Leveraging H3Africa Scholarly Publications for Technology-Enhanced Personalized Bioinformatics Education
Abstract
:1. Introduction
2. Materials and Methods
2.1. Recoding and Reshaping Datasets from an H3Africa Scholarly Article
2.2. Design of Visual Analytics Resources for Personalized Learning of Bioinformatics Knowledge
2.3. Evaluation of Visual Analytics Resources
3. Results
3.1. Curated Datasets and Visual Analytics Resources
- Datasets and visual analytics resources were produced from the content of the H3Africa PubMed Central (PMC) article PMC7759466. A Microsoft Excel file contained 25 datasets that were curated (recoded or reshaped) from the supplementary file of the H3Africa publication. We disseminated the three Tableau packaged workbook files via the Tableau Public website and the GitHub website. The supplementary materials section of this article contains details of the datasets and visual analytics resources. The first visual analytics file (pmc7759466_analytics.tbwx) contained a worksheet, dashboard and story designed for interacting with (1) statements from the PubMed Central article PMC7759466; (2) captions of figures in PMC7759466; (3) PMC7759466 Supplementary Tables 6–8, 19, 20 and 23; and (4) learning principles, namely five content types, adventure learning, data investigation cycle and knowledge visualization. The second visual analytics file (pmc7759466_data_investigations.tbwx) contained the designs for interacting with a list of datasets as well as the contents of (1) two supplementary methods; (2) Supplementary Tables 1–8; and (3) Supplementary Table 19. In some designs, additional fields were added from the content of a data field. In the third visual analytics file (pmc7759466_suppT19_variants.tbwx), the focus was on designs to facilitate the personalized learning of the concept of consequence of sequence variants (e.g., Single-Nucleotide Polymorphisms). Prior to the design in Tableau Desktop, a Tableau Prep flow file (pmc7759466_suppT19_variants.tlfx) was set up to construct a dataset from the Annotation data field in Supplementary Table 19 of PMC7759466. The values in the Ensembl database variables provided in the Annotation field included allele letter, gene identifier, transcript identifier, gene symbol, variant consequences and variant impacts.
3.2. Use Cases of Customization of Instruction and Individualization of Learning of Bioinformatics Content Using H3Africa Datasets
