1. Introduction
Infectious diseases continue to pose threats to public health and social stability, disrupting social and economic order and daily life patterns [
1]. In recent years, a variety of infectious diseases have emerged across different regions and countries, such as the COVID-19 pandemic [
2,
3], Ebola [
4], HIV [
5], and Dengue [
6]. In the context of globalization, international travel and trade have accelerated cross-border population mobility. This connectivity enables infectious diseases to spread rapidly from localized outbreaks to a global scale. The dense urban environment and crowded public places have increased the frequency of interpersonal contact, further facilitating the spread of diseases. The transmission dynamics of infectious diseases are an inherently nonlinear process, which is influenced by complex factors such as vaccination, government intervention, and human mobility [
7]. The large-scale human mobility plays a decisive role in enabling spatial dissemination, as it physically bridges outbreaks across regions. The spatiotemporal heterogeneity caused by large-scale human mobility determines the velocity and geographic reach of epidemics. Quantifying the mobility-driven mode of transmission not only improves predictive accuracy but also formulates intervention strategies. Thus, an effective epidemiological model should account for both localized outbreaks and cross-regional disease spread driven by large-scale human mobility.
However, existing single-region transmission models often fail to capture the impact of dynamic population mobility on interregional disease spread, particularly in a highly interconnected world [
8]. While these transmission models can effectively characterize the evolutionary mechanisms of local epidemics, they struggle to incorporate the complex, real-time effects of human movement across regions, which are critical drivers of disease spread in a globalized context.
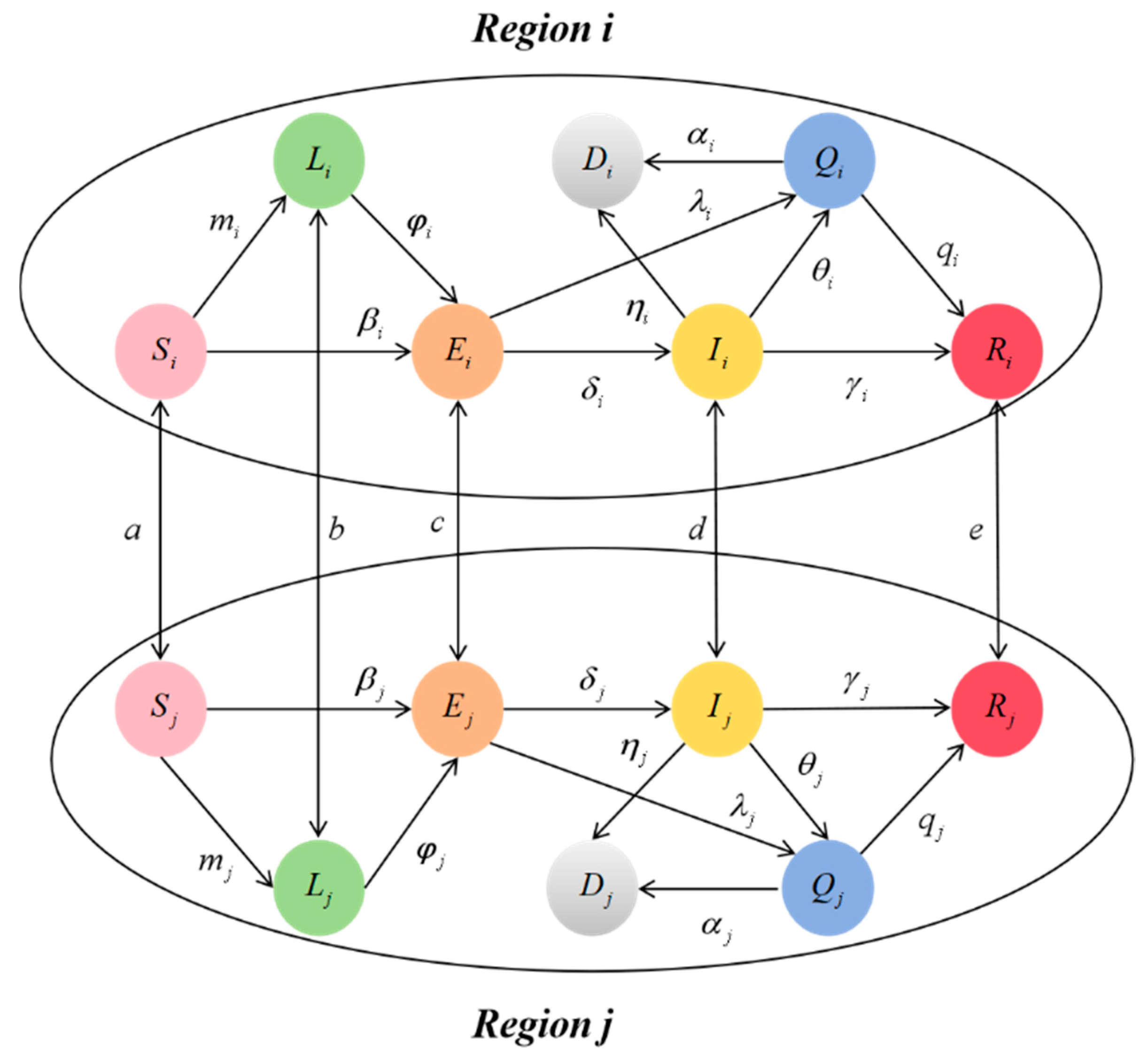
To address these limitations, we propose a nonlinear dynamic model that considers three critical factors: multi-regional population migration, low-risk population, and isolated population. The primary contributions of this paper are described as follows:
- (1)
According to the transmission characteristics of infectious diseases, a multi-regional spatio-temporal transmission dynamic model is proposed, considering low-risk population, isolation factors, and population flow between different regions.
- (2)
We calculate the basic reproduction number, and the relationship between the basic reproduction number of a single region and that of multiple regions is explored. Furthermore, the global stability of the model’s equilibrium points is proved by constructing a Lyapunov function.
- (3)
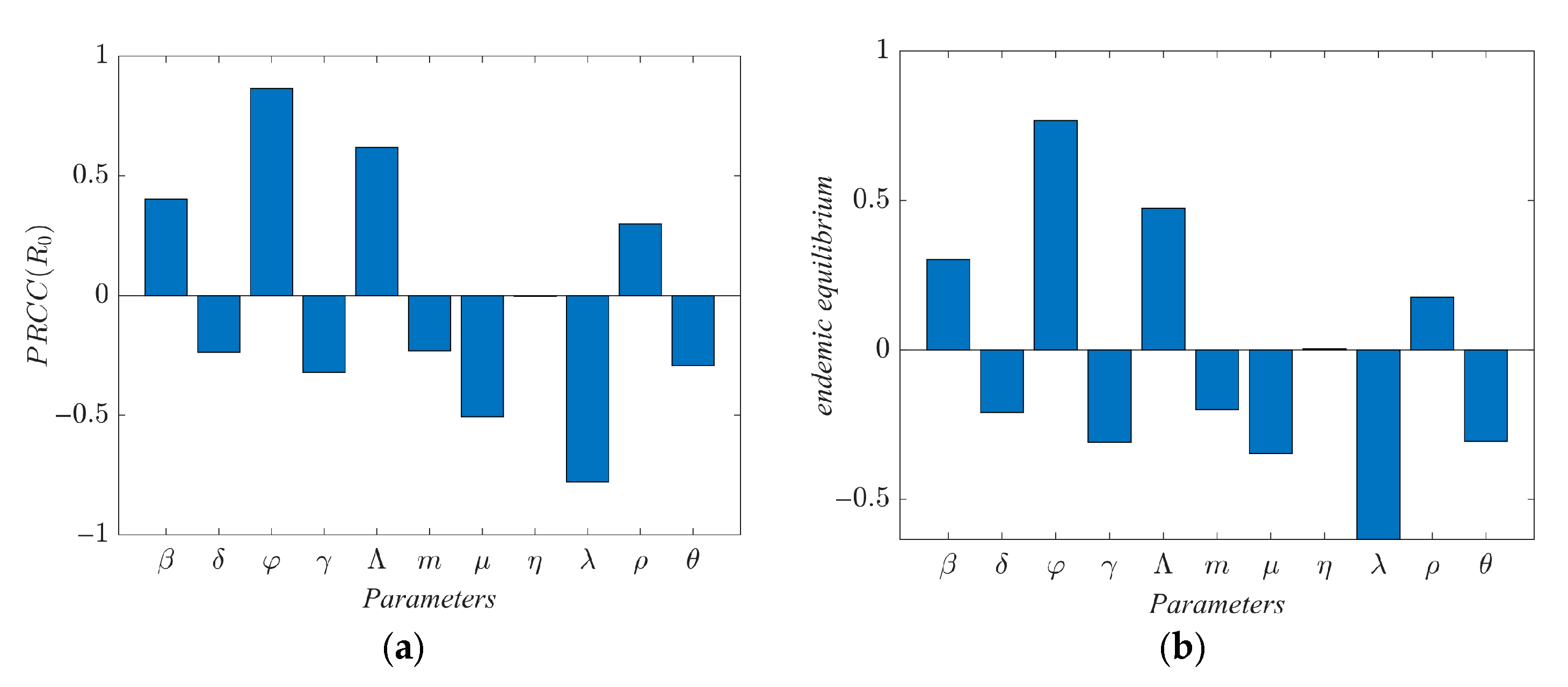
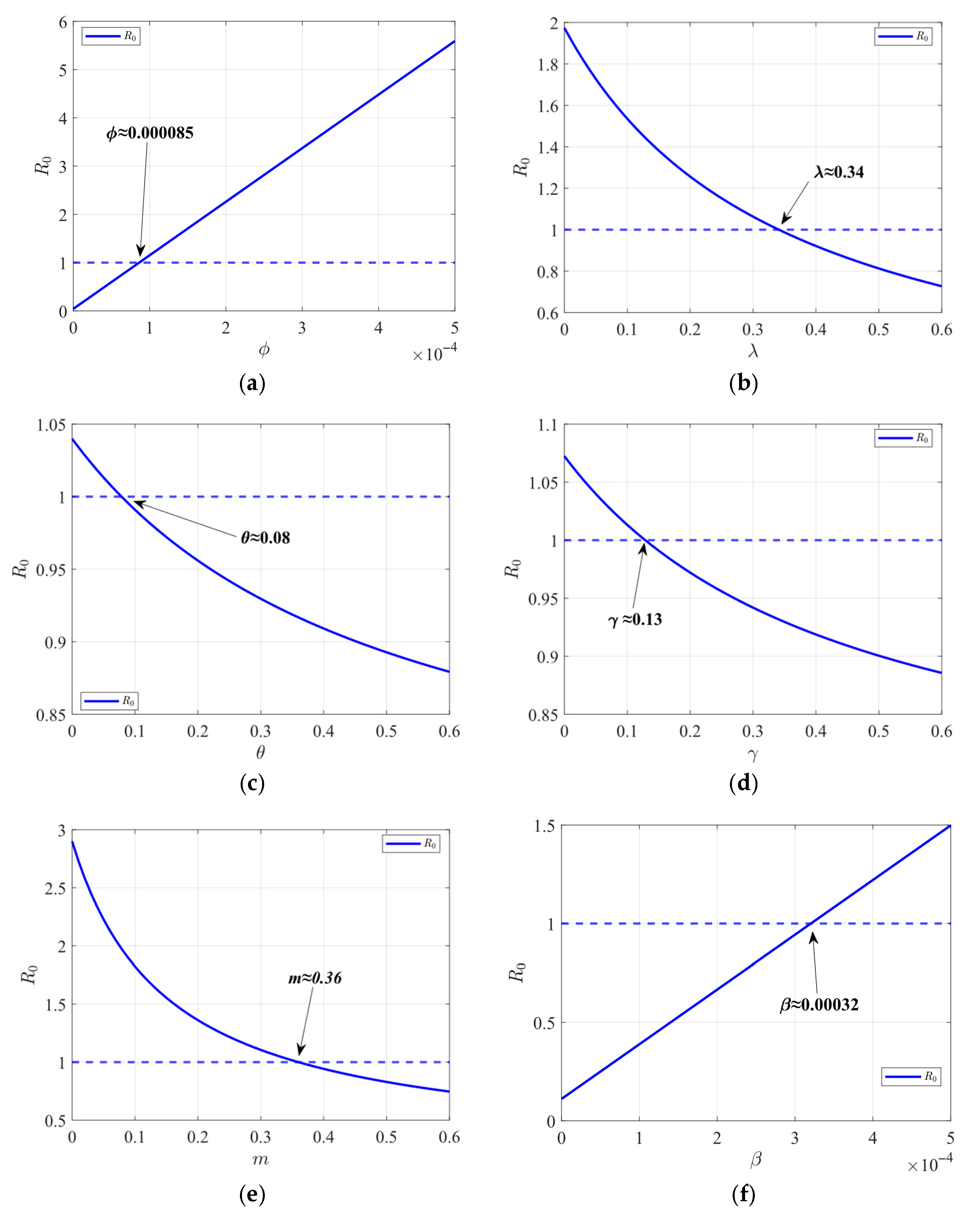
