Towards Intelligent Virtual Clerks: AI-Driven Automation for Clinical Data Entry in Dialysis Care
Abstract
1. Introduction
2. Related Work
2.1. AI in Healthcare Information Management
2.2. Image Processing and Optical Character Recognition in Clinical Contexts
2.3. Agent-Based Systems for Workflow Automation
2.4. Digital Health Transformation and Cybersecurity Constraints
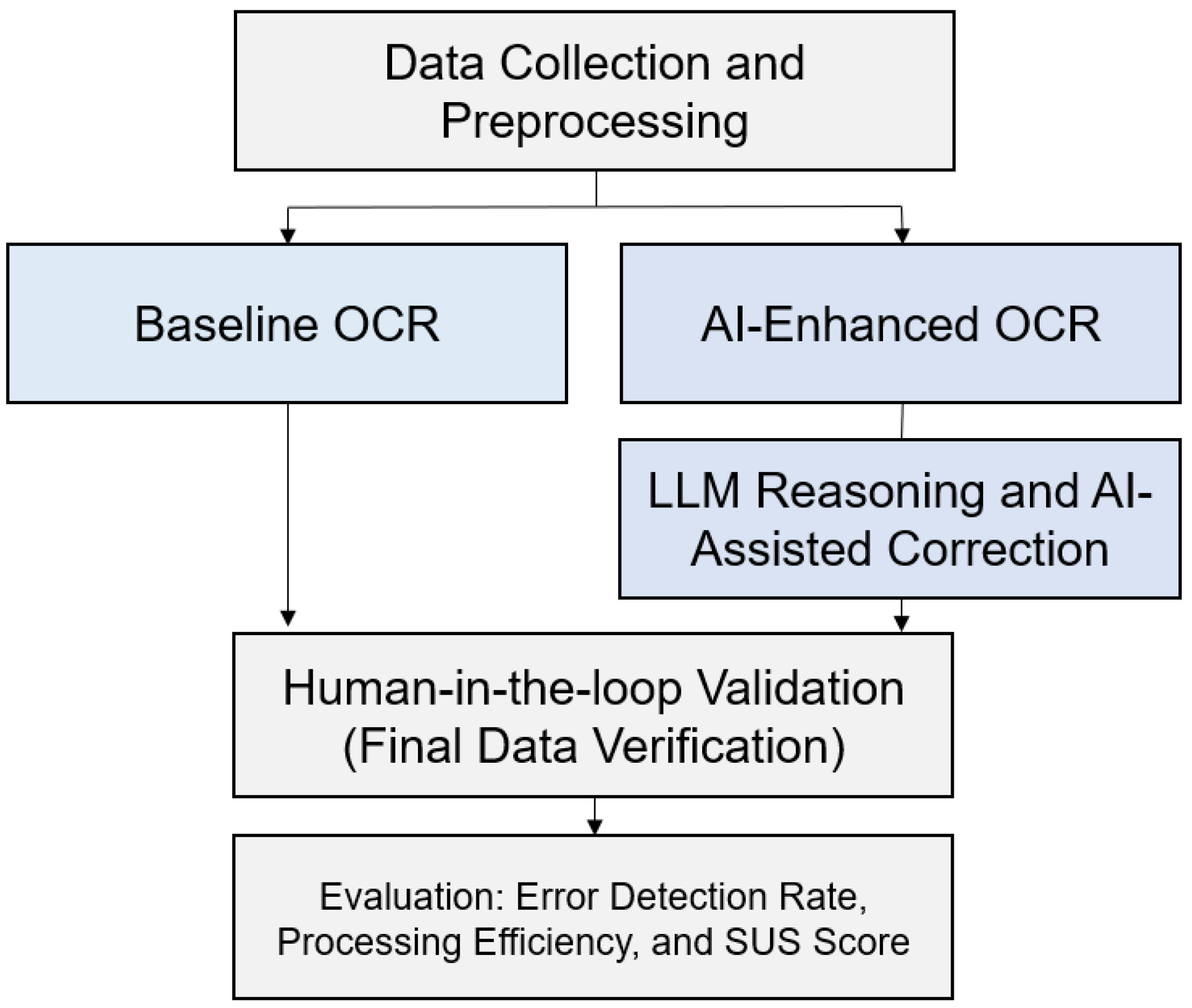
3. Methodology
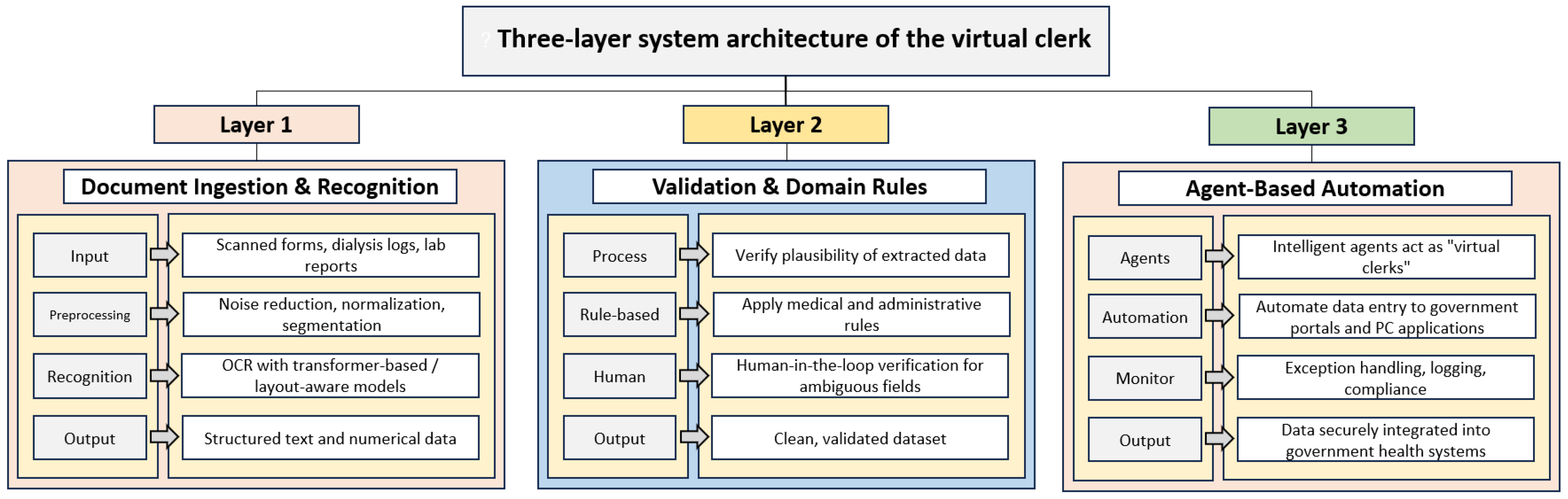
3.1. System Architecture
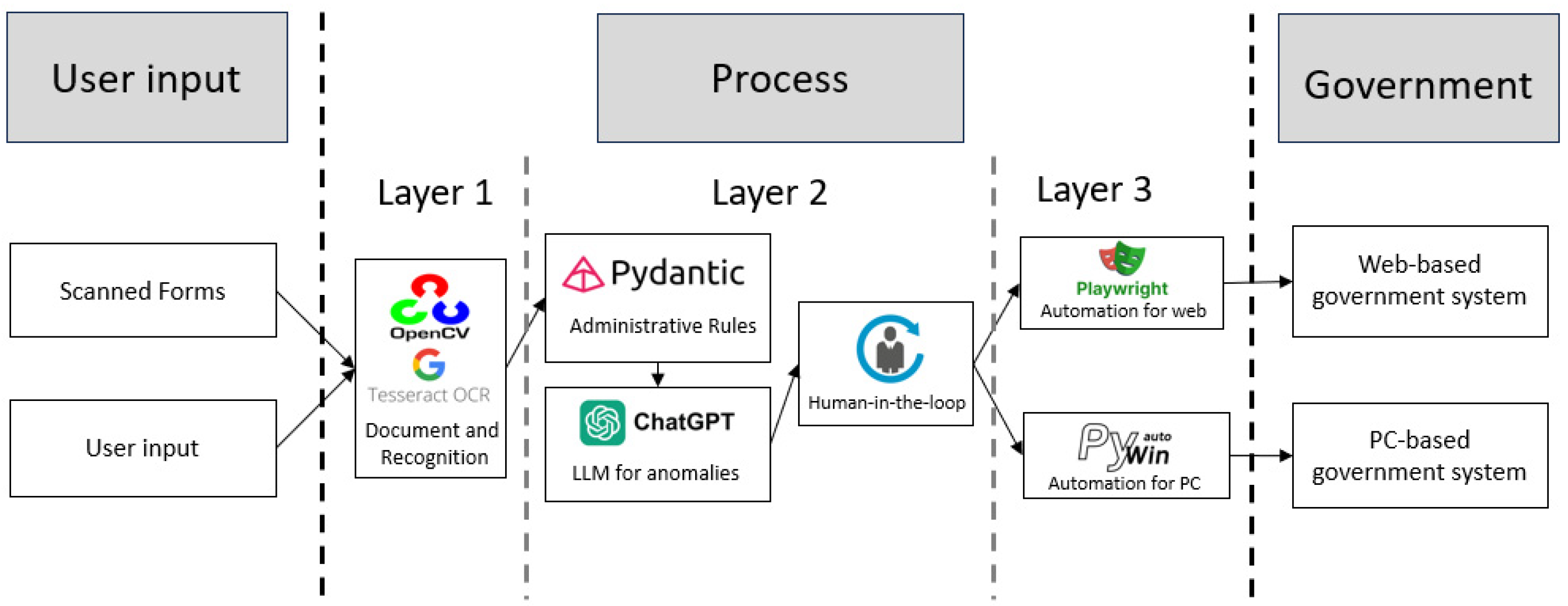
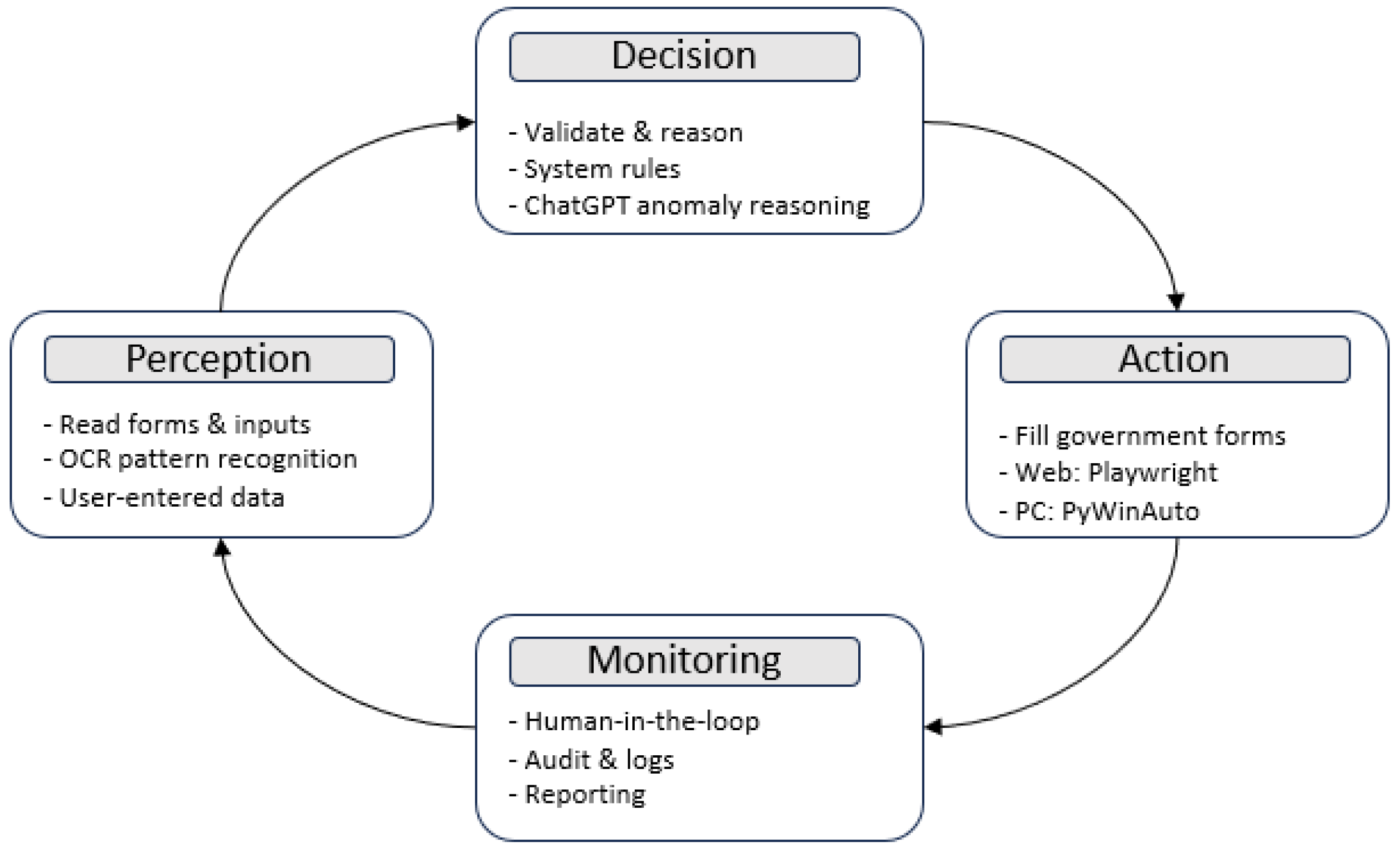
3.2. Agent-Based AI Design
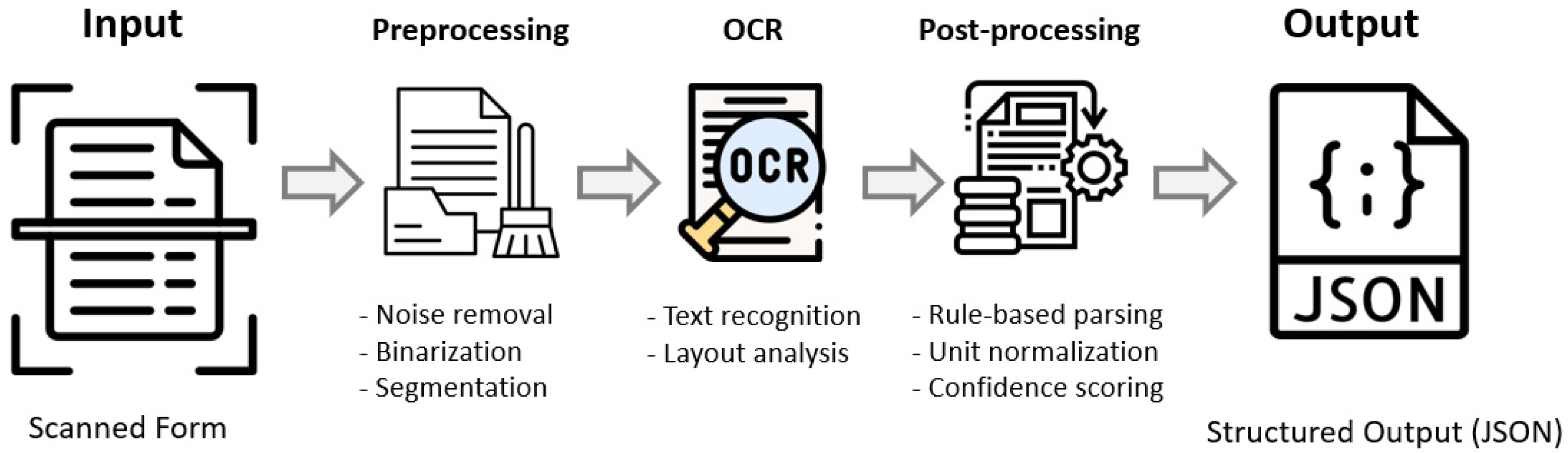
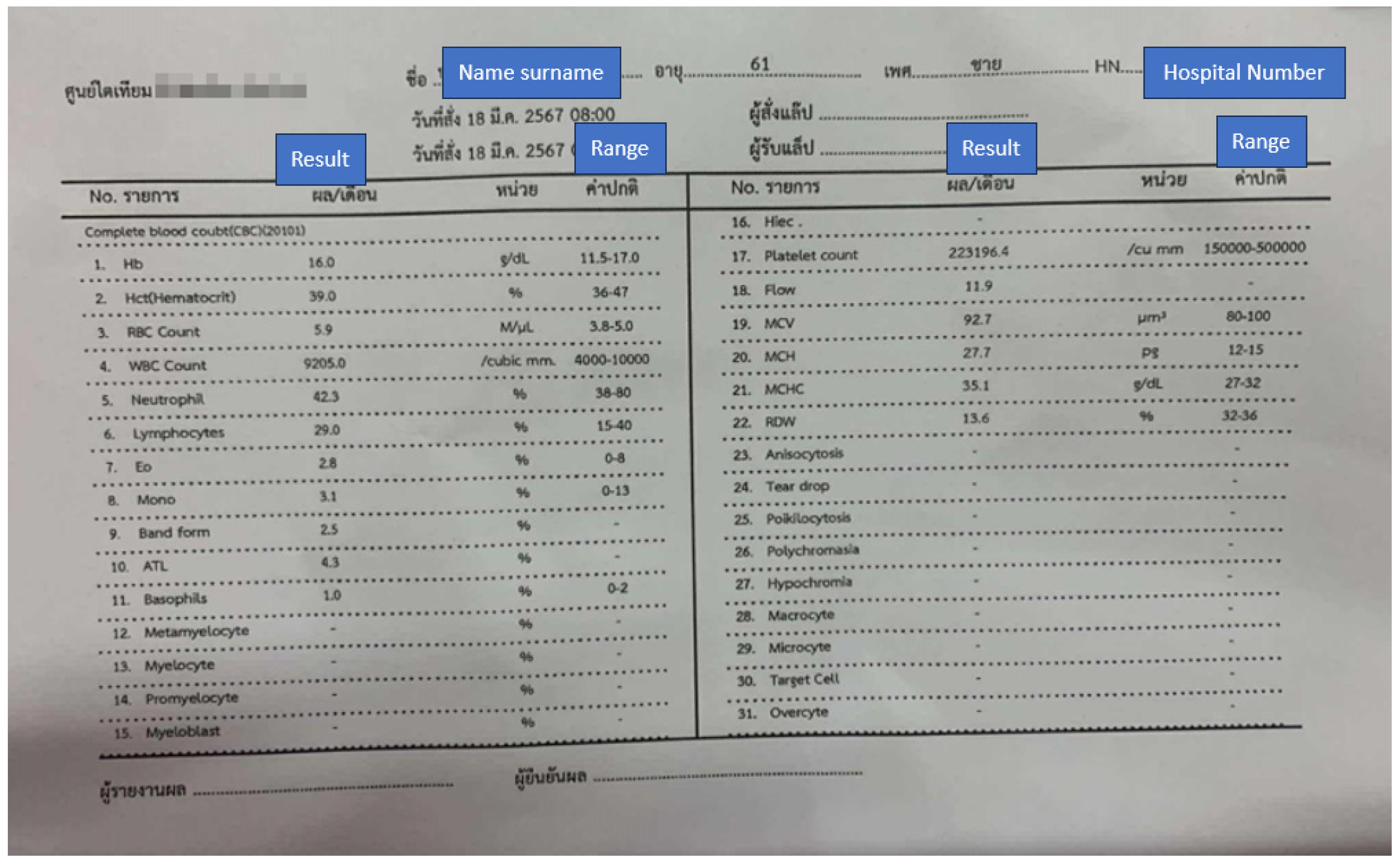
3.3. Image Processing Pipeline
3.4. OCR Configurations and Technical Integration
System Role: You are an AI-based clinical data validator operating within a rule-constrained data entry system.Your task is to analyze structured OCR outputs from nephrology forms, identify anomalies, and propose correctionsonly when they are derivable from contextual or domain-consistent evidence.Instructions:1. Input will be provided as a JSON object containing {field_name, value, confidence, data_type, rule_reference}.2. For each record:- Verify that the value conforms to expected type, unit, and range constraints (as indicated by rule_reference).- If confidence < 0.85 or rule violation is detected:a. Analyze related fields for contextual inference (e.g., Pre_Weight vs. Post_Weight, Urea vs. Creatinine).b. If a correction is logically deducible, output the revised value and reasoning note.c. If ambiguity remains, flag for human review.3. NEVER fabricate or infer data outside the observed record set.4. Return all outputs in strict JSON format:{“field_name”: “ “,“original_value”: “ “,“suggested_value”: “ “,“confidence”: “ “,“status”: “validated|corrected|flagged”,“reason”: “ “}
3.5. Evaluation Metrics
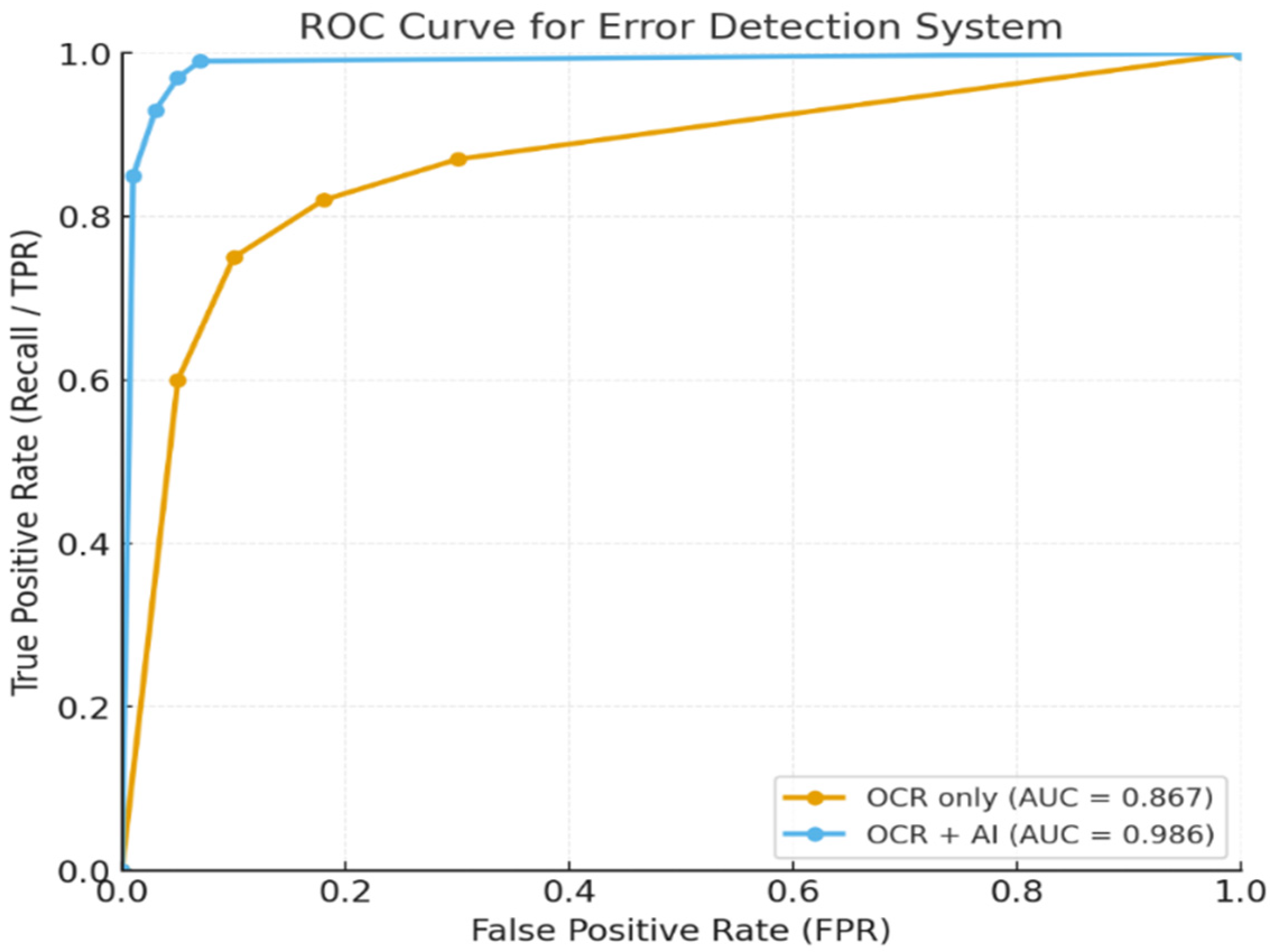