- Principles of Customization Design and Individualization Design in Personalized Educational Technology. The principles of customization and individualization design are based on the framework of personalized education by Kucirkova et al. [8]. In customization design, the system supports the individual’s choices of the content to be learned, while in the individualization design, the individual makes the choice. Within the visual analytics resource, the customization design is performed first by the instructor, and subsequently, the learner performs the individualization design.
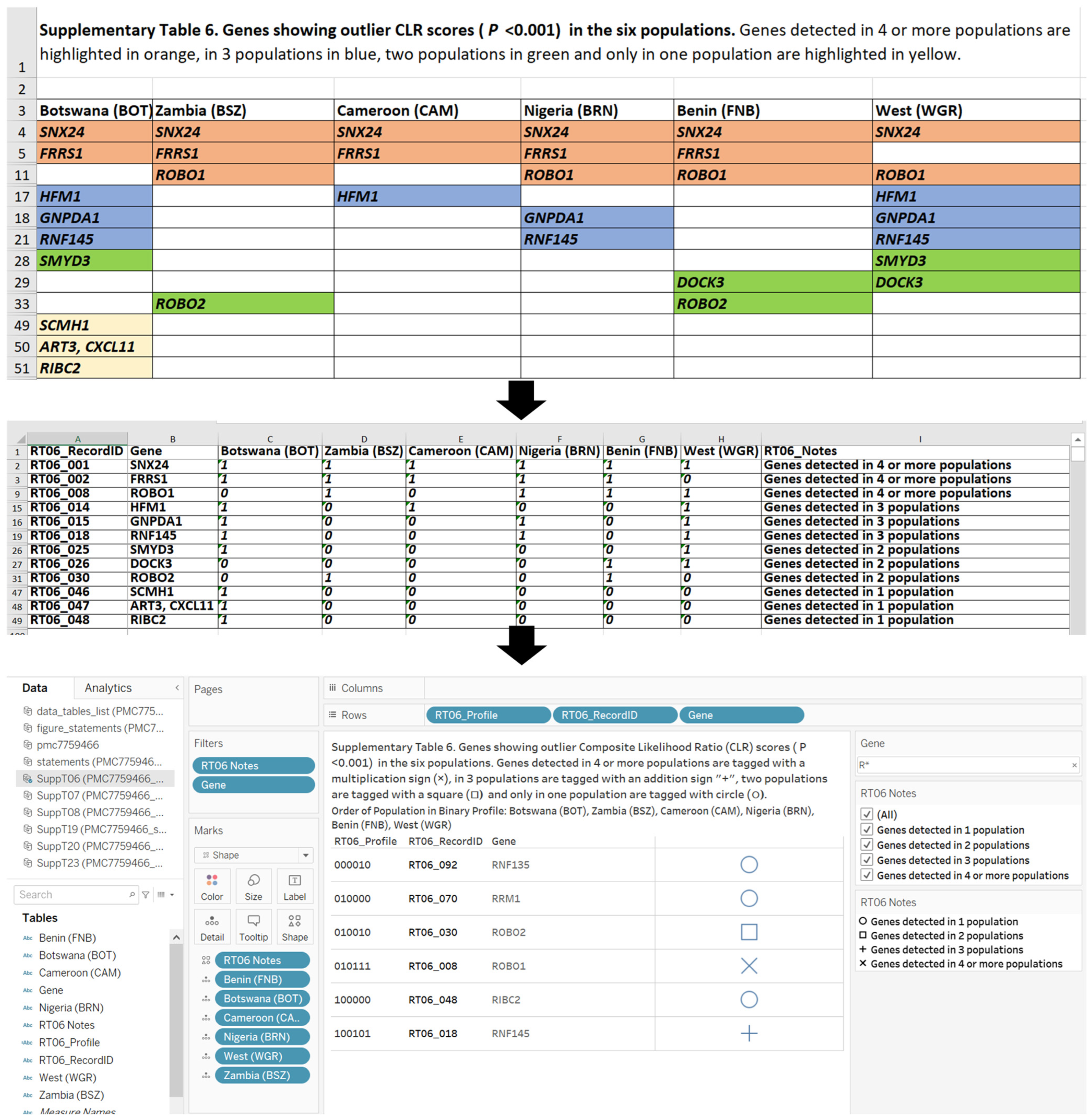
- Customization Design Objective and Results. The objective of the customization design of the visual analytics resource was to present the learner with options to identify key genes contributing to the adaptive evolution of functions in populations as detected in the Composite Likelihood Ratio (CLR) statistic score [40,52]. The data sources were the recoded datasets from Supplementary Tables 6 and 8 of publication PMC7759466. The dashboard design (Figure 2) provides the learner with choices of population and Gene Ontology (GO) function terms as well as connection to the National Center for Biotechnology Information (NCBI) database search. The population groups provide geographic relevance for learning about the key genes. The customization design contains filters that support the choice of six population groups (Zambia, West, Nigeria, Cameroon, Botswana and Benin) and five gene ontology function terms (Immunity, Reproduction, Metabolism, DNA repair and Other).
- Individualization Design Objective and Results. The top image of Figure 2 is an example of individualization, when the learner enters the gene symbol RNF135 in the NCBI search feature and returns 21 databases. The bottom image in Figure 2 is the result when the learner interacts with the visual analytics resource in the top image of Figure 2 and makes the choice to display key genes relevant to a population (BRN) in Nigeria. The result of the interaction includes two GO terms. The learner is interested in the pancreatic lipase gene (PNLIP) based on the NCBI search.
3.3. A Use Case of Adaptation in Personalized Learning by Collecting Performance Metrics during Actions in Visual Analytics Software
- Principle of Adaptation Design in Personalized Educational Technology. In adaptation design in personalized education technology, the system makes the choice based on data patterns such as repeated behaviors and performance measures [8]. The adaptation design can have a diagnostic function to optimize the learning transaction strategies of the learner. The adaptation design requires the completion of both customization and individualization activities.
