected reaction monitoring-based proteomics: Workflows, potential, pitfalls and future directions. Nat. Methods 2012, 9, 555–566. [Google Scholar] [CrossRef]
- Anderson, L.; Hunter, C.L. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteomics 2006, 5, 573–588. [Google Scholar]
- Lange, V.; Picotti, P.; Domon, B.; Aebersold, R. Selected reaction monitoring for quantitative proteomics: A tutorial. Mol. Syst. Biol. 2008, 4, e222. [Google Scholar]
- Yost, R.A.; Enke, C.G. Triple quadrupole mass spectrometry for direct mixture analysis and structure elucidation. Anal. Chem. 1979, 51, 1251–1264. [Google Scholar] [CrossRef]
- Gillette, M.A.; Carr, S.A. Quantitative analysis of peptides and proteins in biomedicine by targeted mass spectrometry. Nat. Methods 2013, 10, 28–34. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Vaudel, M.; Sickmann, A.; Martens, L. Current methods for global proteome identification. Expert Rev. Proteomics 2012, 9, 519–532. [Google Scholar] [CrossRef]
- Eng, J.K.; Searle, B.C.; Clauser, K.R.; Tabb, D.L. A face in the crowd: Recognizing peptides through database search. Mol. Cell. Proteomics 2011, 10, R111.009522. [Google Scholar]
- Matthiesen, R.; Carvalho, A.S. Methods and algorithms for relative quantitative proteomics by mass spectrometry. Methods Mol. Biol. 2010, 593, 187–204. [Google Scholar] [CrossRef]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef]
- Hurley, J.H.; Odorizzi, G. Get on the exosome bus with ALIX. Nat. Cell Biol. 2012, 14, 654–655. [Google Scholar] [CrossRef]
- Old, W.M.; Meyer-Arendt, K.; Aveline-Wolf, L.; Pierce, K.G.; Mendoza, A.; Sevinsky, J.R.; Resing, K.A.; Ahn, N.G. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 2005, 4, 1487–1502. [Google Scholar] [CrossRef]
- Ross, P.L.; Huang, Y.N.; Marchese, J.N.; Williamson, B.; Parker, K.; Hattan, S.; Khainovski, N.; Pillai, S.; Dey, S.; Daniels, S.; et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 2004, 3, 1154–1169. [Google Scholar] [CrossRef]
- Thompson, A.; Schafer, J.; Kuhn, K.; Kienle, S.; Schwarz, J.; Schmidt, G.; Neumann, T.; Johnstone, R.; Mohammed, A.K.; Hamon, C. Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 2003, 75, 1895–1904. [Google Scholar] [CrossRef]
- Li, Z.; Adams, R.M.; Chourey, K.; Hurst, G.B.; Hettich, R.L.; Pan, C. Systematic comparison of label-free, metabolic labeling, and isobaric chemical labeling for quantitative proteomics on LTQ Orbitrap Velos. J. Proteome Res. 2012, 11, 1582–1590. [Google Scholar] [CrossRef]
- Chrysogelos, S.A.; Dickson, R.B. EGF receptor expression, regulation, and function in breast cancer. Breast Cancer Res. Treat. 1994, 29, 29–40. [Google Scholar] [CrossRef]
- Chrysogelos, S.A.; Yarden, R.I.; Lauber, A.H.; Murphy, J.M. Mechanisms of EGF receptor regulation in breast cancer cells. Breast Cancer Res. Treat. 1994, 31, 227–236. [Google Scholar] [CrossRef]
- Danielsen, A.J.; Maihle, N.J. The EGF/ErbB receptor family and apoptosis. Growth Factors 2002, 20, 1–15. [Google Scholar] [CrossRef]