3.5.1. Error Detection Rate of Automation Accuracy
3.5.2. Time Efficiency
3.5.3. System Usability Evaluation
3.6. Experimental Design and Workflow
3.7. Data Sources
4. Results
4.1. Results of Error Detection Rate of Automation Accuracy
4.2. Results of Efficiency of Time
4.3. Results of System Usability and Adoption
5. Discussion
5.1. Summary of Key Findings
5.2. Comparison with Previous Studies
5.3. Practical Implications for Clinical Workflow
5.4. Limitations and Future Work
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| APIs | Application Programming Interfaces |
| EHR | Electronic Health Records |
| OCR | Optical Character Recognition |
| RPA | Robotic Process Automation |
| SUS | System Usability Scale |
References
- Satirapoj, B.; Tantiyavarong, P.; Thimachai, P.; Chuasuwan, A.; Lumpaopong, A.; Kanjanabuch, T.; Ophascharoensuk, V. Thailand Renal Replacement Therapy Registry 2023: Epidemiological Insights into Dialysis Trends and Challenges. Ther. Apher. Dial. 2025, 29, 721–729. [Google Scholar] [CrossRef]
- Spanakis, E.G.; Sfakianakis, S.; Bonomi, S.; Ciccotelli, C.; Magalini, S.; Sakkalis, V. Emerging and Established Trends to Support Secure Health Information Exchange. Front. Digit. Health 2021, 3, 636082. [Google Scholar] [CrossRef]
- Garza, M.Y.; Williams, T.; Ounpraseuth, S.; Hu, Z.; Lee, J.; Snowden, J.; Walden, A.C.; Simon, A.E.; Devlin, L.A.; Young, L.W.; et al. Error Rates of Data Processing Methods in Clinical Research: A Systematic Review and Meta-Analysis of Manuscripts Identified through PubMed. Int. J. Med. Inform. 2025, 195, 105749. [Google Scholar] [CrossRef]
- Zhou, X.; Zeng, T.; Zhang, Y.; Liao, Y.; Smith, J.; Zhang, L.; Wang, C.; Li, Q.; Wu, D.; Chong, Y.; et al. Automated Data Collection Tool for Real-World Cohort Studies of Chronic Hepatitis B: Leveraging OCR and NLP Technologies for Improved Efficiency. New Microbes New Infect. 2024, 62, 101469. [Google Scholar] [CrossRef]