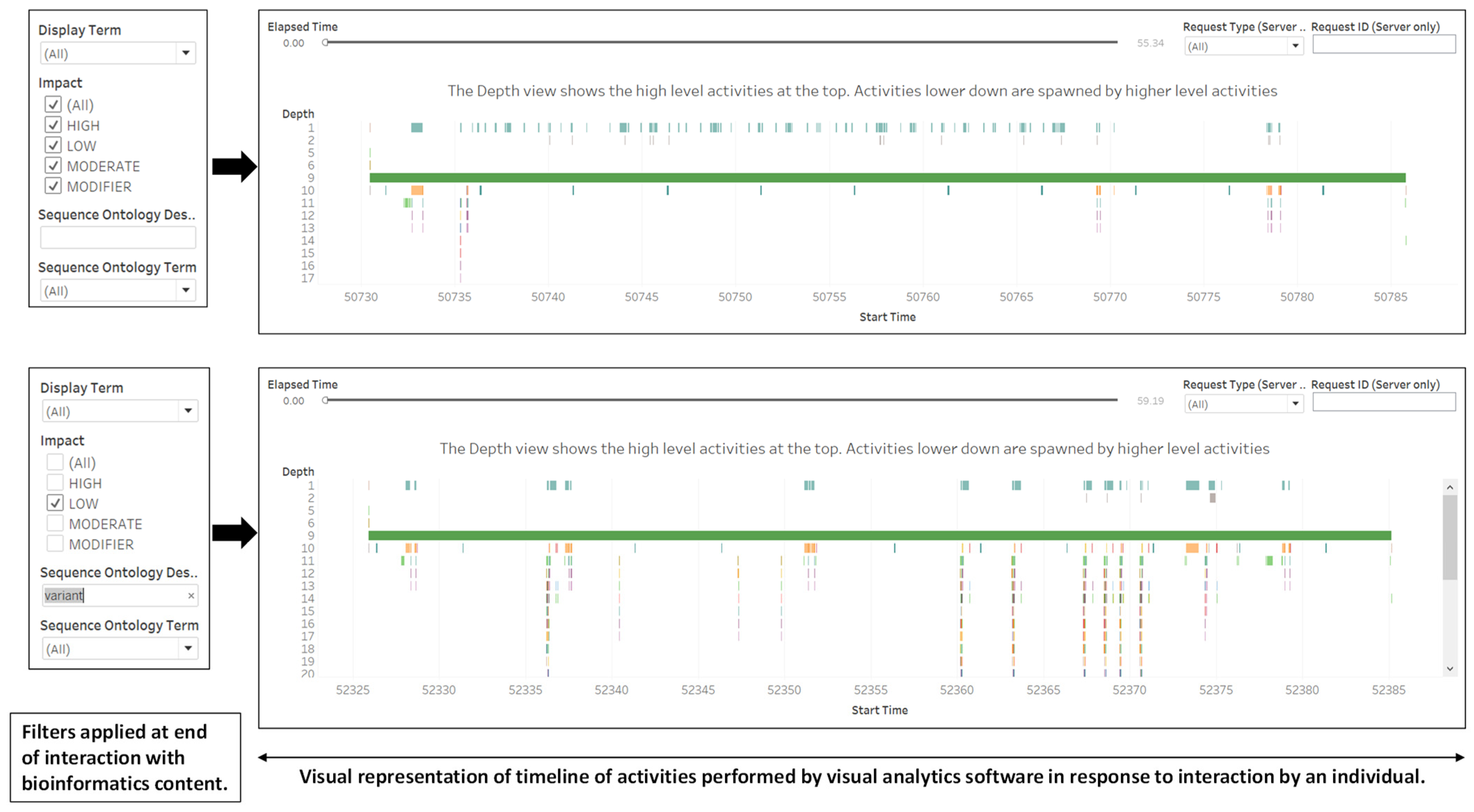
- Adaptation Objective, Procedure and Results. The bioinformatics content for this use case is genomic variants, which are changes (insertions, deletions or replacements) in one allele of a gene of an individual compared with a reference genome [53,54]. There are at least 39 calculated consequences of genomic variations on transcripts or proteins that are located on the transcript structure [55]. The customization design in Figure 3 is a dashboard that consists of (1) a sequence ontology description and other descriptive features for sequence variants; (2) an image from the Ensembl website illustrating the location of sequence variants in the gene structure; (3) filters for specifying the sequence ontology descriptors. We applied the performance-recording function of Tableau Desktop (accessed via Help menu) to acquire data patterns of events performed by software in response to an individual’s requests (Figure 4). Patterns reported include the blending of data, computing layout, compile query, executing query, rendering, computing layout and sorting data. In Figure 4, the differences in the amount of interaction are displayed using performance metrics available in Tableau Desktop Professional [42]. The top image shows activities by the visual analytics resource that reflects that the learner did not make changes to the filters during the period of the interaction. In the bottom image, the performance metrics reflect that the learner changed the value of the impact and included text in the sequence ontology description.
3.4. Visual Analytics Support for Reading Strategies of Scholarly Articles in Personalized Bioinformatics Education
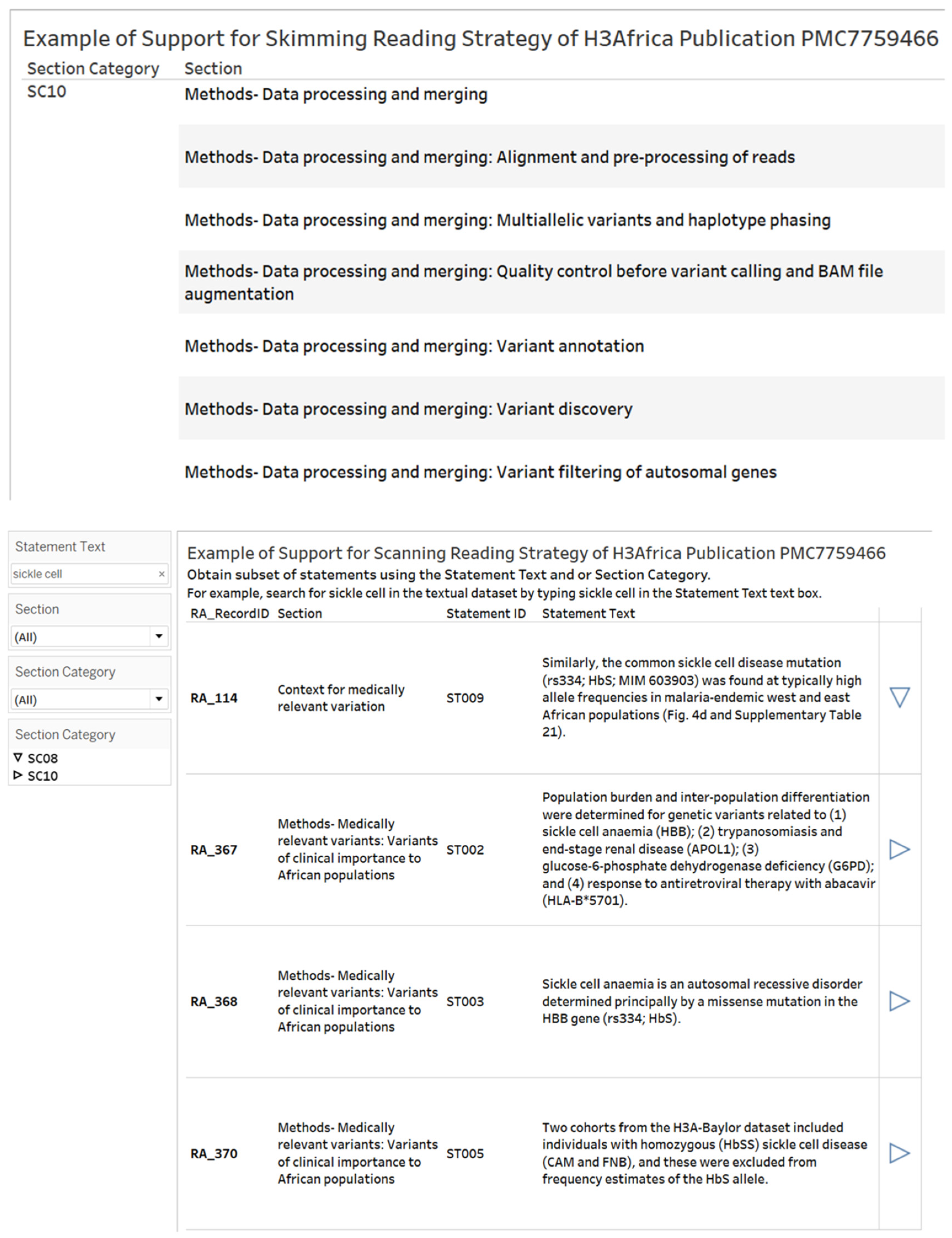
- Principles of Reading Strategies. The reading strategies of skimming (seeking main ideas) and scanning (keyword search) can support the efficient and effective comprehension of online scholarly publications [56]. The growing number and expanding bioinformatics content of H3Africa scholarly publications [27] presents opportunities in personalized bioinformatics education to support readers of H3Africa publications to use reading strategies such as the skimming and scanning of knowledge (content) types [57]. When skimming, the reader seeks the main idea using the outline or table of contents in the text.
