Abstract
The diagnosis of tongue disease is based on the observation of various tongue characteristics, including color, shape, texture, and moisture, which indicate the patient’s health status. Tongue color is one such characteristic that plays a vital function in identifying diseases and the levels of progression of the ailment. With the development of computer vision systems, especially in the field of artificial intelligence, there has been important progress in acquiring, processing, and classifying tongue images. This study proposes a new imaging system to analyze and extract tongue color features at different color saturations and under different light conditions from five color space models (RGB, YcbCr, HSV, LAB, and YIQ). The proposed imaging system trained 5260 images classified with seven classes (red, yellow, green, blue, gray, white, and pink) using six machine learning algorithms, namely, the naïve Bayes (NB), support vector machine (SVM), k-nearest neighbors (KNN), decision trees (DTs), random forest (RF), and Extreme Gradient Boost (XGBoost) methods, to predict tongue color under any lighting conditions. The obtained results from the machine learning algorithms illustrated that XGBoost had the highest accuracy at 98.71%, while the NB algorithm had the lowest accuracy, with 91.43%. Based on these obtained results, the XGBoost algorithm was chosen as the classifier of the proposed imaging system and linked with a graphical user interface to predict tongue color and its related diseases in real time. Thus, this proposed imaging system opens the door for expanded tongue diagnosis within future point-of-care health systems.
1. Introduction
Traditional Chinese medicine (TCM) uses a four-part diagnostic process that includes examination, smell, auditory evaluation, investigation, and physical palpation to obtain valuable information for evaluating diseases. It is worth noting that tongue examination, a practice with a history spanning more than two thousand years, plays a vital role in TCM [1]. Human tongues possess unique characteristics and features connected to the body’s internal organs, which effectively detect illnesses and monitor their progress [2,3,4]. Key features of this evaluation include the color of the tongue, shade of the coating, form of the tongue, depth of the coating, oral moisture, tongue crevices, contusions, red spots, and tooth impressions [5]. Among these, tongue color is of the most importance [3]. A healthy tongue usually shows a pink color and a slender white film [4], and various ailments directly impact this color. For example, diabetes mellitus (DM) often leads to oral complications, resulting in a yellow coating of the tongue [6]. In DM2, the tongue may be blue with a yellow coating [5]. A purple tongue with a thick fatty layer is a symptom of cancer [7], while patients with acute stroke may present with an unusually shaped red tongue [8]. A white tongue may indicate chill syndrome or a lack of iron in the blood, while yellow signifies a condition of increased body heat, hepatic and biliary organ disease [9,10], and vascular or gastrointestinal issues, which may cause the oral organ to turn indigo or violet [11]. Appendicitis can lead to changes in the exterior of the tongue [12], and the tongue can also reflect the intensity of bacterial or viral COVID-19 affliction, appearing faint pink in mild cases, crimson in moderate infections, and deep red (burgundy) in serious cases. These diseases are often accompanied by inflammation and ulceration [10,13]. In cases of Helicobacter pylori infection, the tongue may appear reddish with a touch of white markings [8]. Systems for diagnosing based on the examination of the tongue (TDS) are known to be highly effective without the need for invasive procedures for remote disease diagnosis or direct contact with the patients. These systems play a critical role in clinical decision making, reducing the possibility of human error.
2. Related Work
In recent years, there has been an extensive range of studies focused on computer vision systems to analyze and evaluate the tongue’s color. For example, a study conducted by Park et al. (2016) [14] explored the examination of color features of the tongue, making use of digital cameras combined with distinct digital imaging software. The study involved the use of 200 tongue images from female applicants and employed questionnaires related to phlegm pattern, temperature, and yin deficiency. This project addressed two opposing and balancing elements in TCM (Yin and Yang) through its investigations. Another study by Gabhale et al. (2017) [15] proposed a computer-based analyzer system to detect diabetes by appraising visual variations on the tongue’s surface. This study used statistical mapping of tongue images based on combinations of color and material features with a dataset of 100 tongue images. The study findings demonstrated the method’s effectiveness in classifying diabetes. In another study, Umadevi et al. (2019) [16] performed an implicit association method with patients’ tongue images for diagnosing diabetes based on the dental multi-labeled region approach. The method’s performance was evaluated based on 96 images of patients’ tongues from both a medical facility and a research institution, along with 97 ultraviolet-scanned images of tongues captured using an iPhone equipped with a high-resolution camera. Srividhya and Muthukumaravel (2019) [10] conducted a study employing a classifier based on support vector machines (SVMs) and sophisticated image processing methods to process textures extracted from tongue images. These images were captured using digital cameras without any consideration for lighting conditions. Thirunavukkarasu et al. (2019) [17] presented an infrared tongue thermography system used to assess tongue temperature for diabetes diagnosis. This suggested framework classifies hyperglycemia by analyzing thermal differences in the oral organ based on convolutional neural networks (CNNs). Thermal image tongue information from 140 people was collected by utilizing infrared cameras. Naveed et al. (2020) [18] developed an algorithm based on fractional-order Darwinian particle swarm optimization for hyperglycemia diagnosis based on 700 tongue images taken using digital cameras and smartphones. Another study by Horzof et al. (2021) [13] involved analysis of the tongue based on images from COVID-19 patients using the Cochran–Armitage test based on polymerase chain reaction to drive the anticipated relationship between three elements represented by tongue color, plaque color, and disease severity. A total of 135 patients’ (males and females) tongue images were collected using smartphones. The study found that approximately 64.29% of patients with mild infections, 62.35% of patients with moderate infections, and 99% of patients with severe infections showed light pink tongues, crimson tongues, and deep red (wine-colored) tongues, respectively. Another study conducted by Mansour et al. (2021) [19] illustrated a new model composed, first, of an automated Internet of Things (IoT) system and, second, a deep neural network (DNN) to detect and classify disease from colored tongue images. Recently, a study conducted by Abdullah et al. (2021) [20] utilized classical machine learning algorithms and deep learning techniques for feature fusion in predicting prediabetes and diabetes. The GAXGBT model with fusion features achieved an average F1-score of 0.796 on the test set, demonstrating enhanced prediction accuracy. High precision (0.929) and recall (0.951) were observed in detecting diabetes, indicating the model’s effectiveness in identifying individuals at risk. The GAXGBT model exhibited an average CA of 0.81 and an average AUROC of 0.918 on the test set, reflecting good overall accuracy. The model showed a sensitivity of 0.952 for detecting prediabetes and 0.951 for diabetes, highlighting its ability to correctly identify individuals at risk. A study conducted by Jun Li et al. (2022) [21] utilized a hybrid convolutional neural network with the Deep RBFNN model for diabetes diagnosis. The study was performed using the MATLAB R2019a environment on a system with specific hardware configurations. The main results of the study revealed the successful classification of diabetes based on tongue image features, as the artificial intelligence model achieved high levels of accuracy, sensitivity, and specificity in the diagnostic process. The accuracy of the AI model in diabetes classification was reported to be 0.989 for training and 0.984 for testing, indicating its strong performance in identifying the disease. Sensitivity, which represents the true positive rate, and specificity, which indicates the true negative rate, were also high in the model, further validating its effectiveness in diagnosing diabetes. The research used a dataset containing a specific number of samples, although the exact number is not explicitly stated in the contexts provided. The AI method used in the study of Saritha Balasubramaniyan et al. (2022) [22] involved training deep learning models, such as YOLOv5s6, U-Net, and MobileNetV3, on tongue image datasets to recognize, segment, and classify tongue features. The main results of the study indicated that the tongue diagnosis model achieved satisfactory performance, with an accuracy of 93.33% for tooth marks, 89.60% for stains, and 97.67% for cracks. Evaluation metrics used to evaluate the model included accuracy, sensitivity, and specificity, although they were not explicitly stated in the contexts provided. However, the study showed good detection, segmentation, and classification performance of the AI models. A study conducted by Zibin Yang et al. (2022) [23] presented a live imaging system using a webcam for assessing tongue color and identifying ailments, including hyperglycemia, kidney insufficiency, deficiency in red blood cells, cardiac failure, and respiratory conditions. However, their experimental results were obtained under fixed light conditions, and they utilized “if” and “else if” commands in MATLAB, which constrained the results when input data fell outside the predefined condition range. All the abovementioned studies are promising as computerized tongue color analysis systems. However, they neglected the challenge of considering the intensity and difference of lighting and its effect on the colors of the tongue and thus might have given inaccurate diagnoses in some cases. Therefore, we propose a live imaging system for examining tongue color and diagnosing the related diseases using different machine learning models, namely, the NB, SVM, KNN, DT, RF, and XGBoost algorithms, trained to address the issue of lighting condition intensities during imaging.
3. Materials and Methods
3.1. Data Collection
In this study, two different groups of data were used. The first group consisted of 5260 images classified with seven colors for different color saturations and light conditions, namely, red as class 1 with 1102, yellow as class 2 with 1010 images, green as class 3 with 945, blue as class 4 with 1024, gray as class 5 with 737, and white as class 6 with 300 images to cover the abnormal tongue colors, and pink was set as class 7 with 310 images of normal healthy tongues. Rows of values obtained from the five color spaces (RGB, YCbCr, HSV, LAB, and YIQ) were used as raw features and then saved as an Excel sheet file in CSV (comma delimited) format as input for different machine learning models. For training purposes, 80% of the dataset was employed to train the machine learning algorithms, and 20% of the remaining dataset was employed for testing. The second group of data comprised 60 abnormal tongue images that were collected at Al-Hussein Teaching Hospital in Dhi Qar, Iraq, and Mosul General Hospital in Mosul from January 2022 to December 2023 to test the proposed imaging system in real time. These tongue images included patients with various conditions, including DM, mycotic infection, asthma, COVID-19, fungiform papillae, and anemia. This research adheres to the ethical standards of the Helsinki Declaration (Helsinki, 1964) with the Individual Ethics Protocol ID 201/21 granted by the Ethical Standards for Human Research Commission at the Human Research Ethics Committee at the Ministry of Health and Environment, Training and Human Development Centre, Iraq. Written consent was obtained from all involved subjects after a comprehensive clarification of the experimental methodologies.
3.2. Experimental Configuration
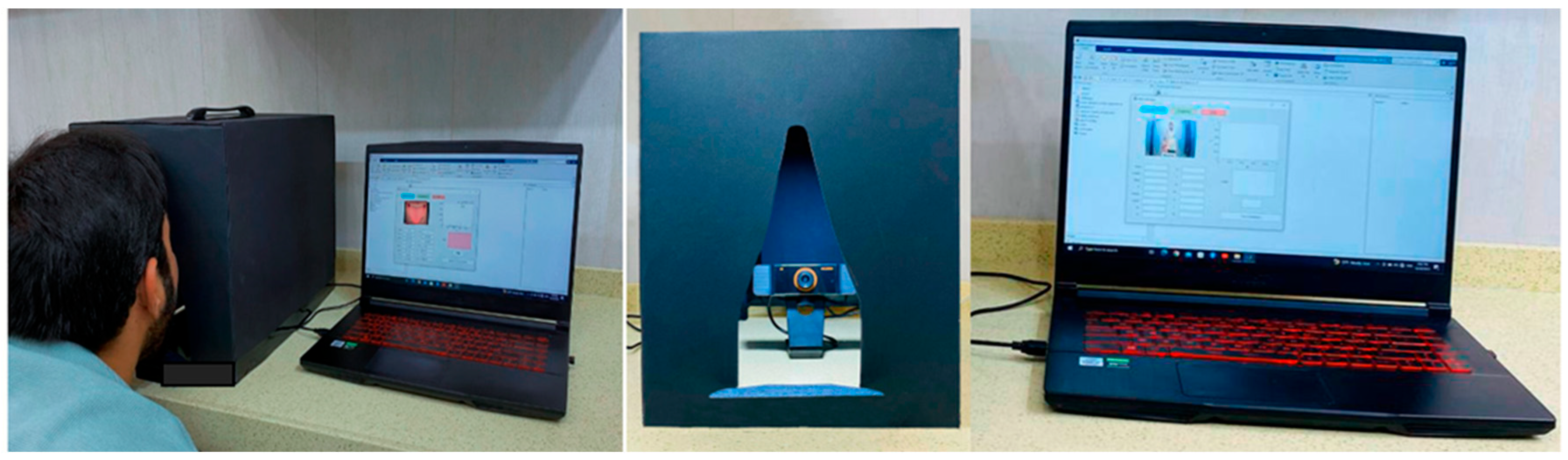
Within this investigation, the patient sat in front of the camera at a distance of 20 cm while the proposed imaging system detected the color of the tongue and predicted health status in real time. A laptop with the MATLAB program installed and a USB webcam (1920 × 1080 pixels) was used to capture and extract color features from the tongue images, as shown in Figure 1. MATLAB App Designer was used as a graphical user interface (GUI) to facilitate real-time analysis of tongue color captured by the webcam. The GUI not only illustrates the values of every hue but also determines the tongue’s color in the image and histogram and identifies possible diseases associated with the observed color.

Figure 1.
Device for image acquisition and experimental configuration.