- Budd, J. Burnout Related to Electronic Health Record Use in Primary Care. J. Prim. Care Community Health 2023, 14, 21501319231166921. [Google Scholar] [CrossRef]
- Gao, S.; Fang, A.; Huang, Y.; Giunchiglia, V.; Noori, A.; Schwarz, J.R.; Ektefaie, Y.; Kondic, J.; Zitnik, M. Empowering Biomedical Discovery with AI Agents. Cell 2024, 187, 6125–6151. [Google Scholar] [CrossRef]
- Rajkomar, A.; Dean, J.; Kohane, I. Machine Learning in Medicine. N. Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef] [PubMed]
- Beam, A.L.; Kohane, I.S. Big Data and Machine Learning in Health Care. JAMA 2018, 319, 1317–1318. [Google Scholar] [CrossRef] [PubMed]
- Shickel, B.; Tighe, P.J.; Bihorac, A.; Rashidi, P. Deep EHR: A Survey of Recent Advances in Deep Learning Techniques for Electronic Health Record (EHR) Analysis. IEEE J. Biomed. Health Inform. 2018, 22, 1589–1604. [Google Scholar] [CrossRef] [PubMed]
- Topol, E.J. High-Performance Medicine: The Convergence of Human and Artificial Intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]
- Sinsky, C.; Colligan, L.; Li, L.; Prgomet, M.; Reynolds, S.; Goeders, L.; Westbrook, J.; Tutty, M.; Blike, G. Allocation of Physician Time in Ambulatory Practice: A Time and Motion Study in 4 Specialties. Ann. Intern. Med. 2016, 165, 753–760. [Google Scholar] [CrossRef]
- Patrício, L.; Varela, L.; Silveira, Z. Integration of Artificial Intelligence and Robotic Process Automation: Literature Review and Proposal for a Sustainable Model. Appl. Sci. 2024, 14, 9648. [Google Scholar] [CrossRef]
- Wang, X.-F.; He, Z.-H.; Wang, K.; Wang, Y.-F.; Zou, L.; Wu, Z.-Z. A Survey of Text Detection and Recognition Algorithms Based on Deep Learning Technology. Neurocomputing 2023, 556, 126702. [Google Scholar] [CrossRef]
- Liu, Z.; Song, R.; Li, K.; Li, Y. From Detection to Understanding: A Systematic Survey of Deep Learning for Scene Text Processing. Appl. Sci. 2025, 15, 9247. [Google Scholar] [CrossRef]