- Reading Strategies Objective and Results. The objective of this use case on reading strategies is to demonstrate skimming and scanning reading strategies with a textual dataset. We used the recoded dataset of 381 statements from the H3Africa publication PMC7759466 [40], which included section headings, statement text and statement identifiers. We designed visual analytics worksheets to support skimming and scanning (Figure 5). When skimming, the reader can display the list of section headings to obtain the main ideas described in the article. The support for scanning allows the reader to search by keywords and retrieve statements that contain keywords as well as details on the context of use in one or more instances. The statement(s) retrieved can be the basis for an intensive reading strategy where the purpose is to obtain the exact understanding of the text [58].
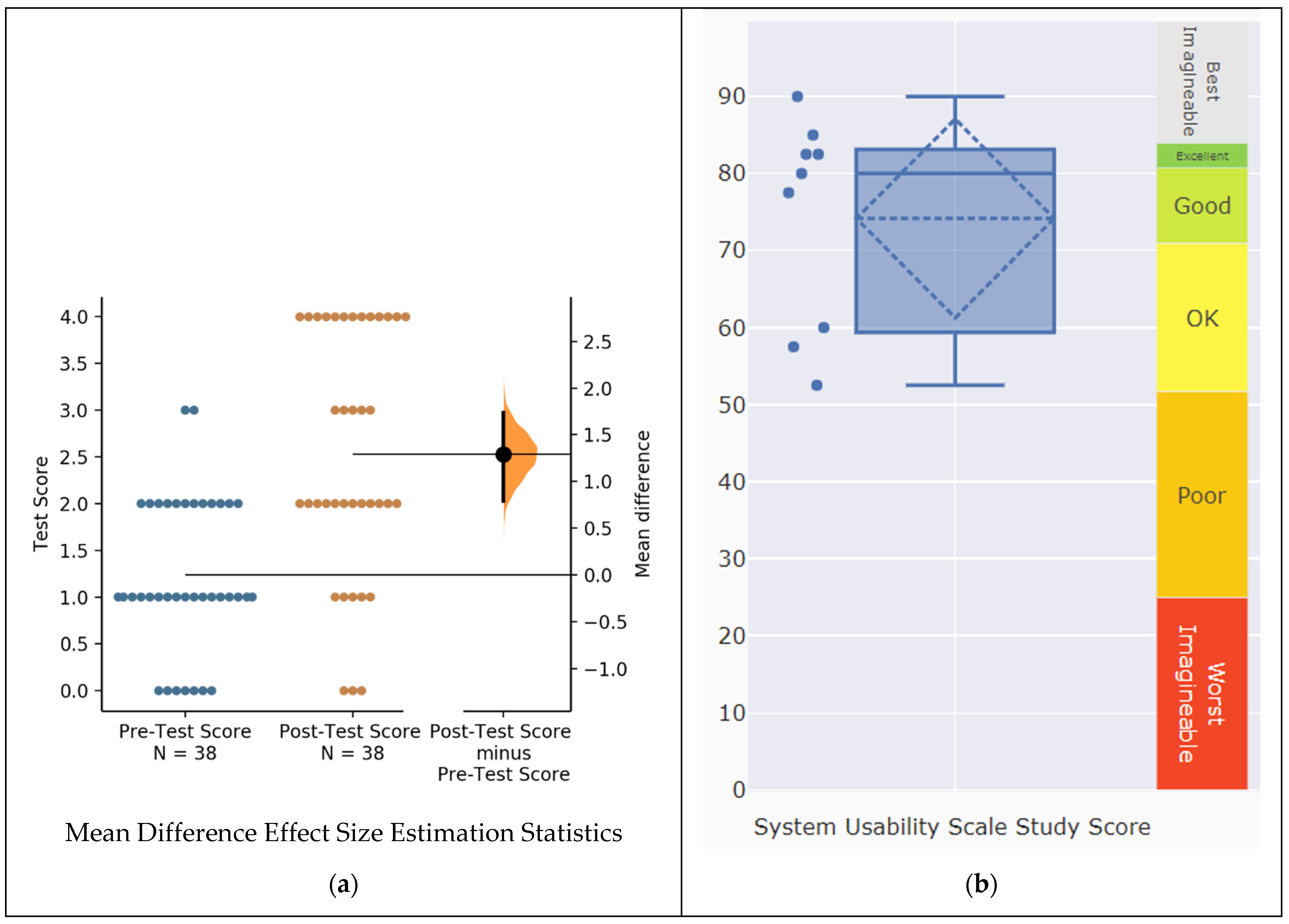
3.5. Evaluation of Visual Analytics Resources
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Question | Multiple-Choice Answers | Correct Answer |
|---|---|---|
| An example of low impact sequence variant is: | A. Frameshift variants B. Start lost C. Protein altering variant D. Stop retained variant | D. Stop retained variant |
| An example of moderate impact sequence variant is: | A. Protein altering variant B. Regulatory region amplification C. Splice region variant D. 3 prime UTR variant | A. Protein altering variant |
| An example of modifier impact sequence variant is: | A. Missense variant B. Coding sequence variant C. Synonymous variant D. Start lost | B. Coding sequence variant |
| An example of high impact sequence variant is: | A. Synonymous variant B. Transcript ablation C. Regulatory region ablation D. Feature elongation | B. Transcript ablation |
| Adapted Questions from the System Usability Scale |
|---|
| 1. I think that I would like to use this visual analytics resource frequently. |
| 2. I found the visual analytics resource unnecessarily complex. |
| 3. I thought the visual analytics resource was easy to use. |
| 4. I think that I would need the support of a technical person to be able to use this visual analytics resource. |
| 5. I found the various functions in this visual analytics resource were well integrated. |
| 6. I thought there was too much inconsistency in this visual analytics resource. |
| 7. I would imagine that most people would learn to use this visual analytics resource very quickly. |
| 8. I found the visual analytics resource very cumbersome to use. |
| 9. I felt very confident using the visual analytics resource. |
| 10. I needed to learn a lot of things before I could get going with this visual analytics resource. |
References
- Gamage, K.A.; Silva, E.K.D.; Gunawardhana, N. Online delivery and assessment during COVID-19: Safeguarding academic integrity. Educ. Sci. 2020, 10, 301. [Google Scholar] [CrossRef]
- Aristovnik, A.; Keržič, D.; Ravšelj, D.; Tomaževič, N.; Umek, L. Impacts of the COVID-19 pandemic on life of higher education students: A global perspective. Sustainability 2020, 12, 8438. [Google Scholar] [CrossRef]
- Ceesay, E.K. Potential impact of COVID-19 outbreak on education, staff development and training in Africa. Res. Glob. 2021, 3, 100049. [Google Scholar] [CrossRef]
- Zancajo, A.; Verger, A.; Bolea, P. Digitalization and beyond: The effects of COVID-19 on post-pandemic educational policy and delivery in Europe. Policy Soc. 2022, 41, 111–128. [Google Scholar] [CrossRef]