The impact of population mobility on the transmission of infectious diseases is studied. Latin hypercube sampling (LHS) and partial rank correlation coefficient (PRCC) are used to analyze the global parameter sensitivity in a single region. A protective isolation strategy is designed on the basis of the optimal path.
The structure of this paper is organized as follows:
Section 2 reviews related works. The SLEIQDR model of multi-regional population migration based on the SEIR model is described in
Section 3.
Section 4 analyzes the global stability of the disease-free equilibrium and the endemic equilibrium of the model. In
Section 5, the theories in
Section 4 are proved by numerical simulations, and the model’s parameter effects on infectious diseases are analyzed through global sensitivity analysis. Finally,
Section 6 summarizes the work of this paper.
2. Related Works
At present, the most representative dynamic models of infectious disease include the susceptible-infected-recovered (SIR) model, the susceptible-infected-recovered-susceptible (SIRS) model, and the susceptible-exposed-infected-recovered (SEIR) model. These dynamic models are based on nonlinear dynamics theory and aim to explore the virus’s evolution, transmission mechanisms, and predict the epidemic’s future trajectory. Alkhazzan et al. [
9] propose a new stochastic SIRS model and analyze the dynamics of two diseases simultaneously. The model includes media coverage, treatment functions, and three types of noise factors. Ishtiaq et al. [
10] propose a delayed stochastic SIRS model that incorporates an exponential birth rate and a saturated incidence function and analyze its dynamics. Yin et al. [
11] integrate the SIR model with multilayer networks and study the joint dynamics of information and epidemics spread across different networks influenced by government policies. Centres et al. [
12] integrate the SIR Model with mobile agent diffusion characteristics and propose two infectious mechanisms–direct contact and close transmission–to simulate disease spread more accurately. Song et al. [
13] combine asynchronous state estimation with a nonlinear reaction-diffusion SIR Model to solve the shortcomings of existing models in multi-stage epidemic propagation modeling and state estimation. Khalifi et al. [
14] introduce the heterogeneity of individual immunity into the SIRS model, which can predict the spread trend of the epidemic more accurately. Guan et al. [
15] use rubella as an example to study a network-based SIR epidemic model that combines saturation infectivity, vertical transmission, and pulse vaccination strategies. The dynamic behavior of the model is analyzed using impulse differential equation theory. Aguiar et al. [
16] studied dengue epidemiology by using an SIR model with temporary immunity and disease-enhancing, and analyzed the characteristics of disease-free and endemic equilibrium transitions by using bifurcation analysis. The adaptability of the SIR model is limited because of its simplistic state group classification, and it does not adequately account for the complexity of real-world scenarios.