3.3. System Design
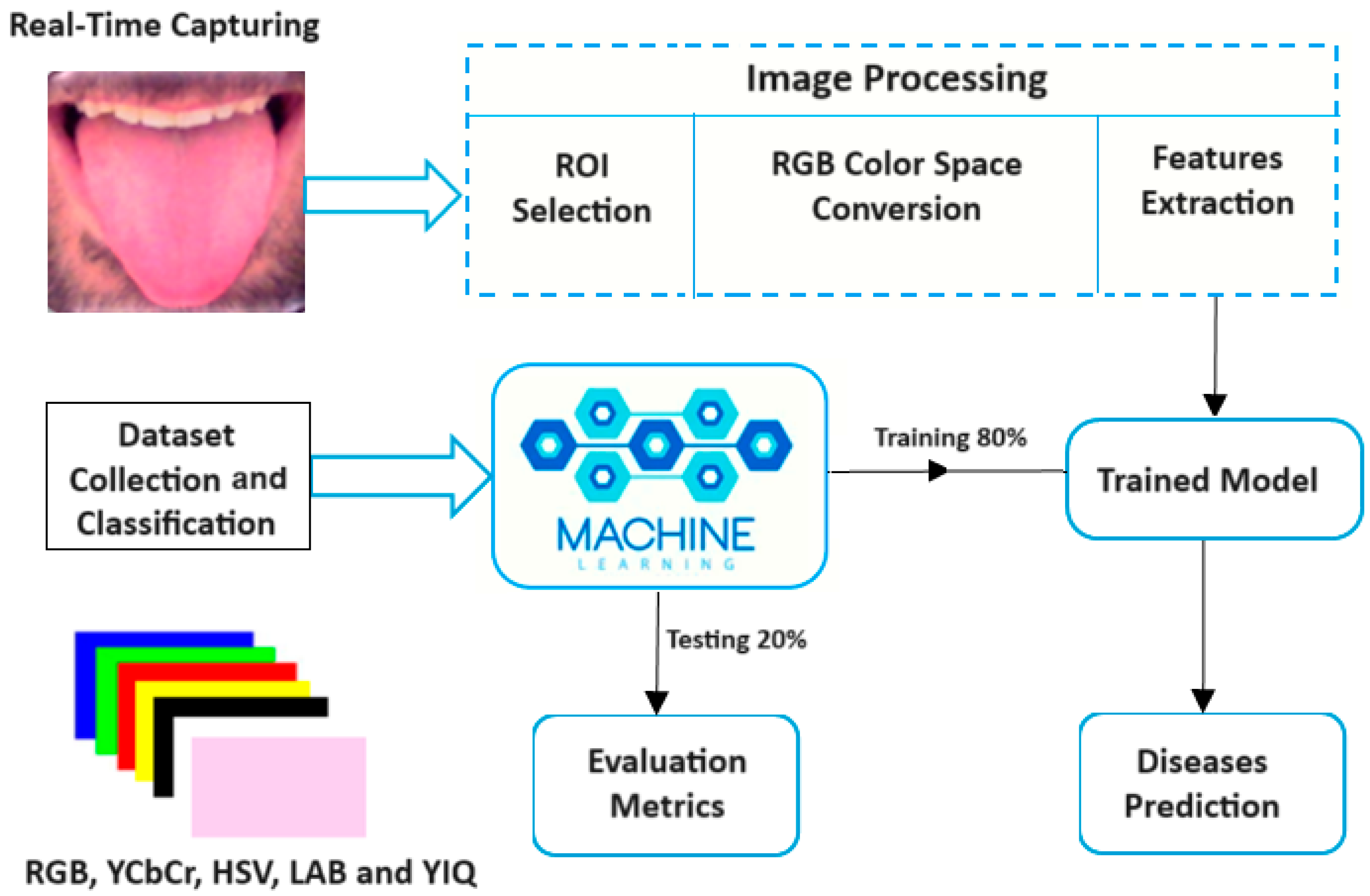
The proposed imaging system is primarily based on image processing methods to analyze tongue color. Several machine learning algorithms were trained to detect tongue color, and the saved trained model was used to diagnose the related diseases in real time, as shown in Figure 2.
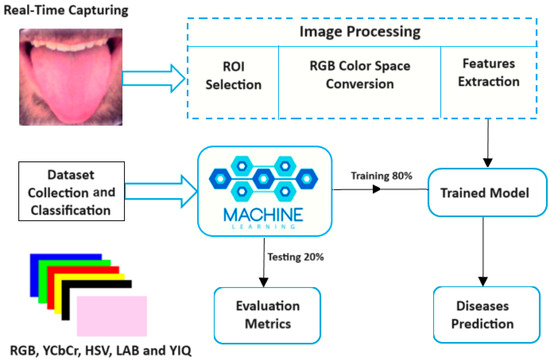
Figure 2.
Block diagram of the tongue color analysis system.
3.3.1. Image Analysis
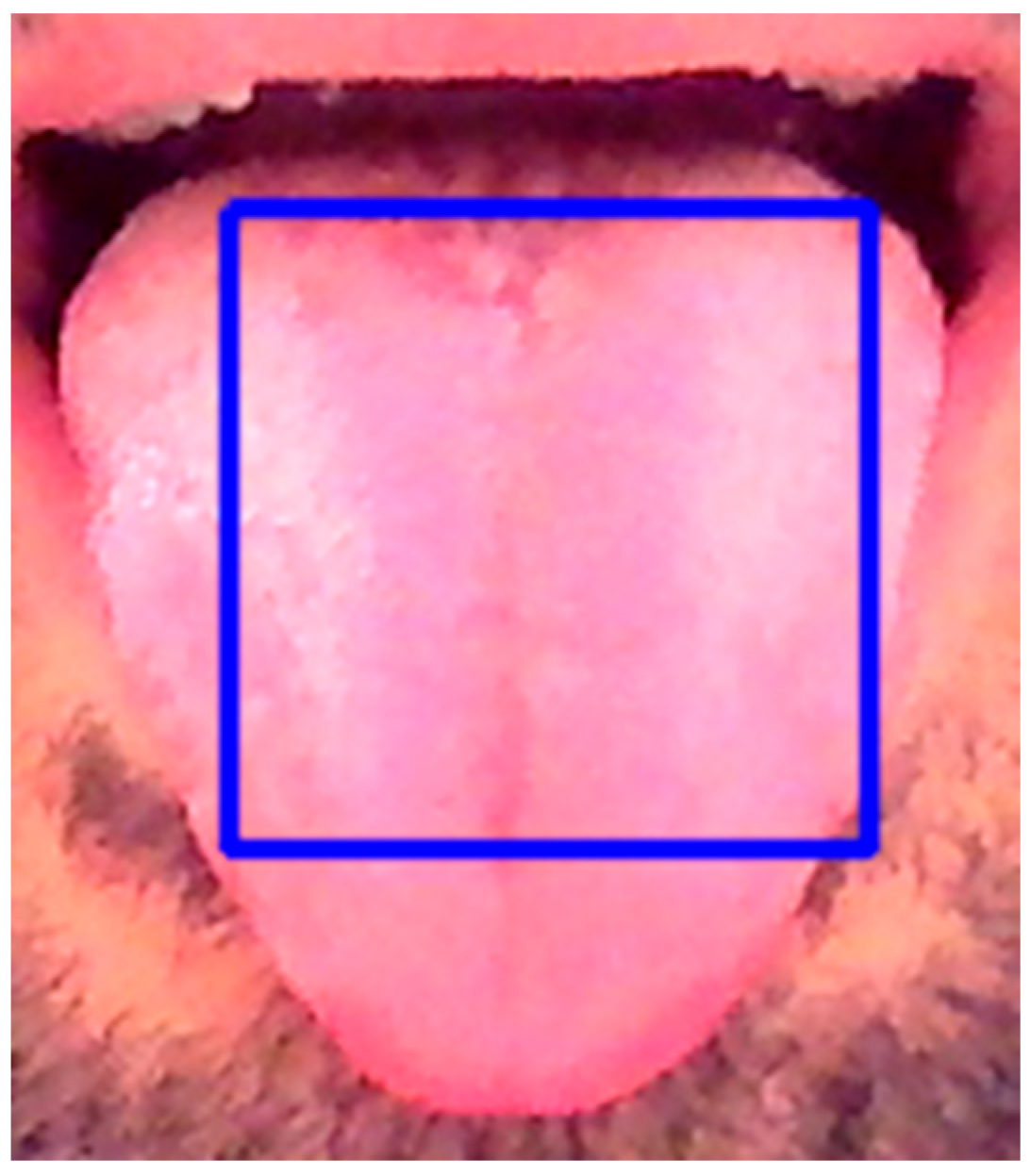
To analyze tongue color after capturing the tongue image, an image segmentation method was used to automatically segment the tongue image and remove the facial background, including the lips, mustache, beard, teeth, etc., and then select the central region of the tongue as a region of interest (ROI), as shown in Figure 3.
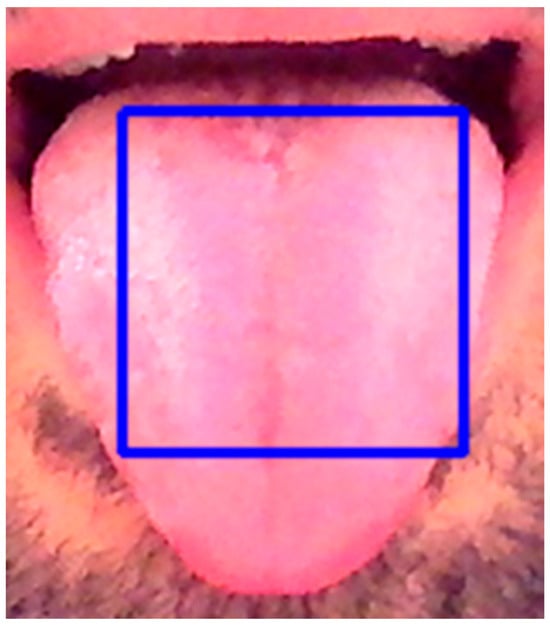
Figure 3.
ROI selection.
The next processing step is color space conversion. The RGB color space generally does not reflect specific color information; therefore, the RGB color space was converted to YCbCr, HVS, LAB, and YIQ models for a more in-depth color information analysis. This process was achieved using a color space conversion function in the MATLAB environment. The intensities from different channel values after classifying their colors were saved in CSV file format and then fed to different machine learning algorithms (NB, SVM, KNN, DT, RF, and XGBoost) to train the model. The color spaces included RGB (red, green, blue), the most common color model used in digital devices like cameras, computer monitors, and TVs. This model represents colors as combinations of red, green, and blue light. Each color channel has a range of intensity from 0 to 255 in an 8-bit system, allowing for a wide range of colors to be displayed. YCbCr is a color space that separates luminance (Y) from chrominance (Cb and Cr). The Y component represents brightness, while the Cb and Cr components represent color information. YCbCr is often used in image and video compression algorithms because it separates color information, making compression more efficient. The HVS (Human Visual System) is a color space designed to mimic the characteristics of the human eye’s perception of color. It takes into account factors, such as color sensitivity and brightness perception, to create a color representation that is closer to how humans see colors. LAB is a color space defined by the International Commission on Illumination (CIE). It consists of three components: L for lightness, A for the green–red axis, and B for the blue–yellow axis. LAB is used in color science and image processing for its perceptual uniformity, meaning that equal distances in the LAB space roughly correspond to equal perceptual differences in a color space used in analog television systems, particularly in the NTSC standard. Similar to YCbCr, it separates luminance (Y) from chrominance (I and Q), with I representing the blue–yellow axis and Q representing the green–magenta axis. YIQ was used in older television systems and is less common in digital image processing compared to RGB or YCbCr.