- Gera, R.; Tick, S.; Saxena, A. CHUNK Learning: A Tool that Supports Personalized Education. In Proceedings of the 15th International Conference on Educational Data Mining, Durham, UK, 24–27 July 2022. [Google Scholar]
- Zhao, Y.; Watterston, J. The changes we need: Education post COVID-19. J. Educ. Chang. 2021, 22, 3–12. [Google Scholar] [CrossRef]
- Polikoff, M.S. Lessons for Improving Curriculum from the COVID–19 Pandemic. Center on Reinventing Public Education. 2022. Available online: https://crpe.org/lessons-for-improving-curriculum-from-the-covid-19-pandemic (accessed on 12 August 2022).
- Kucirkova, N.; Gerard, L.; Linn, M.C. Designing personalised instruction: A research and design framework. Br. J. Educ. Technol. 2021, 52, 1839–1861. [Google Scholar] [CrossRef]
- Reber, R.; Canning, E.A.; Harackiewicz, J.M. Personalized education to increase interest. Curr. Dir. Psychol. Sci. 2018, 27, 449–454. [Google Scholar] [CrossRef]
- Bernacki, M.L.; Walkington, C. The role of situational interest in personalized learning. J. Educ. Psychol. 2018, 110, 864. [Google Scholar] [CrossRef]
- Bayat, A. Science, medicine, and the future: Bioinformatics. Br. Med. J. 2002, 324, 1018–1023. [Google Scholar] [CrossRef]
- Cohen, J.E. Mathematics is biology’s next microscope, only better; biology is mathematics’ next physics, only better. PLoS Biol. 2004, 2, e439. [Google Scholar] [CrossRef]
- Santos, R.A.C.D.; Verli, H.; Minardi, R.C.d.M. Editorial: Original strategies for training and educational initiatives in bioinformatics. Front. Educ. 2022, 7, 1003098. [Google Scholar] [CrossRef]
- Mulder, N.; Schwartz, R.; Brazas, M.D.; Brooksbank, C.; Gaeta, B.; Morgan, S.L.; Pauley, M.A.; Rosenwald, A.; Rustici, G.; Sierk, M. The development and application of bioinformatics core competencies to improve bioinformatics training and education. PLoS Comput. Biol. 2018, 14, e1005772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhan, Y.A.; Wray, C.G.; Namburi, S.; Glantz, S.T.; Laubenbacher, R.; Chuang, J.H. Fostering bioinformatics education through skill development of professors: Big Genomic Data Skills Training for Professors. PLoS Comput. Biol. 2019, 15, e1007026. [Google Scholar] [CrossRef] [Green Version]
- Ali, M.M.; Hamid, M.; Saleem, M.; Malik, S.; Mian, N.A.; Ihsan, M.A.; Tabassum, N.; Mehmood, K.; Awan, F. Status of bioinformatics education in South Asia: Past and present. BioMed Res. Int. 2021, 2021, 5568262. [Google Scholar] [CrossRef]
- 1000 Genomes Project Consortium. A map of human genome variation from population scale sequencing. Nature 2010, 467, 1061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gudmundsson, S.; Singer-Berk, M.; Watts, N.A.; Phu, W.; Goodrich, J.K.; Solomonson, M.; Consortium, G.A.D.; Rehm, H.L.; MacArthur, D.G.; O’Donnell-Luria, A. Variant interpretation using population databases: Lessons from gnomAD. Hum. Mutat. 2022, 43, 1012–1030. [Google Scholar] [CrossRef] [PubMed]
- Gao, P. Using personalized education to take the place of standardized education. J. Educ. Train. Stud. 2014, 2, 44–47. [Google Scholar] [CrossRef]
- Kehrer, J.; Hauser, H. Visualization and visual analysis of multifaceted scientific data: A survey. IEEE Trans. Vis. Comput. Graph. 2012, 19, 495–513. [Google Scholar] [CrossRef]
- H3Africa Consortium. Enabling the genomic revolution in Africa: H3Africa is developing capacity for health-related genomics research in Africa. Sci. (N. Y. NY) 2014, 344, 1346. [Google Scholar] [CrossRef] [Green Version]
- Rotimi, C.N. Enabling Genomic Revolution in Africa. In The Genetics of African Populations in Health and Disease; Ibrahim, M.E., Rotimi, C.N., Eds.; Cambridge University Press: Cambridge, UK, 2019; pp. 320–330. [Google Scholar]