- Xu, Y.; Li, M.; Cui, L.; Huang, S.; Wei, F.; Zhou, M. LayoutLM: Pre-Training of Text and Layout for Document Image Understanding. In Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Virtual, 6–10 July 2020; ACM: New York, NY, USA, 2020. [Google Scholar]
- Chen, X.; Jin, L.; Zhu, Y.; Luo, C.; Wang, T. Text Recognition in the Wild: A Survey. ACM Comput. Surv. 2022, 54, 1–35. [Google Scholar] [CrossRef]
- Nitayavardhana, P.; Liu, K.; Fukaguchi, K.; Fujisawa, M.; Koike, I.; Tominaga, A.; Iwamoto, Y.; Goto, T.; Suen, J.Y.; Fraser, J.F.; et al. Streamlining Data Recording through Optical Character Recognition: A Prospective Multi-Center Study in Intensive Care Units. Crit. Care 2025, 29, 117. [Google Scholar] [CrossRef]
- van der Aalst, W.M.P.; Bichler, M.; Heinzl, A. Robotic Process Automation. Bus. Inf. Syst. Eng. 2018, 60, 269–272. [Google Scholar] [CrossRef]
- Syed, R.; Suriadi, S.; Adams, M.; Bandara, W.; Leemans, S.J.J.; Ouyang, C.; ter Hofstede, A.H.M.; van de Weerd, I.; Wynn, M.T.; Reijers, H.A. Robotic Process Automation: Contemporary Themes and Challenges. Comput. Ind. 2020, 115, 103162. [Google Scholar] [CrossRef]
- Jennings, N.R.; Sycara, K.; Wooldridge, M. A Roadmap of Agent Research and Development. Auton. Agent. Multi. Agent. Syst. 1998, 1, 7–38. [Google Scholar] [CrossRef]
- Maes, P. Agents That Reduce Work and Information Overload. Commun. ACM 1994, 37, 30–40. [Google Scholar] [CrossRef]
- Mandel, J.C.; Kreda, D.A.; Mandl, K.D.; Kohane, I.S.; Ramoni, R.B. SMART on FHIR: A Standards-Based, Interoperable Apps Platform for Electronic Health Records. J. Am. Med. Inform. Assoc. 2016, 23, 899–908. [Google Scholar] [CrossRef]
- Kruse, C.S.; Frederick, B.; Jacobson, T.; Monticone, D.K. Cybersecurity in Healthcare: A Systematic Review of Modern Threats and Trends. Technol. Health Care 2017, 25, 1–10. [Google Scholar] [CrossRef]