The SEIR model is obtained by considering the exposed population in the SIR model. Darabsah [
17] proposes a time-delay SVEIR model with imperfect vaccines. This study uses measles as a case study and performs systematic numerical simulations and comprehensive parameter sensitivity analysis. Yu et al. [
18] propose the SEIMQR model by considering the virus mutation. They analyze the model’s dynamics and use epidemic data from the UK to predict the epidemic. Chebotaeva et al. [
19] propose a new SEIR model for accurately simulating the spread of infectious diseases by more finely distinguishing symptomatic and asymptomatic infected individuals. In the context of seasonal infectious disease transmission, Wang et al. [
20] consider the change in vaccination strategy within the season factors and emphasize the importance of improving vaccine effectiveness, reducing vaccine costs, and considering individual risk perception and herd immunity effects. Khairulbahri [
21] analyzes the impact of asymptomatic infected individuals and various behavioral measures on COVID-19 transmission by using the SEIR model. The results demonstrate that comprehensive lockdown measures in Sweden could significantly reduce the spread. Yang et al. [
22] integrate population migration data into the modified SEIR model and combine a long short-term memory network (LSTM) to predict COVID-19 transmission. Given that official data collection may overlook unreported infections, Chen et al. [
23] propose a segmented SEIUR model. Das et al. [
24] discuss the impact of information dissemination and medical resource saturation on disease dynamics in the process of disease transmission and propose an SEIR model by considering a non-monotonic incidence function influenced by information and a treatment function with saturation. Luebben et al. [
25] studied optimal vaccination strategies for COVID-19 in the U.S. by dividing the population into different age groups based on the SEIR model. Incorporating exposed populations into the SIR model improves the accuracy of simulations, which leads to better predictions of outbreak timing and scale. These studies extend the SEIR model to describe infectious disease transmission in greater detail. Wang et al. [
26] propose a deep epidemiological modeling method based on black box knowledge distillation to accurately predict the spread of an epidemic. By constructing a mixed model as a teacher model, the method uses sequence mix to improve query efficiency and knowledge diversity, and then trains the student network to retain the accuracy of the teacher model while reducing computational costs and data requirements.
However, the models discussed above focus on changes in the spread of infectious diseases over time and largely overlook the spatial spread of the virus. Therefore, population density, traffic networks, geographical characteristics, and social distancing can affect transmission, and these factors are essential for integrating spatial elements into the analysis. Several studies have addressed this gap using diverse approaches. Tu et al. [
27] propose a reaction-diffusion framework by incorporating the vaccination and isolation measures and optimizing strategies to balance social costs and benefits. Policarpo et al. [
28] apply a fractal diffusion model to explore the virus’s scale-free dynamics in Brazil and show how social distancing influences the spread of the virus. Bhouri et al. [
29] integrate human mobility and social behavior into an SEIR model by enhancing LSTM networks to identify infection clusters and simulate key parameters. Mustavee et al. [
30] utilize a Koopman operator framework to estimate early pandemic dynamics with human mobility as a control input. Bekiros et al. [
31] use a stochastic process model based on Brownian motion to track the spatial movements of virus carriers. Other spatial models focus on regional dynamics. Wang et al. [
32] utilize an improved SIRD model with finite element methods to analyze time-varying parameters spatially. Brockmann et al. [
33] present a network-driven model to predict the speed of disease spread, arrival time, and geographical origin. Mahmood et al. [
34] propose a spatiotemporal stochastic model by incorporating contact tracing and spatial risk assessments.