- Fatumo, S.; Yakubu, A.; Oyedele, O.; Popoola, J.; Attipoe, D.A.; Eze-Echesi, G.; Modibbo, F.Z.; Ado-Wanka, N.; Salako, O.; Nashiru, O. Promoting the genomic revolution in Africa through the Nigerian 100K Genome Project. Nat. Genet. 2022, 54, 531–536. [Google Scholar] [CrossRef]
- Mulder, N.; Abimiku, A.l.; Adebamowo, S.N.; de Vries, J.; Matimba, A.; Olowoyo, P.; Ramsay, M.; Skelton, M.; Stein, D.J. H3Africa: Current perspectives. Pharm. Pers. Med. 2018, 11, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramsay, M. African genomic data sharing and the struggle for equitable benefit. Patterns 2022, 3, 100412. [Google Scholar] [CrossRef] [PubMed]
- H3Africa Consortium. H3Africa Publications. Available online: https://h3africa.org/index.php/resource/publication-2/ (accessed on 30 June 2022).
- Choudhury, A.; Sengupta, D.; Aron, S.; Ramsay, M. The H3Africa Consortium: Publication Outputs of a Pan-African Genomics Collaboration (2013 to 2020). In Africa, the Cradle of Human Diversity; Fortes-Lima, C., Mtetwa, E., Schlebusch, C., Eds.; Brill: Leiden, The Netherlands, 2021; pp. 257–304. [Google Scholar]
- Mariano, D.; Martins, P.; Helene Santos, L.; de Melo-Minardi, R.C. Introducing programming skills for life science students. Biochem. Mol. Biol. Educ. 2019, 47, 288–295. [Google Scholar] [CrossRef]
- Nunes, R.; Barbosa de Almeida Júnior, E.; Pessoa Pinto de Menezes, I.; Malafaia, G. Learning nucleic acids solving by bioinformatics problems. Biochem. Mol. Biol. Educ. 2015, 43, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Magana, A.J.; Taleyarkhan, M.; Alvarado, D.R.; Kane, M.; Springer, J.; Clase, K. A survey of scholarly literature describing the field of bioinformatics education and bioinformatics educational research. CBE—Life Sci. Educ. 2014, 13, 607–623. [Google Scholar] [CrossRef] [Green Version]
- Bateman, A.; Lengauer, T. Editorial. Bioinform. Adv. 2021, 1, vbab001. [Google Scholar] [CrossRef]
- Peng, H.; Bateman, A.; Valencia, A.; Wren, J.D. Bioimage informatics: A new category in Bioinformatics. Bioinformatics 2012, 28, 1057. [Google Scholar] [CrossRef] [Green Version]
- Michael, J.A. Mental models and meaningful learning. J. Vet. Med. Educ. 2004, 31, 1–5. [Google Scholar] [CrossRef]
- Mayer, R.E. Rote versus meaningful learning. Theory Into Pract. 2002, 41, 226–232. [Google Scholar] [CrossRef]
- Karpicke, J.D.; Grimaldi, P.J. Retrieval-based learning: A perspective for enhancing meaningful learning. Educ. Psychol. Rev. 2012, 24, 401–418. [Google Scholar] [CrossRef]
- Fan, K.-K.; Xiao, P.-w.; Su, C. The effects of learning styles and meaningful learning on the learning achievement of gamification health education curriculum. Eurasia J. Math. Sci. Technol. Educ. 2015, 11, 1211–1229. [Google Scholar]
- Carvalho, P.; Rachatasumrit, N.; Koedinger, K.R. Learning depends on knowledge: The benefits of retrieval practice vary for facts and skills. In Proceedings of the 44th Annual Meeting of the Cognitive Science Society, Toronto, ON, Canada, 27–30 July 2022; pp. 170–175. [Google Scholar]
- Tawfik, A.A.; Gatewood, J.; Gish-Lieberman, J.J.; Hampton, A.J. Toward a definition of learning experience design. Technol. Knowl. Learn. 2022, 27, 309–334. [Google Scholar] [CrossRef]