- Kuo, T.-T.; Kim, H.-E.; Ohno-Machado, L. Blockchain Distributed Ledger Technologies for Biomedical and Health Care Applications. J. Am. Med. Inform. Assoc. 2017, 24, 1211–1220. [Google Scholar] [CrossRef]
- Sheller, M.J.; Edwards, B.; Reina, G.A.; Martin, J.; Pati, S.; Kotrotsou, A.; Milchenko, M.; Xu, W.; Marcus, D.; Colen, R.R.; et al. Federated Learning in Medicine: Facilitating Multi-Institutional Collaborations without Sharing Patient Data. Sci. Rep. 2020, 10, 12598. [Google Scholar] [CrossRef] [PubMed]
- Otsu, N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Sauvola, J.; Pietikäinen, M. Adaptive Document Image Binarization. Pattern Recognit. 2000, 33, 225–236. [Google Scholar] [CrossRef]
- Smith, R. An Overview of the Tesseract OCR Engine. In Proceedings of the Ninth International Conference on Document Analysis and Recognition (ICDAR 2007), Curitiba, Brazil, 23–26 September 2007; IEEE: Piscataway, NJ, USA, 2007; Volume 2. [Google Scholar]
- Hsu, E.; Malagaris, I.; Kuo, Y.-F.; Sultana, R.; Roberts, K. Deep Learning-Based NLP Data Pipeline for EHR-Scanned Document Information Extraction. JAMIA Open 2022, 5, ooac045. [Google Scholar] [CrossRef]
- Powers, D.M.W. Evaluation: From Precision, Recall and F-Measure to ROC, Informedness, Markedness and Correlation. arXiv 2020. [Google Scholar] [CrossRef]
- Weiskopf, N.G.; Weng, C. Methods and Dimensions of Electronic Health Record Data Quality Assessment: Enabling Reuse for Clinical Research. J. Am. Med. Inform. Assoc. 2013, 20, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, T. An Introduction to ROC Analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Brooke, J. SUS—A Quick and Dirty Usability Scale. Ahrq.gov. Available online: https://digital.ahrq.gov/sites/default/files/docs/survey/systemusabilityscale%2528sus%2529_comp%255B1%255D.pdf (accessed on 26 September 2025).