The multi-patch model integrates dynamic systems with discrete spatial units to characterize infectious disease at multiple scales, which is of great significance in spatial epidemiology. Jia et al. [
35] analyze provincial COVID-19 transmission patterns in China using a dynamic model with mobility data. Guo et al. [
36] examine nonlocal infection dynamics and analyze how delays in infection dissemination reduce local concentrations. Allman et al. [
37] developed an SEIQRD model for Belgium, and the results show that reducing social contact is more effective than limiting mobility in controlling outbreaks. Das et al. [
38] studied the SIS infectious disease model of nonlocal transmission mechanism and revealed the important influence of nonlocal transmission on the formation of space-time patterns of diseases. Wang et al. [
39] consider the two ways of foot-and-mouth disease infection through contact and airborne transmission and use the integral kernel function to describe the nonlocal transmission characteristics of foot-and-mouth disease in space and time, to simulate the transmission of foot-and-mouth disease more comprehensively. Sun et al. [
40] use a two-patch SIR Model and combine it with the regulatory effect of historical monitoring data on infectious diseases to reveal the threshold behavior of disease transmission and the periodic fluctuations caused by the delayed effect, providing a theoretical basis for the prevention and control strategy of infectious diseases. Wu et al. [
41] provide a new method to study spatial heterogeneity of syphilis transmission by transforming the reactive-diffusion model into a patch model under a discrete spatial framework and using actual case data for parameter estimation. Wang et al. [
42] consider both the high and low infectivity of Vibrio cholerae and the infection of the human host. To better capture the transmission dynamics during the incubation period, a nonlocal time delay is introduced to describe individual movement and infection. Akuno et al. [
43] propose a multi-patch SEIRS epidemic model that considers partial mobility, residence time, and demographic factors. By separating the mobility ratio from the residence time matrix, the realistic applicability of the model is enhanced. Zhao et al. [
44] propose the first systematic theoretical framework for epidemic models driven by Lévy flights, revealing the profound implications of nonlocal diffusion on spatiotemporal transmission dynamics. Their work provides a rigorous mathematical foundation for understanding the role of long-distance mobility in the geographical spread of disease. Gross et al. [
45] analyze the spatio-temporal spread of COVID-19 using scaling laws. Spatially, it reveals the power-law relationship between the number of infections and the distance from the source of the epidemic, population size, and the number of people who left Hubei, confirming the effectiveness of inter-city isolation. Temporally, it proposes a two-stage model: a constant infection rate in the early stage and an exponential decline in the later stage.