- Smith, M.A.; Karpicke, J.D. Retrieval practice with short-answer, multiple-choice, and hybrid tests. Memory 2014, 22, 784–802. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, A.; Aron, S.; Botigué, L.R.; Sengupta, D.; Botha, G.; Bensellak, T.; Wells, G.; Kumuthini, J.; Shriner, D.; Fakim, Y.J. High-depth African genomes inform human migration and health. Nature 2020, 586, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Isokpehi, R.D.; Johnson, C.P.; Tucker, A.N.; Gautam, A.; Brooks, T.J.; Johnson, M.O.; Cozart, T.; Wathington, D.J. Integrating datasets on public health and clinical aspects of sickle cell disease for effective community-based research and practice. Diseases 2020, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Beard, L.; Aghassibake, N. Tableau (version 2020.3). J. Med. Libr. Assoc. JMLA 2021, 109, 159. [Google Scholar] [CrossRef]
- Saragih, W.; Natalia, C.H. Types of sentences used by male and female writers in journal article abstracts. J. BAHAS. 2020, 31, 227–244. [Google Scholar] [CrossRef]
- Isokpehi, R.; Johnson, M.O.; Campos, B.; Sanders, A.; Cozart, T.; Harvey, I.S. Knowledge visualizations to inform decision making for improving food accessibility and reducing obesity rates in the United States. Int. J. Environ. Res. Public Health 2020, 17, 1263. [Google Scholar] [CrossRef] [Green Version]
- Sedig, K.; Parsons, P. Interaction design for complex cognitive activities with visual representations: A pattern-based approach. AIS Trans. Hum.-Comput. Interact. 2013, 5, 84–133. [Google Scholar] [CrossRef] [Green Version]
- Cunningham, F.; Moore, B.; Ruiz-Schultz, N.; Ritchie, G.R.; Eilbeck, K. Improving the sequence ontology terminology for genomic variant annotation. J. Biomed. Semant. 2015, 6, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Hunt, S.E.; Moore, B.; Amode, R.M.; Armean, I.M.; Lemos, D.; Mushtaq, A.; Parton, A.; Schuilenburg, H.; Szpak, M.; Thormann, A. Annotating and prioritizing genomic variants using the Ensembl Variant Effect Predictor—A tutorial. Hum. Mutat. 2022, 43, 986–997. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.; Tumkaya, T.; Aryal, S.; Choi, H.; Claridge-Chang, A. Moving beyond p values: Data analysis with estimation graphics. Nat. Methods 2019, 16, 565–566. [Google Scholar] [CrossRef] [PubMed]
- Vlachogianni, P.; Tselios, N. Perceived usability evaluation of educational technology using the System Usability Scale (SUS): A systematic review. J. Res. Technol. Educ. 2022, 54, 392–409. [Google Scholar] [CrossRef]
- Brooke, J. SUS: A retrospective. J. Usability Stud. 2013, 8, 29–40. [Google Scholar]
- Blattgerste, J.; Behrends, J.; Pfeiffer, T. A Web-Based Analysis Toolkit for the System Usability Scale. In Proceedings of the 15th International Conference on PErvasive Technologies Related to Assistive Environments, Corfu, Greece, 29 June–1 July 2022; pp. 237–246. [Google Scholar]
- Vy, H.M.T.; Kim, Y. A composite-likelihood method for detecting incomplete selective sweep from population genomic data. Genetics 2015, 200, 633–649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jamuar, S.S.; Walsh, C.A. Genomic variants and variations in malformations of cortical development. Pediatr. Clin. 2015, 62, 571–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geraci, F.; Massidda, R.; Pisanti, N. Technology and species independent simulation of sequencing data and genomic variants. In Proceedings of the 2019 IEEE 19th International Conference on Bioinformatics and Bioengineering (BIBE), Athens, Greece, 28–30 October 2022; pp. 138–145. [Google Scholar]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The ensembl variant effect predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.M.N. The Essential Roles of Skimming and Scanning Techniques in Teaching Reading Comprehension. Available online: http://nnkt.ueh.edu.vn/wp-content/uploads/2015/07/20.pdf (accessed on 12 August 2022).