- Bangor, A.; Kortum, P.; Miller, J. Determining What Individual SUS Scores Mean: Adding an Adjective Rating Scale. J. Usability stud. 2009, 4, 114–123. [Google Scholar]
- Lewis, J.R. The System Usability Scale: Past, Present, and Future. Int. J. Hum. Comput. Interact. 2018, 34, 577–590. [Google Scholar] [CrossRef]
- Wu, Y.; Dalianis, H.; Velupillai, S. Errors in Clinical Text Processing and Their Impact on Decision-Making: A Review. Artif. Intell. Med. 2020, 104, 101833. [Google Scholar]
- Nguyen, P.A.; Shim, J.S.; Ho, T.B.; Li, W. Machine Learning-Based Approaches for Clinical Text Error Detection: A Systematic Review. J. Biomed. Inform. 2022, 127, 104018. [Google Scholar]
- Luo, Y.; Thompson, W.K.; Herr, T.M.; Zeng, Z.; Berendsen, M.A.; Jonnalagadda, S.R.; Carson, M.B.; Starren, J. Natural Language Processing for EHR-Based Pharmacovigilance: A Structured Review. Drug Saf. 2017, 40, 1075–1089. [Google Scholar] [CrossRef]
- Amann, J.; Blasimme, A.; Vayena, E.; Frey, D.; Madai, V.I.; Precise4Q consortium. Explainability for Artificial Intelligence in Healthcare: A Multidisciplinary Perspective. BMC Med. Inform. Decis. Mak. 2020, 20, 310. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.J.; Karthikesalingam, A.; Suleyman, M.; Corrado, G.; King, D. Key Challenges for Delivering Clinical Impact with Artificial Intelligence. BMC Med. 2019, 17, 195. [Google Scholar] [CrossRef]
- Small, W.R.; Wang, L.; Horng, S. EHR-Embedded Large Language Models for Hospital-Course Summarization. JAMA Netw. Open 2025, 8, e250112. [Google Scholar] [CrossRef]
- Kernberg, A.; Gold, J.A.; Mohan, V. Using ChatGPT-4 to Create Structured Medical Notes from Audio Recordings of Physician-Patient Encounters: Comparative Study. J. Med. Internet Res. 2024, 26, e54419. [Google Scholar] [CrossRef]
- World Health Organization. Ethics and Governance of Artificial Intelligence for Health: Guidance on Large Multi-Modal Models. Who.int. Available online: https://www.who.int/publications/i/item/9789240084759 (accessed on 26 September 2025).