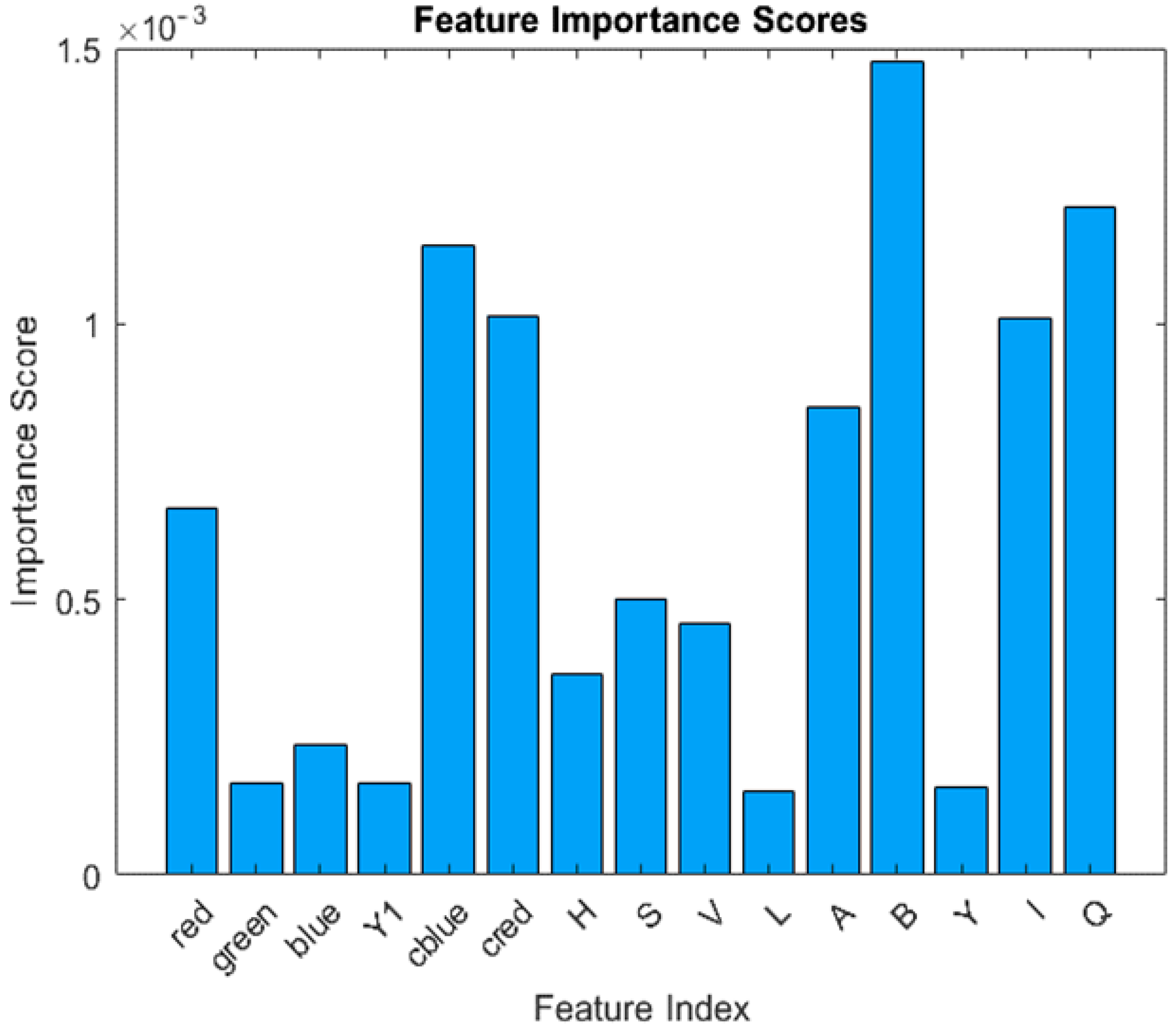
The feature importance used in the decision making of machine learning algorithms is shown in Figure 4.
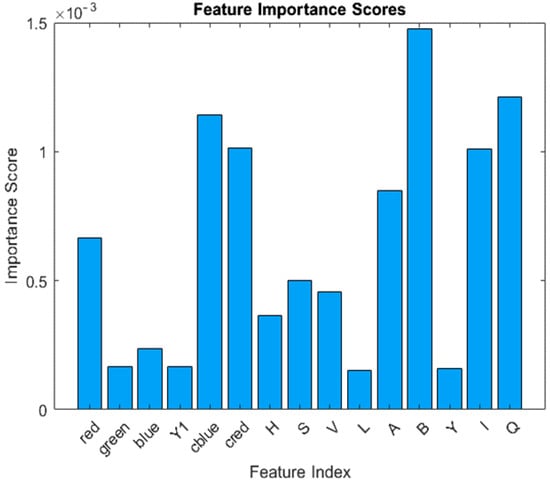
Figure 4.
Feature importance for RGB, YCbCr, HVS, LAB, and YIQ channels.
The proposed imaging system based on the trainable model obtained from the best-performing algorithm can detect tongue color and classify it as normal or abnormal in real time. The intensity values obtained from RGB YCbCr, HVS, LAB, and YIQ channels in real time were compared to the trainable model to predict tongue color and possible related diseases.
3.3.2. Tongue Color as a Diagnostic Indicator for Diseases
The convergence of important artificial intelligence techniques in tongue diagnosis research aims to enhance the reliability and accuracy of diagnosis and address the long-term outlook for large-scale artificial intelligence applications in healthcare. The emergence of artificial intelligence has not only created a distinct and rapidly developing niche in the field of TCM but has also generated urgent and widespread demand for advances in tongue diagnostic technology [24]. In the clinical practice of TCM, practitioners rely on their observations pertaining to various tongue attributes, including color, contours, coating, texture, and amount of saliva, as indicators for diseases [25]. However, this approach is often influenced by subjective evaluation arising from individual visual perception, color analysis, and individual experience. Thus, this reliance on independence leads to subjectivity in diagnosis and challenges in replication. Traditional tongue diagnosis mostly focuses on the visual aspects of the tongue, including color, shape, and coating [24]. A change in tongue color may indicate several diseases, as shown in Table 1.

Table 1.
Tongue color as a diagnostic indicator for diseases.
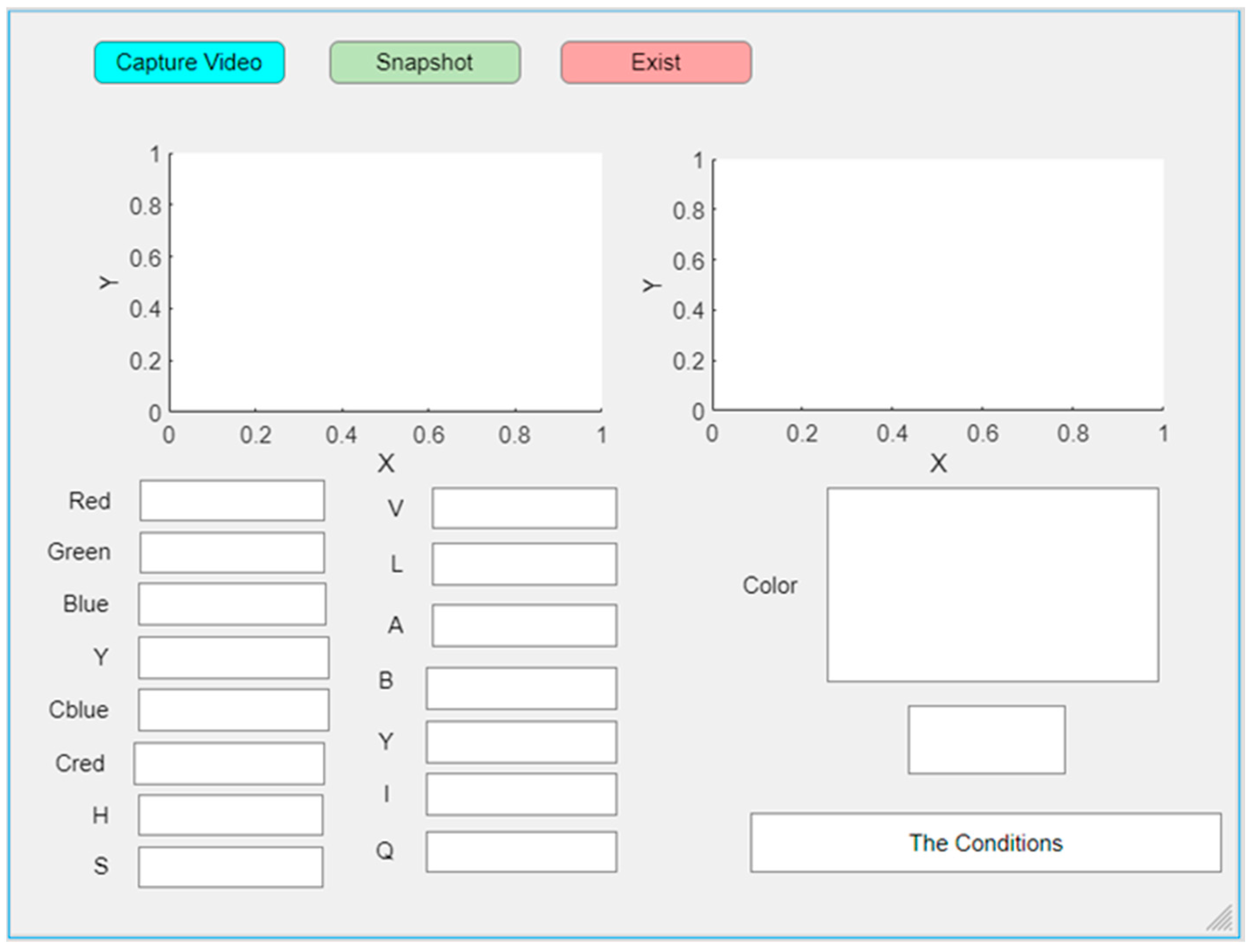
3.3.3. MATLAB App Designer
The graphical interface of this program was designed to simplify the use of MATLAB for the purpose of determining the patient’s condition. First, we connected the camera to the computer and ran the MATLAB program to display the graphical interface, as shown in Figure 5, by pressing the “Capture Video” button. An image of the patient’s tongue appeared, after which we pressed the “Snapshot” button. The program took a picture of the oral organ and displayed the color of the tongue in addition to all the features of the tongue, including RGB, YCbCr, HVS, LAB, and YIQ color models, and displayed whether the person is in a pathological or normal condition.
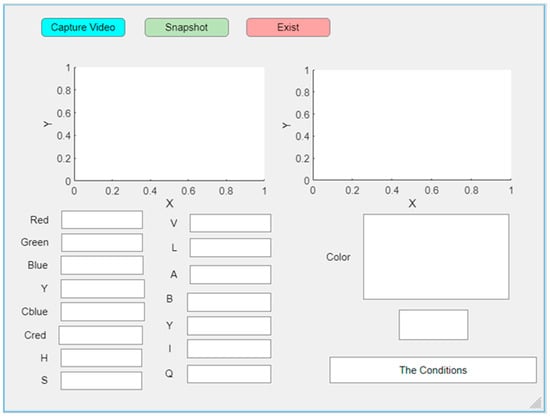
Figure 5.
Designed GUI of the proposed system using MATLAB App Designer.
3.4. Implementation of Machine Learning Algorithms as Classifiers
3.4.1. Naïve Bayes Categorizer
The naïve Bayes classifier is an approach to classification based on Bayes’ theorem, employing probability and statistical principles [26].
where forecasts for the future are possibly derived from past encounters. The naïve Bayes classifier additionally possesses distinctive attributes that make strong assumptions of independence for any occurrence or state [27,28]. As shown in Figure 6, some benefits of employing the naïve Bayes approach are as follows:
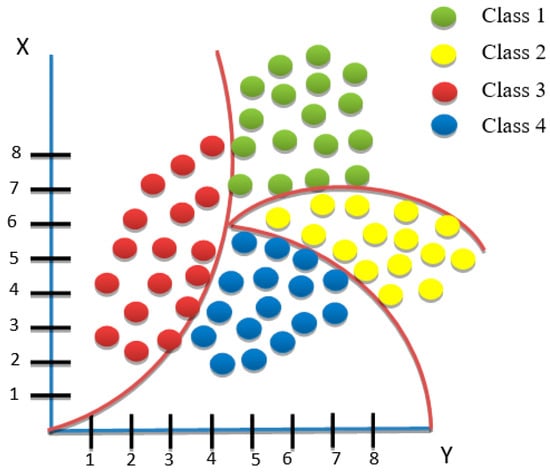
Figure 6.
An example of naïve Bayes classification.
- The model leverages polynomial or binary data.
- The approach permits the application of different datasets.
- The method avoids matrix computations, mathematical optimization, etc., so its application is comparatively straightforward.