- Bucci, B.; D'Agnano, I.; Botti, C.; Mottolese, M.; Carico, E.; Zupi, G.; Vecchione, A. EGF-R expression in ductal breast cancer: Proliferation and prognostic implications. Anticancer Res. 1997, 17, 769–774. [Google Scholar]
- Buchholz, T.A.; Tu, X.; Ang, K.K.; Esteva, F.J.; Kuerer, H.M.; Pusztai, L.; Cristofanilli, M.; Singletary, S.E.; Hortobagyi, G.N.; Sahin, A.A. Epidermal growth factor receptor expression correlates with poor survival in patients who have breast carcinoma treated with doxorubicin-based neoadjuvant chemotherapy. Cancer 2005, 104, 676–681. [Google Scholar] [CrossRef]
- Navolanic, P.M.; Steelman, L.S.; McCubrey, J.A. EGFR family signaling and its association with breast cancer development and resistance to chemotherapy. Int. J. Oncol. 2003, 22, 237–252. [Google Scholar]
- Peng, X.H.; Karna, P.; Cao, Z.; Jiang, B.H.; Zhou, M.; Yang, L. Cross-talk between epidermal growth factor receptor and hypoxia-inducible factor-1alpha signal pathways increases resistance to apoptosis by up-regulating survivin gene expression. J. Biol. Chem. 2006, 281, 25903–25914. [Google Scholar]
- Tu, C.; Ortega-Cava, C.F.; Winograd, P.; Stanton, M.J.; Reddi, A.L.; Dodge, I.; Arya, R.; Dimri, M.; Clubb, R.J.; Naramura, M.; et al. Endosomal-sorting complexes required for transport (ESCRT) pathway-dependent endosomal traffic regulates the localization of active Src at focal adhesions. Proc. Natl. Acad. Sci. USA 2010, 107, 16107–16112. [Google Scholar] [CrossRef]
- Tu, C.; Ahmad, G.; Mohapatra, B.; Bhattacharyya, S.; Ortega-Cava, C.F.; Chung, B.M.; Wagner, K.U.; Raja, S.M.; Naramura, M.; Band, V.; et al. ESCRT proteins: Double-edged regulators of cellular signaling. Bioarchitecture 2011, 1, 45–48. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Thomas, S.N.; Liao, Z.; Clark, D.; Chen, Y.; Samadani, R.; Mao, L.; Ann, D.K.; Baulch, J.E.; Shapiro, P.; Yang, A.J. Exosomal Proteome Profiling: A Potential Multi-Marker Cellular Phenotyping Tool to Characterize Hypoxia-Induced Radiation Resistance in Breast Cancer. Proteomes 2013, 1, 87-108. https://doi.org/10.3390/proteomes1020087
Thomas SN, Liao Z, Clark D, Chen Y, Samadani R, Mao L, Ann DK, Baulch JE, Shapiro P, Yang AJ. Exosomal Proteome Profiling: A Potential Multi-Marker Cellular Phenotyping Tool to Characterize Hypoxia-Induced Radiation Resistance in Breast Cancer. Proteomes. 2013; 1(2):87-108. https://doi.org/10.3390/proteomes1020087
Chicago/Turabian StyleThomas, Stefani N., Zhongping Liao, David Clark, Yangyi Chen, Ramin Samadani, Li Mao, David K. Ann, Janet E. Baulch, Paul Shapiro, and Austin J. Yang. 2013. "Exosomal Proteome Profiling: A Potential Multi-Marker Cellular Phenotyping Tool to Characterize Hypoxia-Induced Radiation Resistance in Breast Cancer" Proteomes 1, no. 2: 87-108. https://doi.org/10.3390/proteomes1020087
APA StyleThomas, S. N., Liao, Z., Clark, D., Chen, Y., Samadani, R., Mao, L., Ann, D. K., Baulch, J. E., Shapiro, P., & Yang, A. J. (2013). Exosomal Proteome Profiling: A Potential Multi-Marker Cellular Phenotyping Tool to Characterize Hypoxia-Induced Radiation Resistance in Breast Cancer. Proteomes, 1(2), 87-108. https://doi.org/10.3390/proteomes1020087