- Howell, M.D. Generative Artificial Intelligence, Patient Safety and Healthcare Quality: A Review. BMJ Qual. Saf. 2024, 33, 748–754. [Google Scholar] [CrossRef] [PubMed]
- Reddy, S. Generative AI in Healthcare: An Implementation Science Informed Translational Path on Application, Integration and Governance. Implement. Sci. 2024, 19, 27. [Google Scholar] [CrossRef] [PubMed]
- Bakken, S. AI in Health: Keeping the Human in the Loop. J. Am. Med. Inform. Assoc. 2023, 30, 1225–1226. [Google Scholar] [CrossRef] [PubMed]
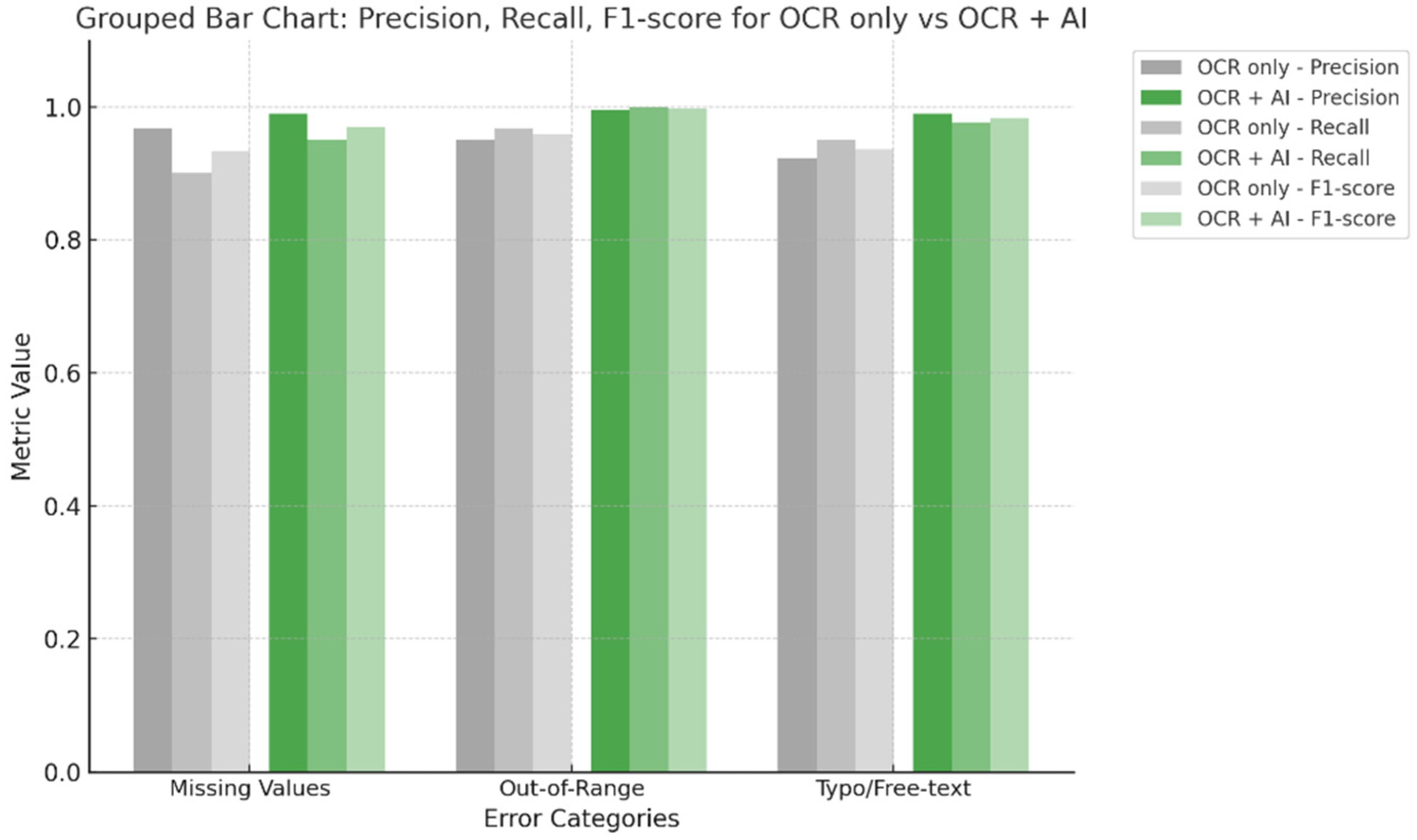
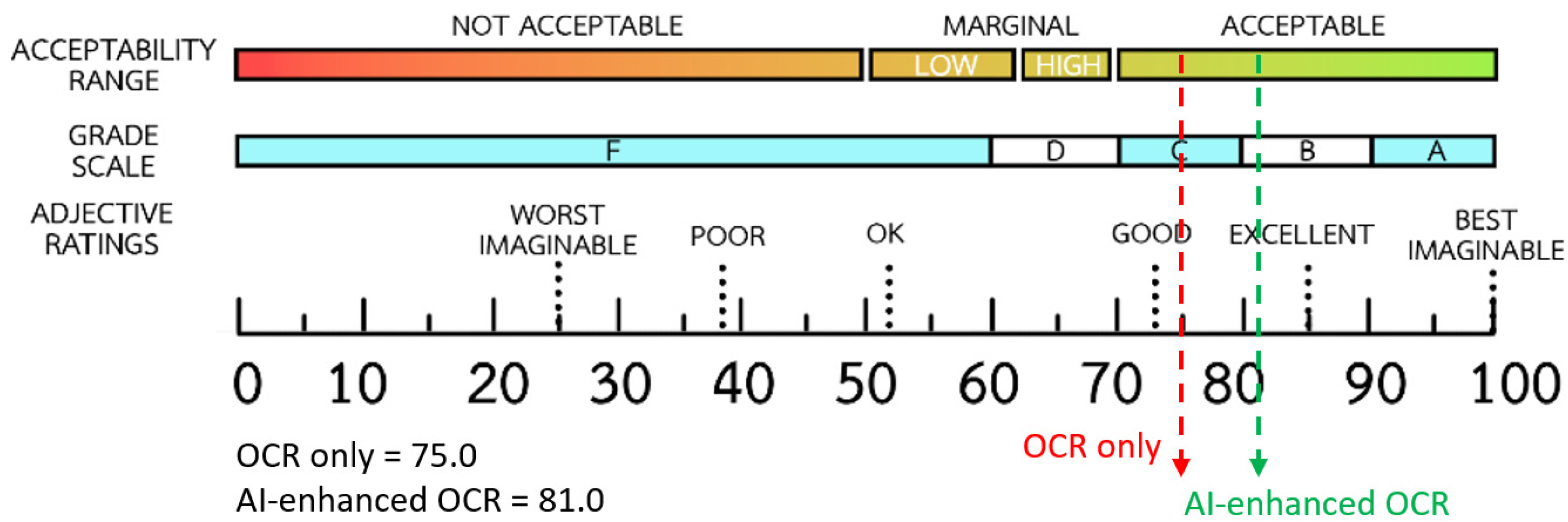
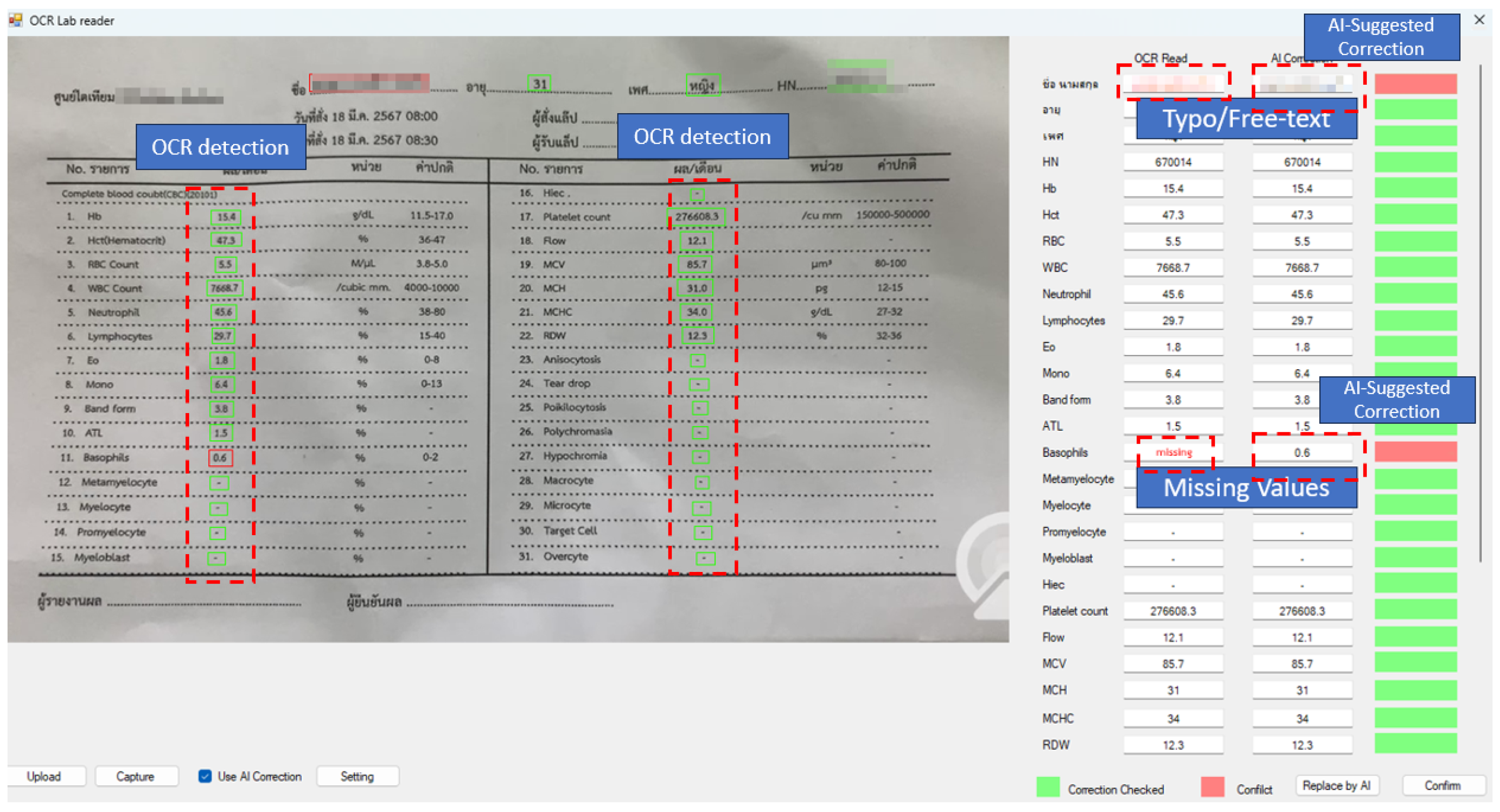
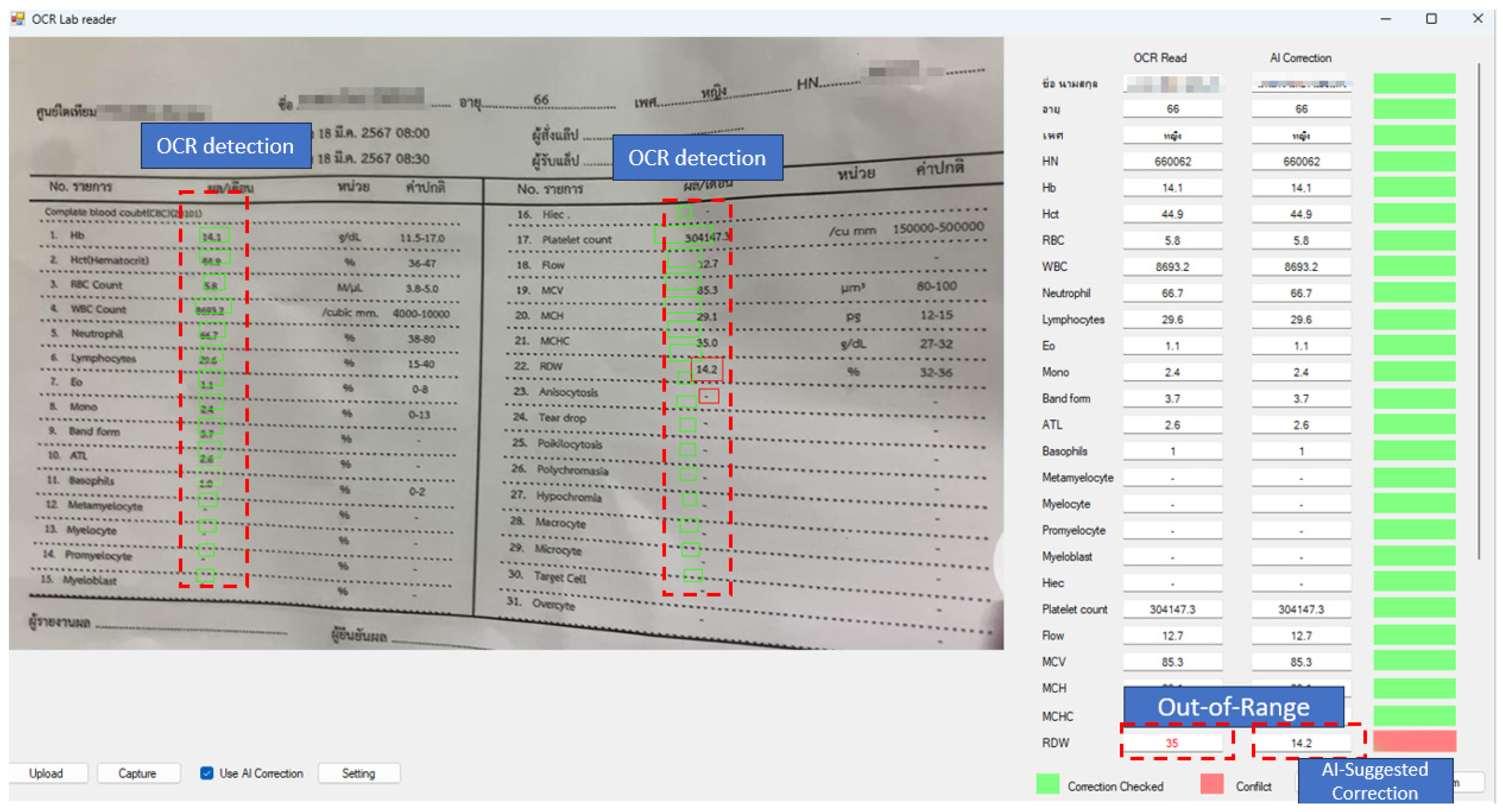
| Error Category | Detect Method | Precision | Recall (EDR) | F1-Score |
|---|---|---|---|---|
| Missing Values | OCR only | 0.968 | 0.900 | 0.933 |
| OCR + AI | 0.990 | 0.950 | 0.969 | |
| Out-of-Range | OCR only | 0.951 | 0.967 | 0.959 |
| OCR + AI | 0.995 | 0.999 | 0.997 | |
| Typo/Free-text | OCR only | 0.922 | 0.950 | 0.936 |
| OCR + AI | 0.990 | 0.977 | 0.983 |
| Metric | OCR Only | OCR + AI | Average Time Difference |
|---|---|---|---|
| Average Time per Record (sec) | 85.2 | 42.1 | 43.1 |
| Total Completion Time (min) | 92.3 | 45.6 | 46.7 |
| No. | Type | Questions | N | OCR Only Mean (SD) | OCR + AI Mean (SD) |
|---|---|---|---|---|---|
| 1 | Positive | I think I would like to use the OCR system/AI-enhanced OCR system regularly for completing healthcare data entry tasks. | 5 | 4.68 (0.47) | 4.74 (0.42) |
| 2 | Negative | I found the OCR system/AI-enhanced OCR system unnecessarily complex. | 5 | 1.92 (0.58) | 1.64 (0.51) |
| 3 | Positive | I thought the OCR system/AI-enhanced OCR system was easy to use. | 5 | 4.28 (0.52) | 4.52 (0.46) |
| 4 | Negative | I think I would need support from a technical expert to effectively use the OCR system/AI-enhanced OCR system. | 5 | 2.08 (0.63) | 1.78 (0.55) |