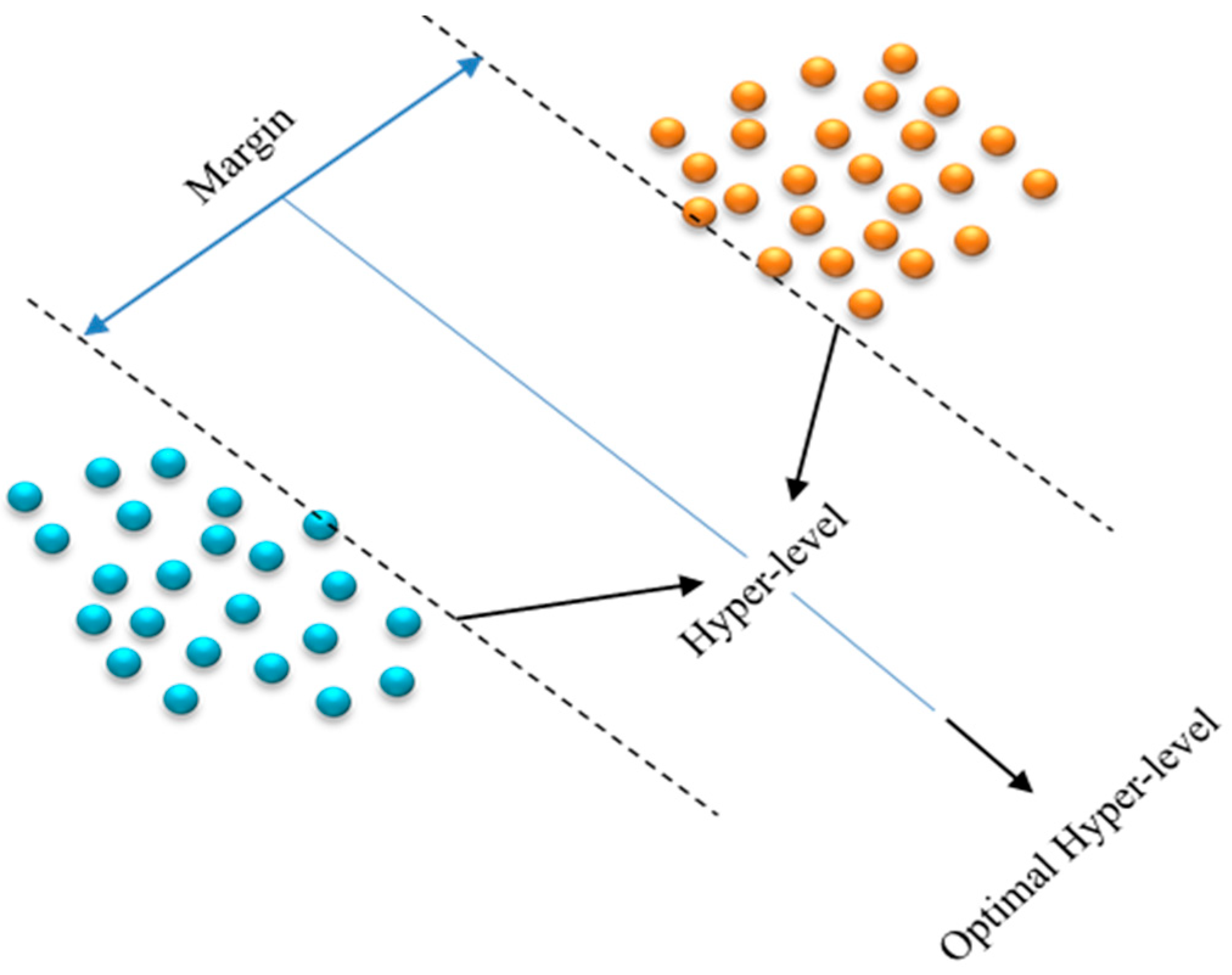
3.4.2. Support Vector Machine (SVM)
In general, the SVM is a new and effective type of machine learning based on the statistical learning principle developed by Vapnik [29]. SVMs are supervised learning algorithms with associated learning algorithms that analyze data, recognize patterns, and are used for classification and regression analysis [30]. The SVM is mainly based on the principle of hyper-level classifiers and is able to display the hyper-level that maximizes the margin between two classes. With a provided set of training examples, each labeled as belonging to one of two categories, the SVM training algorithm constructs an algorithm capable of assigning new instances to either of the two classes, rendering it a non-probabilistic binary linear classifier, as shown in Figure 7. An SVM algorithm is a depiction of examples as points in space, transformed such that instances of discrete classes are partitioned by as wide a distinct separation as feasible. Subsequently, new instances are transformed into the same space and are forecasted to be part of a particular class based on their position relative to the gap.
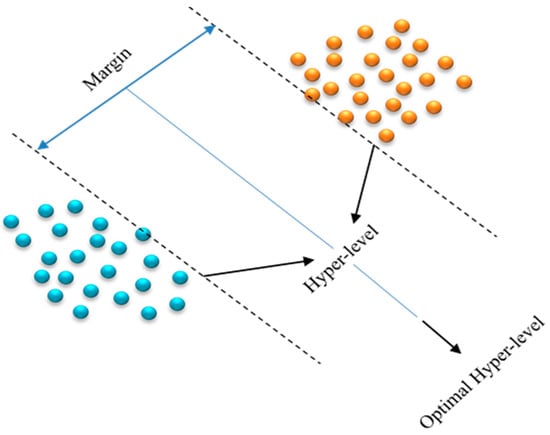
Figure 7.
Hyper-level in SVM.
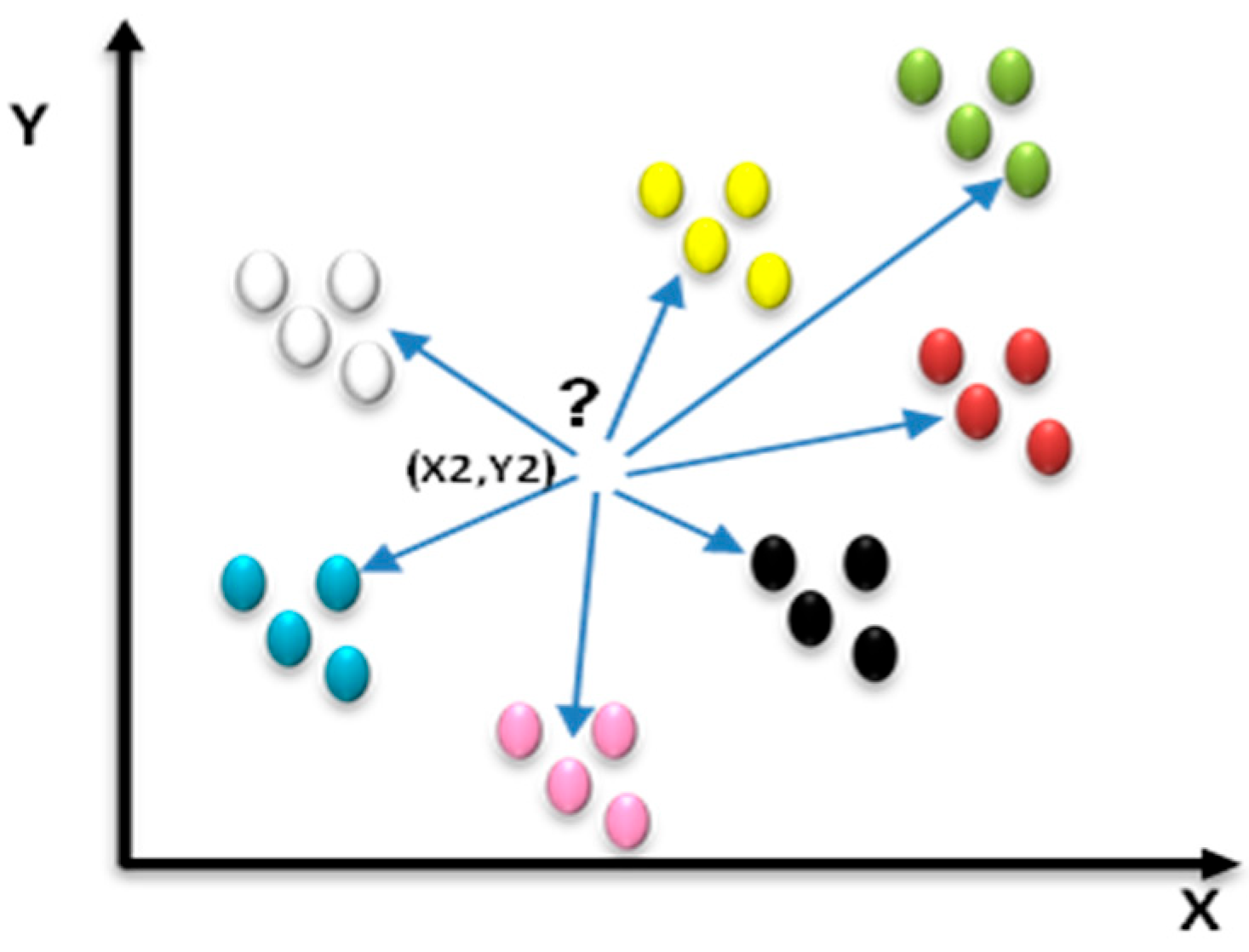
3.4.3. K-Nearest Neighbors Classification (KNN)
KNN categorization [31] identifies a set of k-selected training rows (k-nearest neighbors) from the training set that are in proximity to the unknown instance row. In the categorization of an unclassified ulcer image, the proximity of this image to the categorized image is calculated to determine the k-nearest one, as shown in Figure 8.
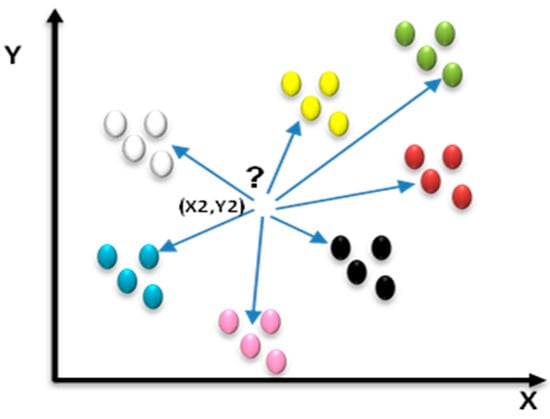
Figure 8.
Example of k-nearest neighbors.
The most common neighbors and class labels of these nearest neighbors are then used to determine the class label for the unknown row [32]. Unclassified images can be classified according to their proximity to the categorized ones based on a designated distance or similarity metric [33]. Euclidean distance serves as a metric in this process. The Euclidean distance () separating two rows is calculated using:
Once the list of nearest neighbors is derived, the test row is categorized according to the prevalent class among its closest neighbors. If k = 1, the class of the uncategorized image is merely allocated to its closest neighbor [34].
The KNN algorithm is detailed below:
- Input: The collection of training rows and the unlabeled test picture.
- Procedure: The proximity between the uncategorized test picture and every training image is calculated. The collection of nearest training images (k-nearest neighbors) to the uncategorized row is chosen.
- Output: A label is assigned to the test row based on the predominant class among its closest neighbors.
- The choice: The choice of the k value is crucial in the KNN classifier.