These multi-patch models have made significant progress in spatial epidemiology [
35,
36,
37,
38,
39,
40,
41,
42,
43,
44,
45], but their theoretical frameworks still have some limitations. In terms of migration pattern modeling, most models assume homogeneous migration [
43], which fails to reflect the mobility differences in different infection status groups in real scenarios. Secondly, when characterizing susceptible populations, traditional models lack a risk stratification mechanism and fail to describe the impact of protective behaviors (such as wearing masks) on transmission dynamics. Furthermore, there are still limited results on the global stability of equilibrium points in multi-patch models, as previous work is usually restricted to local analysis or threshold conditions [
37]. To address these problems, we propose the SLEIQDR model for multi-regional population migration.
6. Conclusions
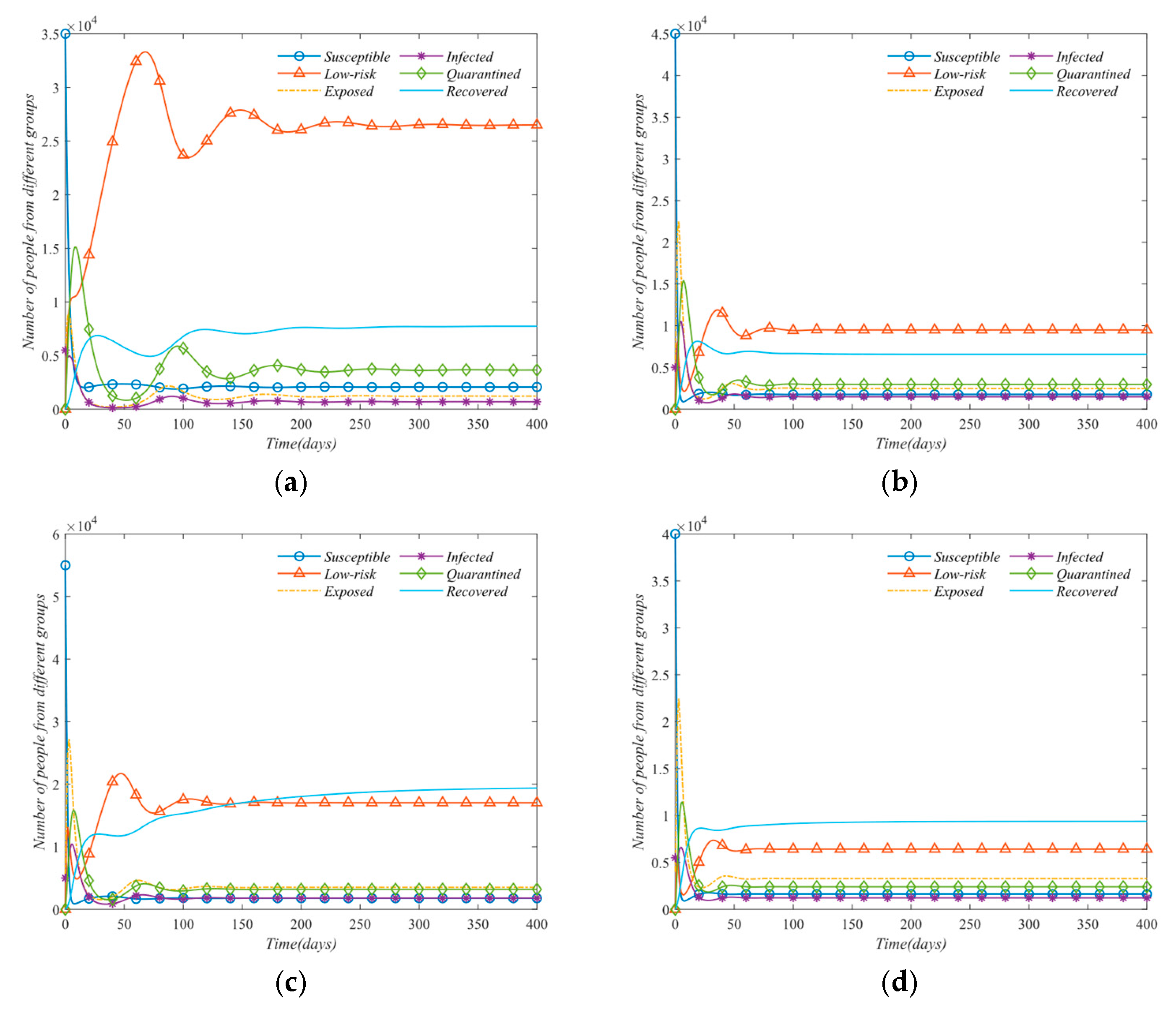
In this paper, on the basis of the traditional SEIR model, a SLEIQDR model of multi-regional population migration is established by combining the factors of population migration, low-risk individuals, and isolated individuals. First, the model’s uniqueness of the disease-free equilibrium is proved, the basic reproduction number is calculated, and the relationship between the single region and multiple regions is analyzed. Furthermore, a Lyapunov function is constructed to prove the stability of the endemic equilibrium and the disease-free equilibrium, and the correctness of the theory is proved by numerical analysis. To explore the impact of population migration on the number of infected, changes in infection numbers across different regions under varying migration rates are simulated. The results show that population migration facilitates the spread of the virus from high-infection areas to low-infection areas, exacerbating the situation in the latter. However, strong public health measures implemented in low-infection areas can effectively prevent the spread and reduce the overall basic reproduction number. In addition, the number of infected increases in areas where the immigration rate is greater than the emigration rate. Finally, a parameter sensitivity analysis is conducted for the single region case, and an optimal protection isolation strategy is proposed. The results indicate that increasing isolation and recovery rates, along with reducing the infection rate, will significantly reduce the spread of the disease. This study will help decision-makers better grasp the dynamics of the epidemic and optimize the response measures to effectively mitigate the epidemic’s impact on society. However, due to the complexity of the model, it is difficult to estimate its parameters with real datasets. Future work should focus on developing parameter estimation methods for this model and calibrating the model using real datasets.