| 5 | Positive | I found the functions of the OCR system/AI-enhanced OCR system to be well integrated into the existing workflow. | 5 | 4.40 (0.49) | 4.46 (0.48) |
| 6 | Negative | I thought there was too much inconsistency in the OCR system/AI-enhanced OCR system. | 5 | 3.18 (0.81) | 2.86 (0.77) |
| 7 | Positive | I imagine most healthcare staff would learn to use the OCR system/AI-enhanced OCR system very quickly. | 5 | 3.58 (0.69) | 3.92 (0.66) |
| 8 | Negative | I found the OCR system/AI-enhanced OCR system cumbersome to use. | 5 | 1.84 (0.54) | 1.66 (0.53) |
| 9 | Positive | I felt confident using the OCR system/AI-enhanced OCR system. | 5 | 4.32 (0.51) | 4.62 (0.47) |
| 10 | Negative | I needed to learn many things before I could start using the OCR system/AI-enhanced OCR system. | 5 | 2.24 (0.60) | 1.92 (0.56) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Worragin, P.; Chernbumroong, S.; Puritat, K.; Julrode, P.; Intawong, K. Towards Intelligent Virtual Clerks: AI-Driven Automation for Clinical Data Entry in Dialysis Care. Technologies 2025, 13, 530. https://doi.org/10.3390/technologies13110530
Worragin P, Chernbumroong S, Puritat K, Julrode P, Intawong K. Towards Intelligent Virtual Clerks: AI-Driven Automation for Clinical Data Entry in Dialysis Care. Technologies. 2025; 13(11):530. https://doi.org/10.3390/technologies13110530
Chicago/Turabian StyleWorragin, Perasuk, Suepphong Chernbumroong, Kitti Puritat, Phichete Julrode, and Kannikar Intawong. 2025. "Towards Intelligent Virtual Clerks: AI-Driven Automation for Clinical Data Entry in Dialysis Care" Technologies 13, no. 11: 530. https://doi.org/10.3390/technologies13110530
APA StyleWorragin, P., Chernbumroong, S., Puritat, K., Julrode, P., & Intawong, K. (2025). Towards Intelligent Virtual Clerks: AI-Driven Automation for Clinical Data Entry in Dialysis Care. Technologies, 13(11), 530. https://doi.org/10.3390/technologies13110530