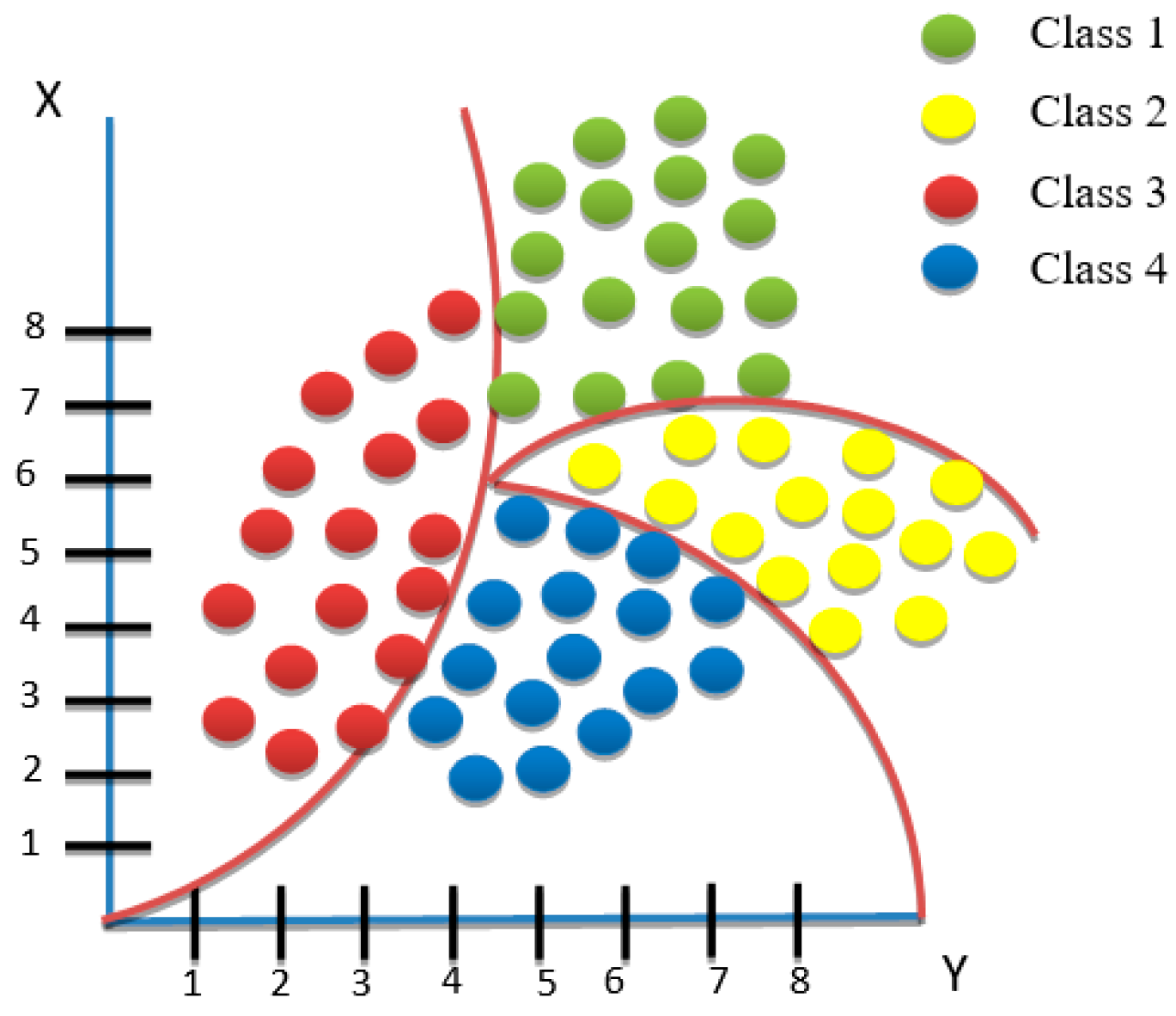
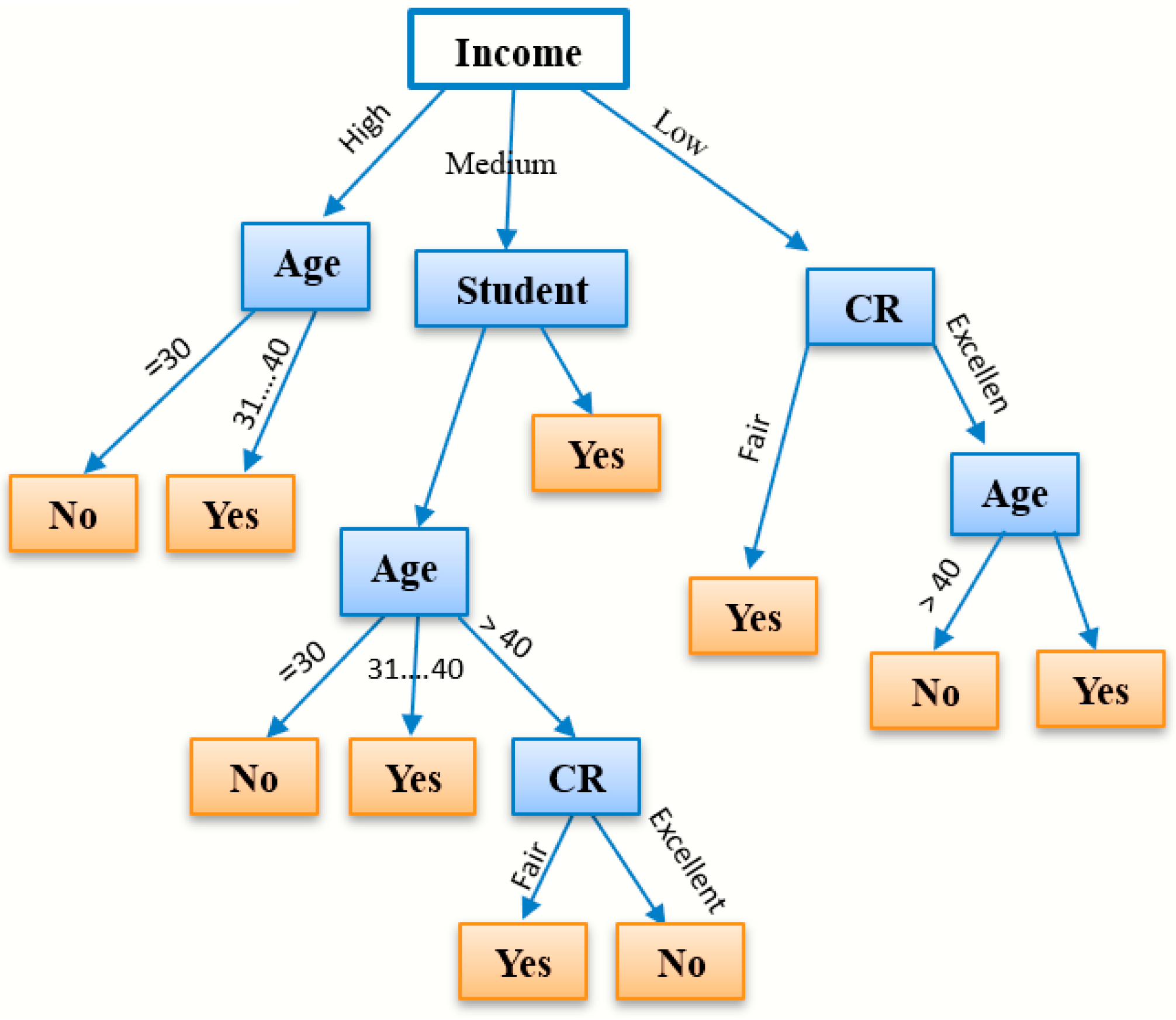
3.4.4. Decision Tree (DT) Classification
DT classification is a commonly utilized algorithm in data mining systems that generates classification models [35]. In data mining, classification algorithms can manage a large amount of data, as shown in Figure 9. Decision trees can be employed to make inferences about categorical class labels, to categorize information based on training datasets and class labels, and to categorize newly acquired data. Machine learning classification algorithms encompass various methods, and decision tree classification is one of the influential methods frequently employed in different domains, including machine learning, image processing, and pattern identification [36]. DT classification is a sequential model that efficiently and logically unifies a sequence of basic testing where the numeric feature is contrasted with the threshold value in each assessment [37]. Conceptual rules are much easier to construct than numerical weights in a neural network for connections between nodes. DT classification is mainly used for assembly purposes and in data mining. Each tree consists of nodes and branches. Each node represents features in a class to be classified, and each subset defines a value that the node can take. Due to its simple analysis and accuracy in multiple data models, DT classification has been implemented in many areas [36].
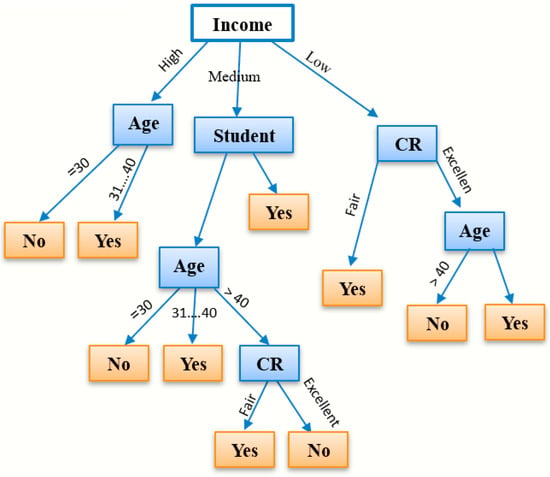
Figure 9.
An example of a decision tree algorithm.
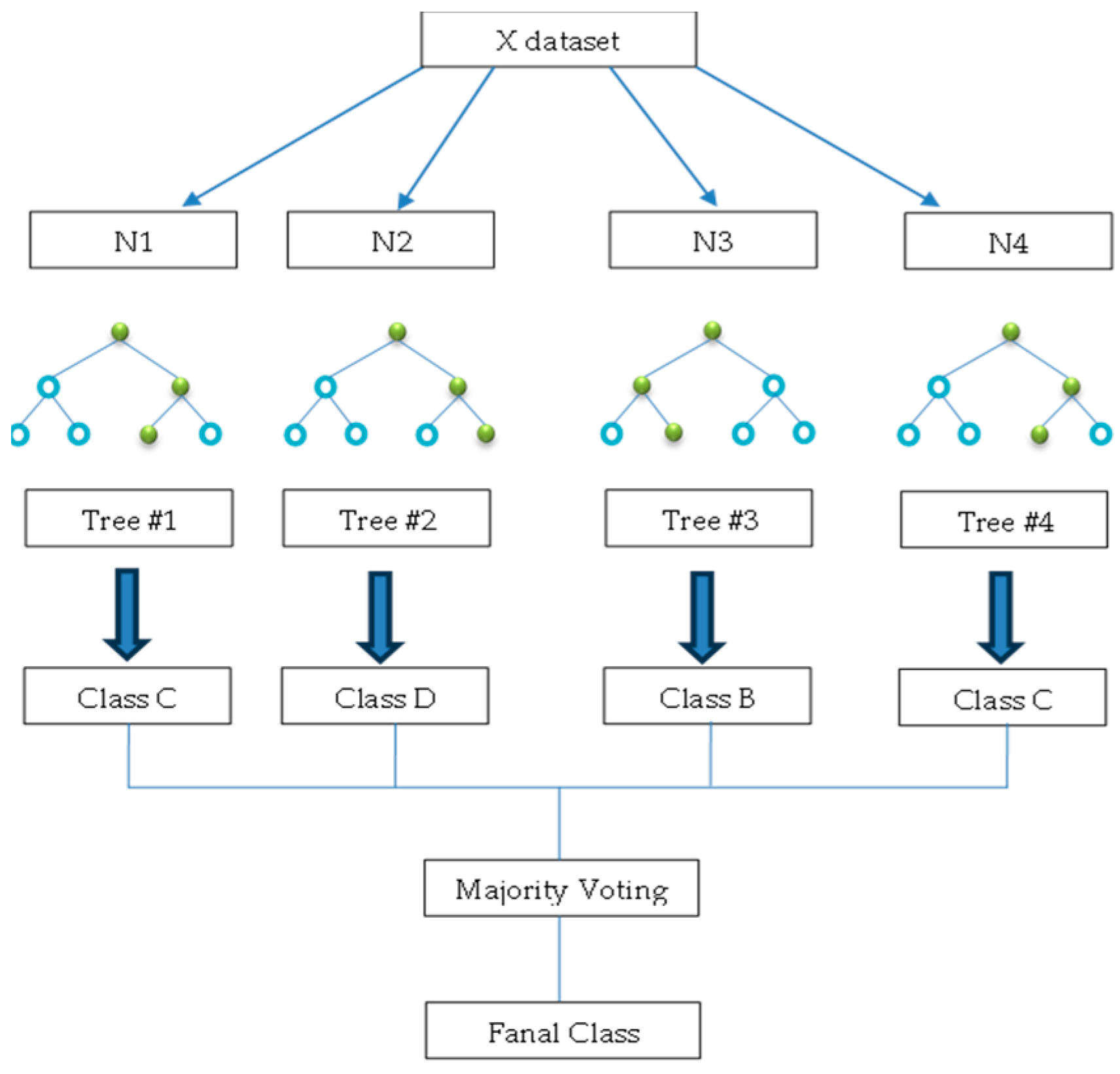
3.4.5. Random Forest (RF)
The RF model is a classification method in ensemble learning and regression techniques suitable for addressing issues related to grouping data into categories [38]. The algorithm was devised by Breiman [38]. A group of decision trees that give a classification decision for a set of data and the average of these decisions is given by the random forest [39], as shown in Figure 10. Therefore, this approach deals with a large amount of data and has greater accuracy than DT classification [40]. The classification process is slower than that of the decision tree method, as it reflects stability in prediction.
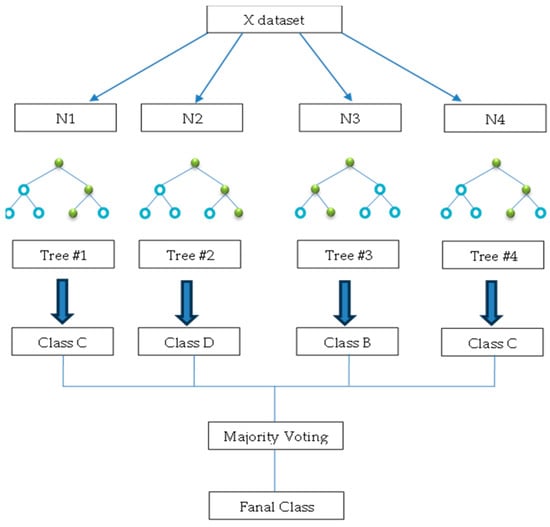
Figure 10.
An example of a random forest algorithm (where the blue circles indicate non-terminal nodes (internal nodes) and green circles indicate terminal nodes (leaf nodes).