- Maxwell, M.J. Skimming and scanning improvement: The needs, assumptions and knowledge base. J. Read. Behav. 1972, 5, 47–59. [Google Scholar] [CrossRef] [Green Version]
- Berardo, S.A. The use of authentic materials in the teaching of reading. Read. Matrix 2006, 6, 60–69. [Google Scholar]
- Clark, R.; Harrelson, G.L. Designing instruction that supports cognitive learning processes. J. Athl. Train. 2002, 37, S-152–S-159. [Google Scholar]
- Bhutoria, A. Personalized education and artificial intelligence in United States, China, and India: A systematic review using a Human-In-The-Loop model. Comput. Educ. Artif. Intell. 2022, 3, 100068. [Google Scholar] [CrossRef]
- Koedinger, K.R.; Corbett, A.T.; Perfetti, C. The Knowledge-Learning-Instruction framework: Bridging the science-practice chasm to enhance robust student learning. Cogn. Sci. 2012, 36, 757–798. [Google Scholar] [CrossRef]
- Koedinger, K.R.; Booth, J.L.; Klahr, D. Instructional complexity and the science to constrain it. Science 2013, 342, 935–937. [Google Scholar] [CrossRef] [PubMed]
- Karpicke, J.D.; Butler, A.C.; Roediger, H.L., III. Metacognitive strategies in student learning: Do students practise retrieval when they study on their own? Memory 2009, 17, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Ware, C. Perception & Data Visualization: The Foundations of Experimental Semiotics. In Proceedings of the Graphics Interface ’98, Vancouver, BC, Canada, 18–20 June 1998; pp. 92–98. [Google Scholar]
- Sacha, D.; Stoffel, A.; Stoffel, F.; Kwon, B.C.; Ellis, G.; Keim, D.A. Knowledge generation model for visual analytics. IEEE Trans. Vis. Comput. Graph. 2014, 20, 1604–1613. [Google Scholar] [CrossRef] [PubMed]
- Jern, M.; Franzen, J. “GeoAnalytics”—Exploring spatio-temporal and multivariate data. In Proceedings of the Tenth International Conference on Information Visualisation (IV’06), London, UK, 5–7 July 2006; pp. 25–31. [Google Scholar]
- Oladejo, A.I.; Okebukola, P.A.; Olateju, T.T.; Akinola, V.O.; Ebisin, A.; Dansu, T.V. In search of culturally responsive tools for meaningful learning of chemistry in Africa: We stumbled on the culturo-techno-contextual approach. J. Chem. Educ. 2022, 99, 2919–2931. [Google Scholar] [CrossRef]
- Renninger, K.A.; Hidi, S.E. To level the playing field, develop interest. Policy Insights Behav. Brain Sci. 2020, 7, 10–18. [Google Scholar] [CrossRef]
- Ainsworth, S. DeFT: A conceptual framework for considering learning with multiple representations. Learn. Instr. 2006, 16, 183–198. [Google Scholar] [CrossRef]
- Nelms, A.A.; Segura-Totten, M. Expert–novice comparison reveals pedagogical implications for students’ analysis of primary literature. CBE—Life Sci. Educ. 2019, 18, ar56. [Google Scholar] [CrossRef] [PubMed]
- Stengel, A.; Stanke, K.M.; McPherson, M.R.; Drijber, R.A. Virtually engaging students through collaborative investigation of scientific literature, a case study. Nat. Sci. Educ. 2021, 50, e20051. [Google Scholar] [CrossRef]
- Dammann, O. Data, information, evidence, and knowledge: A proposal for health informatics and data science. Online J. Public Health Inform. 2018, 10, e224. [Google Scholar] [CrossRef] [PubMed]
- Eldridge, A. Best Practices for Designing Efficient Tableau Workbooks. Available online: https://www.tableau.com/learn/whitepapers/designing-efficient-workbooks (accessed on 12 August 2022).
- Dent, H.L.; Jo White, B.; Kapakos, W.A.; Fulk, H.K. Widen the big tent for big data: Understanding business student perceptions of Tableau. Issues Inf. Syst. 2021, 22, 49–58. [Google Scholar]
- Richardson, J.; Sallam, R.; Schlegel, K.; Kronz, A.; Sun, J. Magic Quadrant for Analytics and Business Intelligence Platforms; Gartner: Stamford, CT, USA, 2020; Available online: https://b2bsalescafe.files.wordpress.com/2020/04/gartner-magic-quadrant-for-analytics-and-business-intelligence-platforms-feb-2020.pdf (accessed on 12 August 2022).


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Makolo, A.U.; Smile, O.; Ezekiel, K.B.; Destefano, A.M.; McCall, J.L.; Isokpehi, R.D. Leveraging H3Africa Scholarly Publications for Technology-Enhanced Personalized Bioinformatics Education. Educ. Sci. 2022, 12, 859. https://doi.org/10.3390/educsci12120859
Makolo AU, Smile O, Ezekiel KB, Destefano AM, McCall JL, Isokpehi RD. Leveraging H3Africa Scholarly Publications for Technology-Enhanced Personalized Bioinformatics Education. Education Sciences. 2022; 12(12):859. https://doi.org/10.3390/educsci12120859
Chicago/Turabian StyleMakolo, Angela U., Olubukola Smile, Kehinde B. Ezekiel, Antoinette M. Destefano, Junell L. McCall, and Raphael D. Isokpehi. 2022. "Leveraging H3Africa Scholarly Publications for Technology-Enhanced Personalized Bioinformatics Education" Education Sciences 12, no. 12: 859. https://doi.org/10.3390/educsci12120859
APA StyleMakolo, A. U., Smile, O., Ezekiel, K. B., Destefano, A. M., McCall, J. L., & Isokpehi, R. D. (2022). Leveraging H3Africa Scholarly Publications for Technology-Enhanced Personalized Bioinformatics Education. Education Sciences, 12(12), 859. https://doi.org/10.3390/educsci12120859