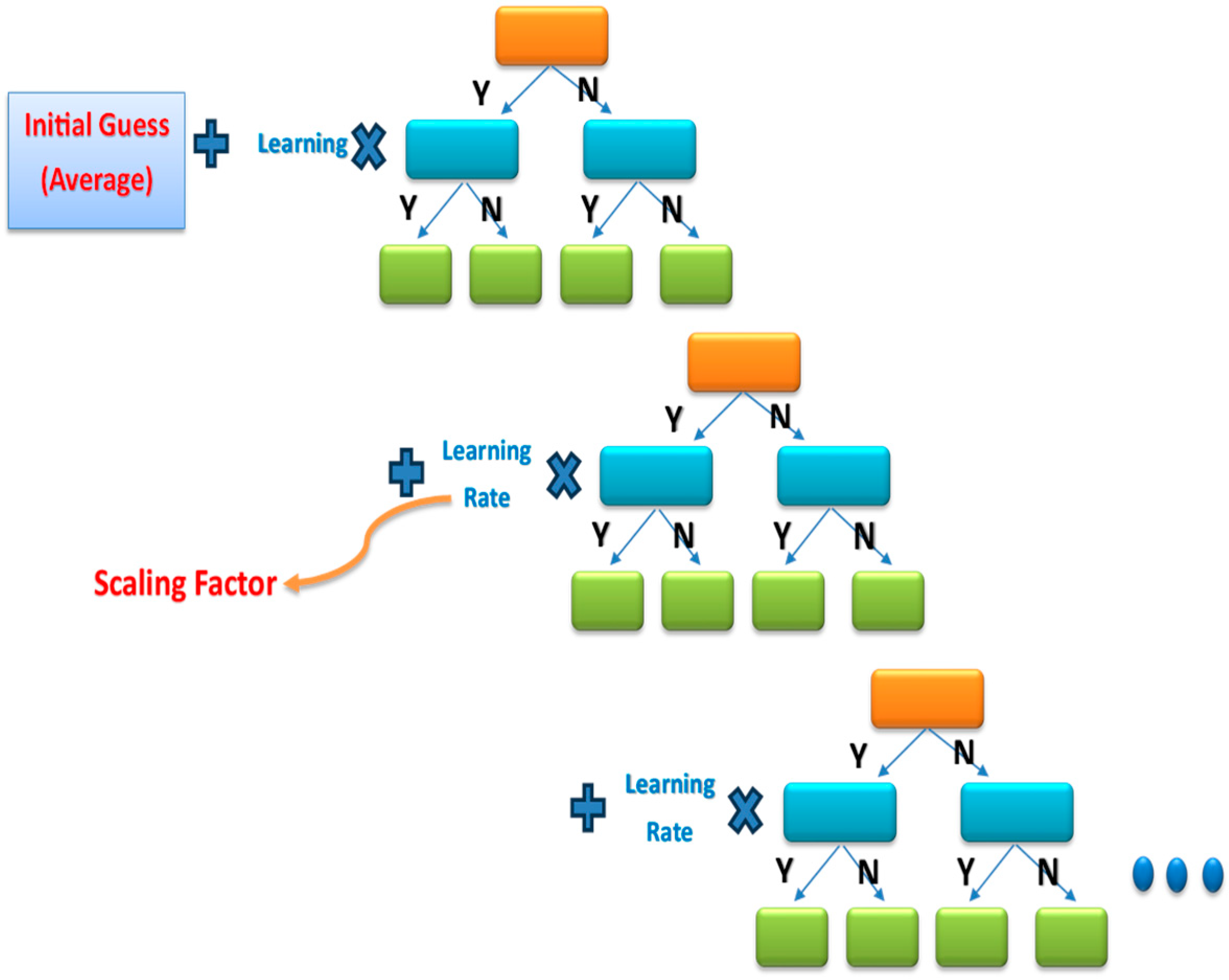
3.4.6. Extreme Gradient Boost (XGBoost)
XGBoost has been extensively utilized in various domains to achieve cutting-edge results in numerous data challenges and is a highly efficient and scalable machine learning framework for tree boosting [41]. Developed by Chen et al., it demonstrates scalability in every scenario [42]. It provides an initial guess in the first stage, after which the error rate is calculated. Thereafter, it adds the learning rate to the guess gradually, as shown in Figure 11, until it reaches the closest result to the desired output. Thus, the accuracy of this model is very high.
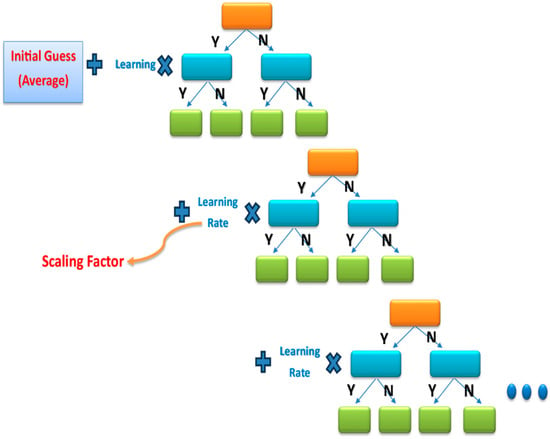
Figure 11.
An example of the XGBoost classifier with a gradient tree.
The parameters used for different machine learning algorithms are shown in Table 2.

Table 2.
The parameters used for different machine learning algorithms.
3.5. Evaluation Metrics
To assess the effectiveness of the classifiers and select which model provides the best evaluation metrics values, this study used 12 metrics, namely, the accuracy, precision, recall, F1-score, Jaccard index, zero-one loss, G-score, Hamming loss, Cohen’s kappa, MCC, and Fowlkes–Mallows index. These metrics offer quantitative means of appraising models’ predictive capacity by comparing their predictions to actual values.
where , and are true positives (correctly predicted positive instances), true negatives (correctly predicted negative instances), false positives (incorrectly predicted positive instances), and false negatives (incorrectly predicted negative instances), respectively. is the total number of instances, is the total number of labels, is the true label of the ith instance, is the predicted label of the ith instance, is the true label of the jth class for the ith instance, is the predicted label of the jth class for the ith instance, and is an indicator function, which returns 1 if the condition inside is true and 0 otherwise.
4. Experimental Results and Discussion
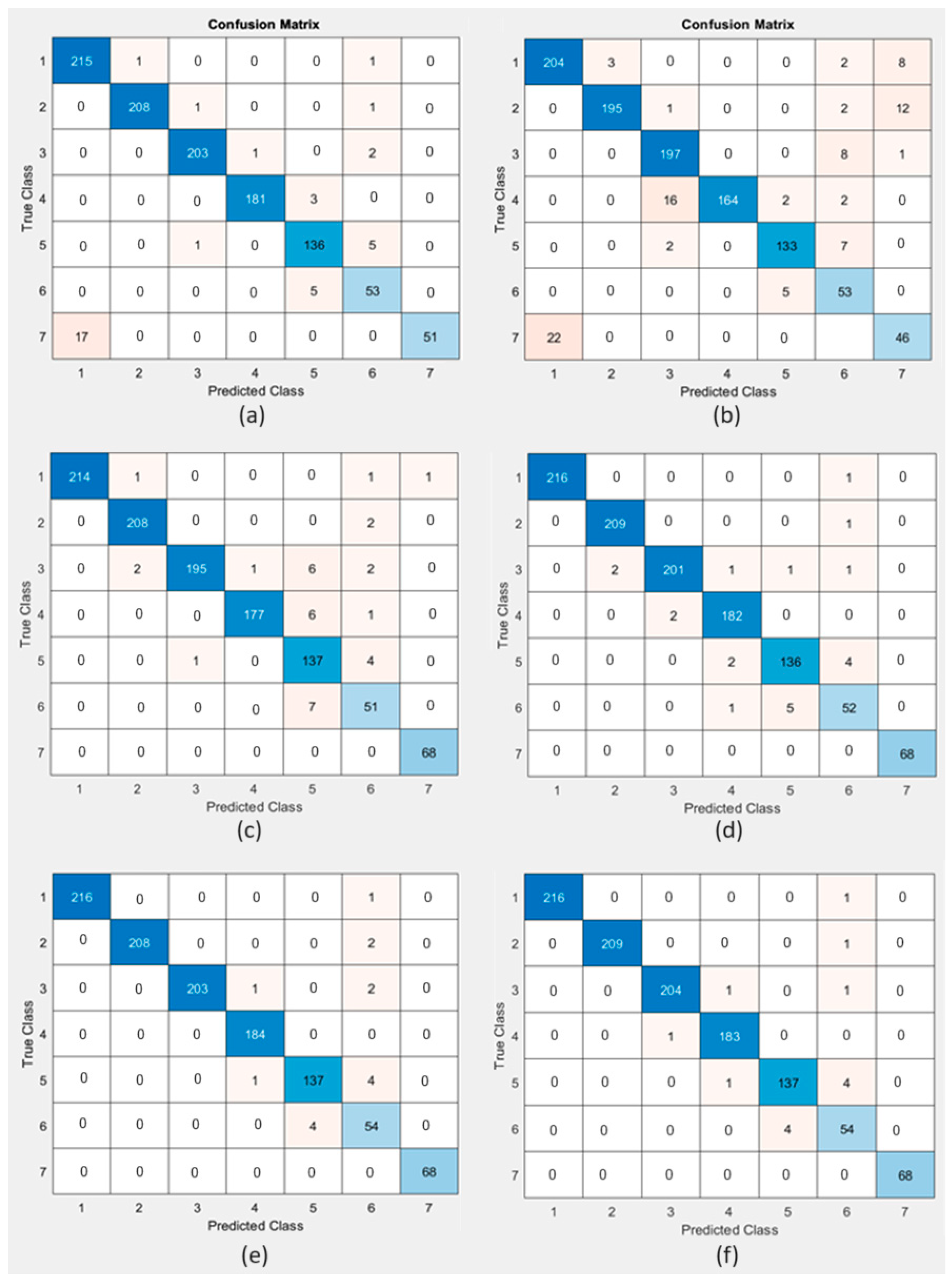
The performance of six machine learning algorithms was evaluated to determine which algorithm has the best performance in detecting tongue colors. The performance comparison of the used algorithms based on the confusion matrix for each algorithm, which provides a comprehensive view of the efficiency of the classification model in accurately predicting each class, is illustrated in Figure 12.
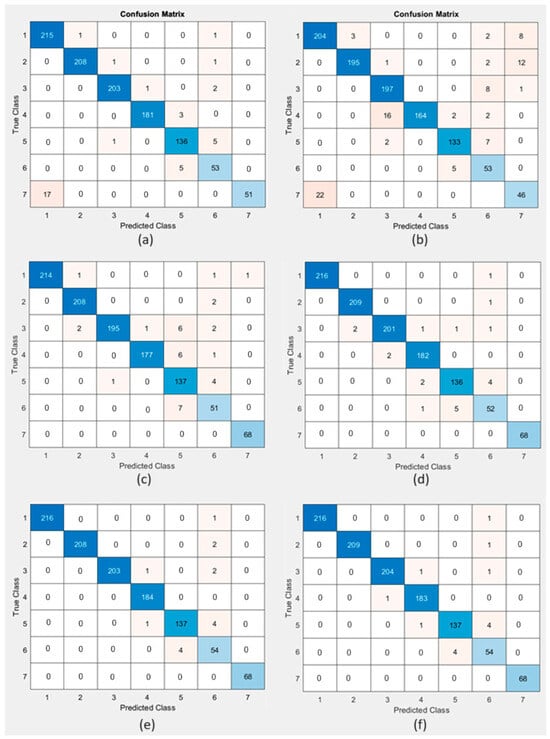
Figure 12.
Confusion matrix for (a) SVM, (b) NB, (c) KNN, (d) DT, (e) RF, and (f) XGBoost.
The data evaluation process also used 12 metrics to test the classification performance, namely, the accuracy, precision, recall, F1-score, Jaccard index, zero-one loss, G-score, Hamming loss, Cohen’s kappa, MCC, and Fowlkes–Mallows index, as shown in Table 3.

Table 3.
Results obtained from the machine learning algorithms used in the proposed study.
It is clear from Table 3 that the NB algorithm had the lowest accuracy of 91.43% with a balanced precision, recall, F1-score, and Jaccard index of 0.88, 0.89, 0.88, and 0.9273, respectively, with a zero-one loss of 0.0857, G-score of 0.6154, Hamming loss of 0.0857, Cohen’s kappa of 0.97, MCC of 0.32, and Fowlkes–Mallows index of 0.8892. The SVM algorithm had an accuracy of 96.50% with a balanced precision, recall, F1-score, and Jaccard index of 0.96, 0.94, 0.95, and 0.9406, respectively, with a zero-one loss of 0.0350, G-score of 0.8563, Hamming loss of 0.0350, Cohen’s kappa of 0.99, MCC of 0.34, and Fowlkes–Mallows index of 0.9482. The KNN algorithm had accuracy of 96.77% with a balanced precision, recall, F1-score, and Jaccard index of 0.95, 0.96, 0.96, and 0.9643, respectively, with a zero-one loss at 0.0323, G-score of 0.8400, Hamming loss of 0.0323, Cohen’s kappa of 0.99, MCC of 0.35, and Fowlkes–Mallows index of 0.9583. The DT algorithm had an accuracy of 98.06% with a balanced precision, recall, F1-score, and Jaccard index of 0.97, 0.97, 0.97, and 0.9927, respectively, with a zero-one loss of 0.0194, G-score of 0.9000, Hamming loss of 0.0194, Cohen’s kappa of 0.99, MCC of 0.35, and Fowlkes–Mallows index of 0.9727. The RF algorithm had an accuracy of 98.62% with a balanced precision, recall, F1-score, and Jaccard index of 0.97, 0.98, 0.98, and 0.9826, respectively, with a zero-one loss of 0.0138, G-score of 0.9077, Hamming loss of 0.0138, Cohen’s kappa of 1.00, MCC of 0.35, and Fowlkes–Mallows index of 0. 9799. XGBoost demonstrated outstanding performance, with the highest accuracy (98.71%), precision (98%), and recall (98%) among the techniques. The F1-score of 98% indicated an excellent balance between precision and recall, while the Jaccard index of 0.9895 with a zero-one loss of 0.0129, G-score of 0.9202, Hamming loss of 0.0129, Cohen’s kappa of 1.00, MCC of 0.35, and Fowlkes–Mallows index of 0. 9799 suggested an almost perfect positive correlation, indicating that XGBoost was highly effective and reliable for classification tasks. All the techniques performed well, but XGBoost was the top performer in terms of accuracy, precision, recall, F1-score, and MCC. Following the abovementioned results, the GUI was generated using the trained model from XGBoost (via the XGBoost MATLAB interface), as it showed superior performance compared to other techniques used in this study.
The proposed imaging system underwent real-time testing, and promising results were observed after implementation. This test included multiple images of healthy and affected oral organs. The framework evaluation included 60 images collected at the Al-Hussein teaching hospital in Dhi Qar and the Mosul general hospital, including individuals experiencing different conditions, such as diabetes, kidney failure, and deficiency in red blood cells, along with normal individuals. The framework accurately diagnosed 58 images out of 60, with a detection accuracy rate of 96.6%.
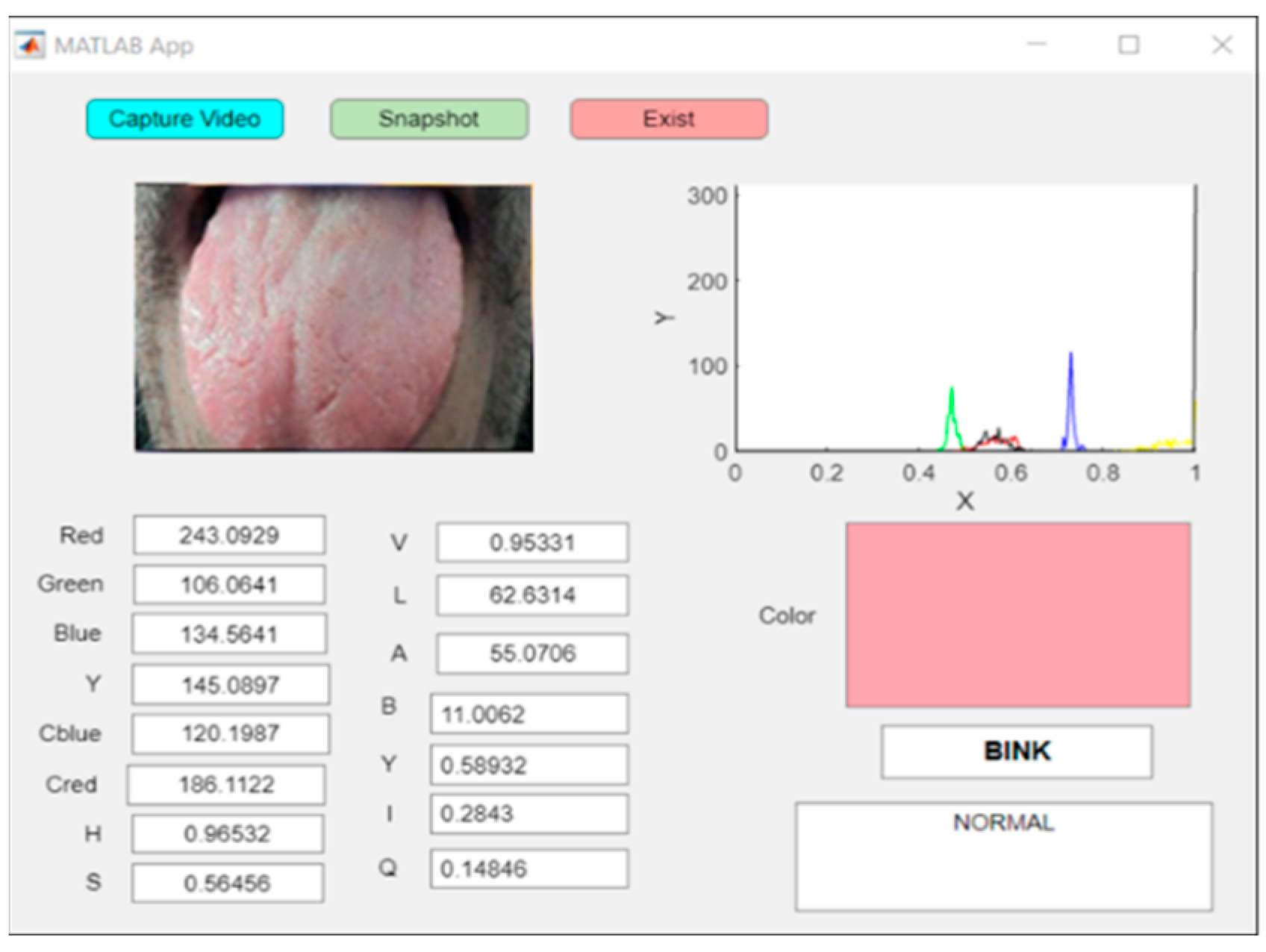
In this system, a pink tongue indicates health, while other colors indicate infection, and each color corresponds to specific diseases programmed into the GUI. A real-time image showing a normal scan of a pink tongue is shown in Figure 13.
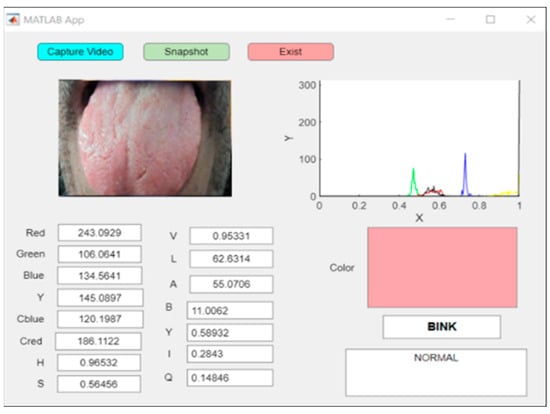
Figure 13.
An example of real-time capturing for a pink tongue (healthy case).
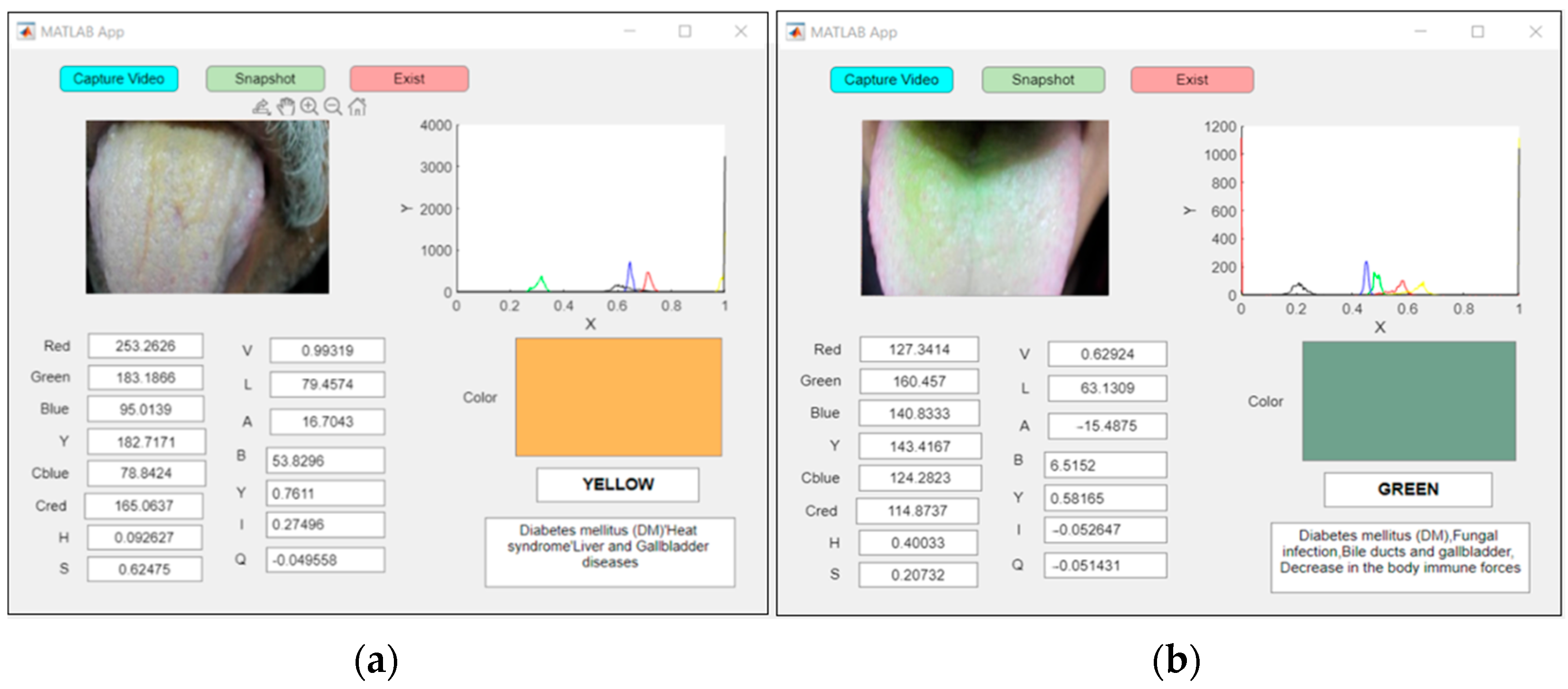
The proposed system’s real-time examination of two patients with abnormal yellow and green tongues classified the patient with a yellow tongue as a diabetic patient, as shown in Figure 14a, and the patient with a green tongue as having a mycotic infection, as shown in Figure 14b.
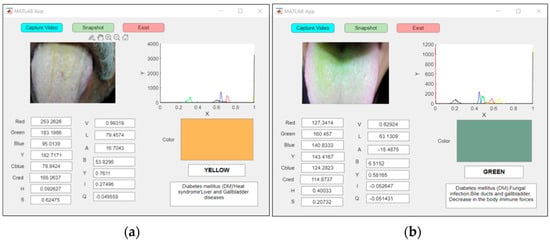
Figure 14.
An example of real-time capturing for a (a) yellow tongue (DM) and (b) green tongue (mycotic infection).
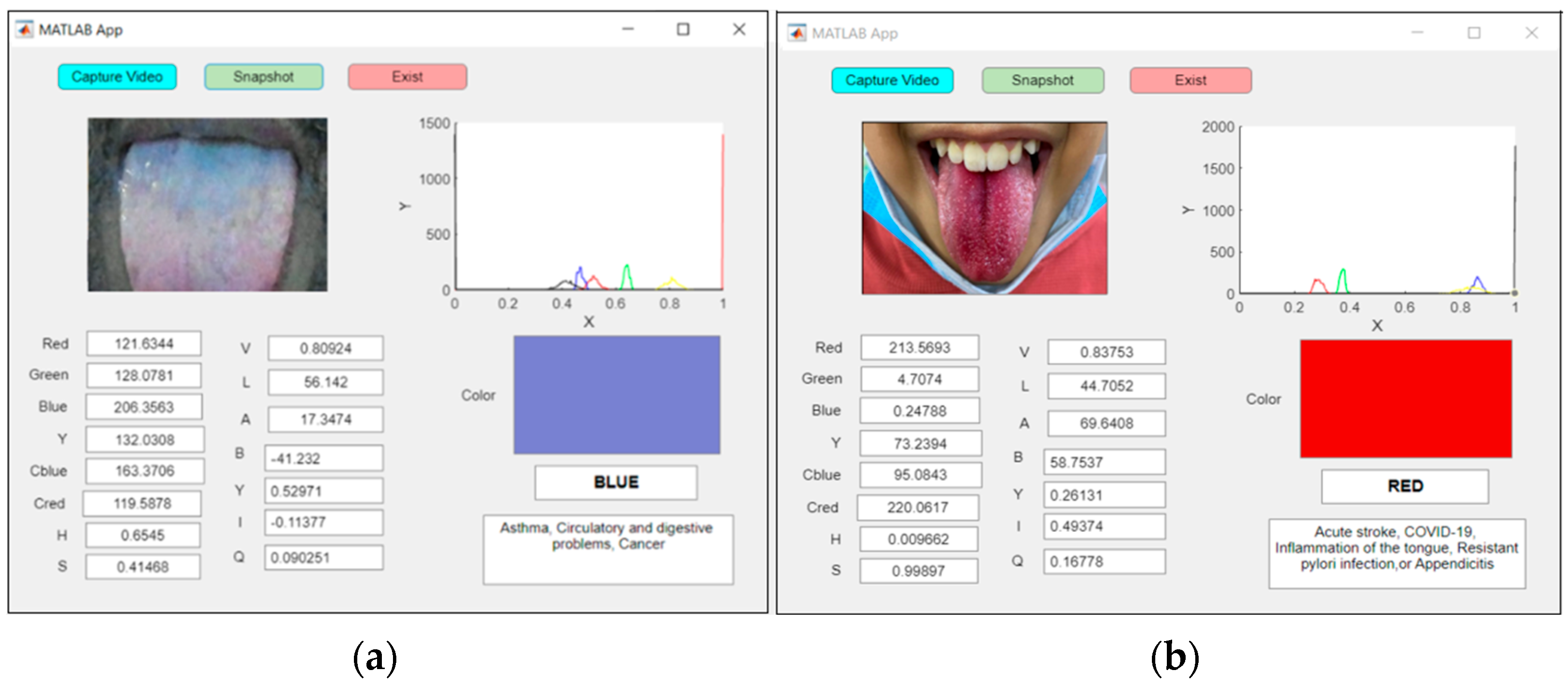
Figure 15a shows an examination of an abnormal blue tongue in real time, indicating asthma, whereas Figure 15b shows an examination of a red tongue for a patient with COVID-19.
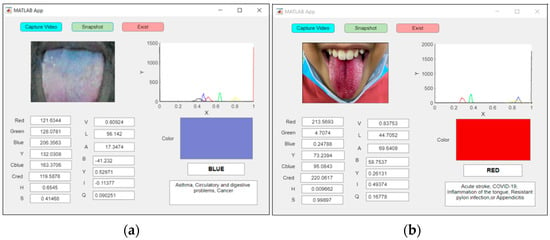
Figure 15.
An example of real-time capturing for a (a) blue tongue (asthma) and (b) red tongue (COVID-19).
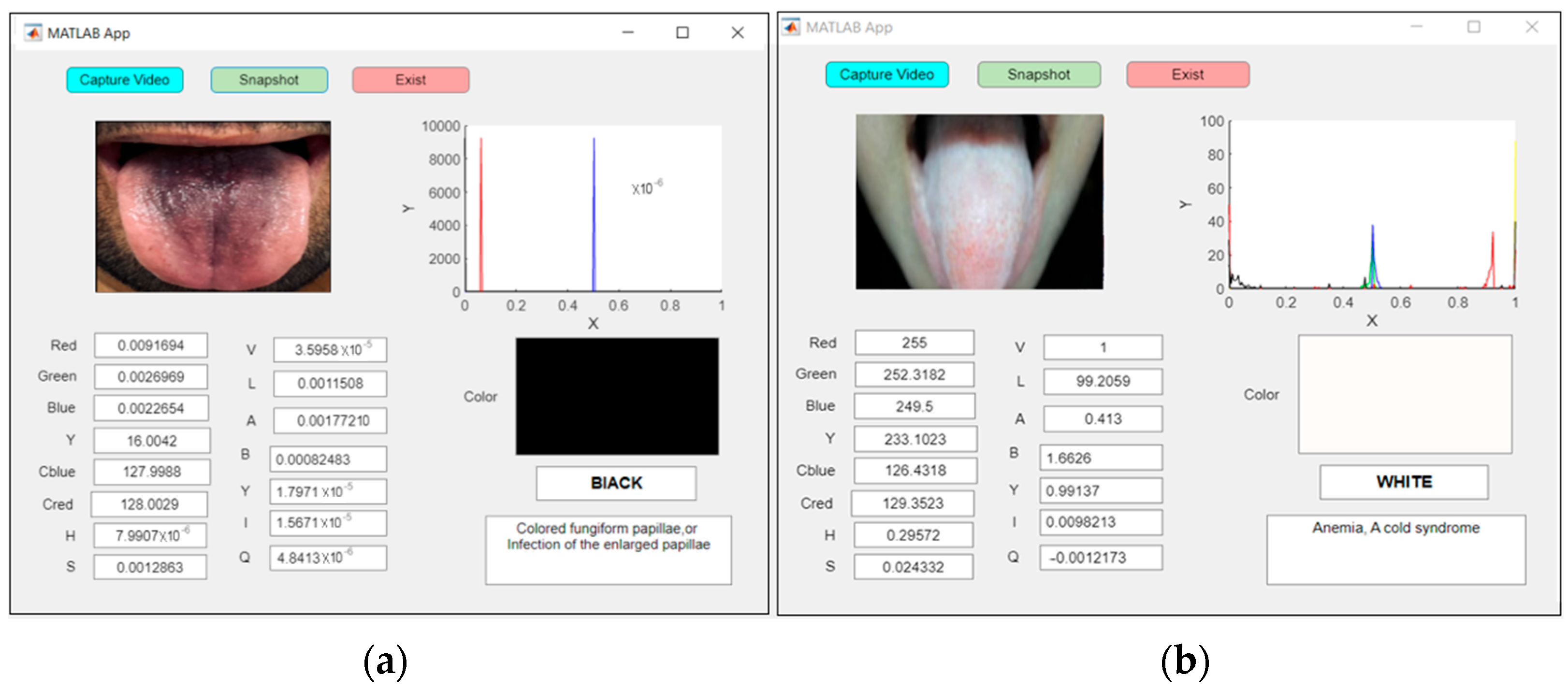
Figure 16a shows an examination of an abnormal tongue in real time for a patient with fungiform papillae with a black tongue, whereas anemia was detected for the patient with a white tongue, as shown in Figure 16b.
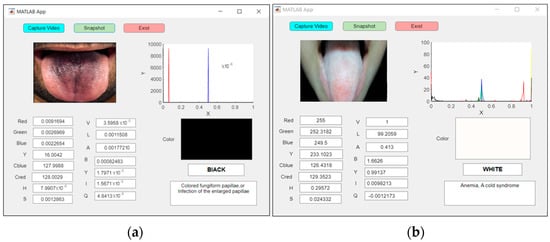
Figure 16.
An example of real-time capturing for a (a) black tongue (fungiform papillae) and (b) white tongue (anemia).
Table 4 illustrates a comparison of the literature that supports each of the hypotheses with the results obtained in this pilot study.

Table 4.
Comparison of the literature that supports each of the hypotheses with the results obtained in the pilot study.
One of the limitations that we faced during the research was the reluctance of patients to consent to data collection. Also, camera reflections are among the problems leading to variation in detected colors and thus affect the diagnosis. Future research will account for reflections captured by the camera using advanced image processors and filters as well as deep learning techniques to classify and distinguish between colors to obtain higher accuracy.
Comparative analysis indicates a clear development in the field of tongue diagnosis using artificial intelligence, with the trend towards more advanced methods of extracting features, larger and more diverse data groups, artificial intelligence algorithms, and advanced deep learning. These developments collectively contribute to high rates of accuracy and more reliable diagnostic capabilities, indicating the capabilities of artificial intelligence in enhancing traditional Chinese medicine practices and other medical applications, as shown in Table 5.

Table 5.
Comparative analysis with previous studies based on features, data collection, or algorithms.
5. Conclusions
Automated systems for analyzing tongue color show remarkable precision in differentiating between healthy and diseased individuals. Additionally, they demonstrate the ability to diagnose various diseases and determine the phases of their evolution, particularly considering the remarkable progress in artificial intelligence and camera technologies. The proposed system could efficiently detect different ailments that show apparent changes in tongue color, with the accuracy rate of the trained models exceeding 98%. In this study, a webcam was employed to capture images of the tongue in real time using MATLAB GUI software. The proposed system was tested using 60 images of both patients and healthy individuals captured in real time with a diagnostic accuracy rate reaching 96.6%. These results confirm the feasibility of AI systems for tongue diagnosis in the field of medical applications and indicate that this approach is a secure, efficient, user-friendly, comfortable and cost-effective method for disease screening.
Author Contributions
Conceptualization, A.A.-N.; methodology, A.R.H., A.A.-N., G.A.K. and J.C.; software, A.R.H., G.A.K. and A.A.-N.; validation, A.R.H., G.A.K. and A.A.-N.; formal analysis, A.R.H., G.A.K. and A.A.-N.; investigation, A.R.H., G.A.K. and A.A.-N.; resources, A.R.H.; data curation, A.R.H.; writing—original draft preparation, A.R.H. and A.A.-N.; writing—review and editing, G.A.K. and J.C.; visualization, G.A.K. and J.C.; supervision, A.A.-N. and G.A.K.; project administration, A.A.-N. and G.A.K. and J.C.; funding acquisition, J.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Human Research Ethics Committee at the Ministry of Health and Environment, Training and Human Development Centre, Iraq (Protocol ID 201/21) for studies involving humans.
Informed Consent Statement
Written informed consent has been obtained from the patient(s) to publish this paper.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
The research team expresses its sincere thanks and gratitude to Al-Hussein Teaching Hospital in Nasiriyah and Mosul General Hospital for their kind cooperation and providing the necessary data to complete this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Velanovich, V. Correlations with “Non-Western” Medical Theory and Practice: Traditional Chinese Medicine, Traditional Indigenous Medicine, Faith Healing, and Homeopathy. In Medical Persuasion: Understanding the Impact on Medical Argumentation; Velanovich, V., Ed.; Springer International Publishing: Cham, Switzerland, 2023; pp. 271–306. [Google Scholar]
- Wiseman, N. Chinese medical dictionaries: A guarantee for better quality literature. Clin. Acupunct. Orient. Med. 2001, 2, 90–98. [Google Scholar] [CrossRef]
- Kirschbaum, B. Atlas of Chinese Tongue Diagnosis; Eastland Press: Seattle, WA, USA, 2000. [Google Scholar]
- Li, J.; Huang, J.; Jiang, T.; Tu, L.; Cui, L.; Cui, J.; Ma, X.; Yao, X.; Shi, Y.; Wang, S.; et al. A multi-step approach for tongue image classification in patients with diabetes. Comput. Biol. Med. 2022, 149, 105935. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, A.K.; Mohammed, S.L.; Al-Naji, A. Tongue Color Analysis and Disease Diagnosis Based on a Computer Vision System. In Proceedings of the 2022 4th International Conference on Advanced Science and Engineering (ICOASE), Zakho, Iraq, 21–22 September 2022; pp. 25–30. [Google Scholar]
- Zhang, N.; Jiang, Z.; Li, J.; Zhang, D. Multiple color representation and fusion for diabetes mellitus diagnosis based on back tongue images. Comput. Biol. Med. 2023, 155, 106652. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Yang, X.; Qi, Q.; Pan, Y.; Chen, Y.; Shen, J.; Liao, H.; Ji, Z. Potential screening and early diagnosis method for cancer: Tongue diagnosis. Int. J. Oncol. 2016, 48, 2257–2264. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Jing, C.; Zhang, Z.; Xu, J.; Duan, Y.; Xu, D. Digital tongue image analyses for health assessment. Med. Rev. 2021, 1, 172–198. [Google Scholar] [CrossRef]
- Jiang, B.; Liang, X.; Chen, Y.; Ma, T.; Liu, L.; Li, J.; Jiang, R.; Chen, T.; Zhang, X.; Li, S. Integrating next-generation sequencing and traditional tongue diagnosis to determine tongue coating microbiome. Sci. Rep. 2012, 2, 936. [Google Scholar] [CrossRef] [PubMed]
- Srividhya, E.; Muthukumaravel, A. Diagnosis of Diabetes by Tongue Analysis. In Proceedings of the 2019 1st International Conference on Advances in Information Technology (ICAIT), Chikmagalur, India, 25–27 July 2019; pp. 217–222. [Google Scholar]
- Kanawong, R.; Obafemi-Ajayi, T.; Ma, T.; Xu, D.; Li, S.; Duan, Y. Automated Tongue Feature Extraction for ZHENG Classification in Traditional Chinese Medicine. Evid.-Based Complement. Altern. Med. 2012, 2012, 912852. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Q.; Gan, S.; Zhang, L. Human-computer interaction based health diagnostics using ResNet34 for tongue image classification. Comput. Methods Programs Biomed. 2022, 226, 107096. [Google Scholar] [CrossRef] [PubMed]
- Horzov, L.; Goncharuk-Khomyn, M.; Hema-Bahyna, N.; Yurzhenko, A.; Melnyk, V. Analysis of Tongue Color-Associated Features among Patients with PCR-Confirmed COVID-19 Infection in Ukraine. Pesqui. Bras. Odontopediatria Clínica Integr. 2021, 21, e0011. [Google Scholar] [CrossRef]
- Park, Y.-J.; Lee, J.-M.; Yoo, S.-Y.; Park, Y.-B. Reliability and validity of tongue color analysis in the prediction of symptom patterns in terms of East Asian Medicine. J. Tradit. Chin. Med. 2016, 36, 165–172. [Google Scholar] [CrossRef][Green Version]
- Umadevi, G.; Malathy, V.; Anand, M. Diagnosis of Diabetes from Tongue Image Using Versatile Tooth-Marked Region Classification. TEST Eng. Manag. 2019, 81, 5953–5965. [Google Scholar]
- Srividhya, E.; Muthukumaravel, A. Feature Extraction of Tongue Diseases Diagnosis Using SVM Classifier. In Proceedings of the 2019 International Conference on Computational Intelligence and Knowledge Economy (ICCIKE), Dubai, United Arab Emirates, 11–12 December 2019; pp. 260–263. [Google Scholar]
- Thirunavukkarasu, U.; Umapathy, S.; Krishnan, P.T.; Janardanan, K. Human Tongue Thermography Could Be a Prognostic Tool for Prescreening the Type II Diabetes Mellitus. Evid.-Based Complement. Altern. Med. 2020, 2020, 3186208. [Google Scholar] [CrossRef] [PubMed]
- Naveed, S. Early Diabetes Discovery From Tongue Images. Comput. J. 2022, 65, 237–250. [Google Scholar] [CrossRef]
- Mansour, R.F.; Althobaiti, M.M.; Ashour, A.A. Internet of Things and Synergic Deep Learning Based Biomedical Tongue Color Image Analysis for Disease Diagnosis and Classification. IEEE Access 2021, 9, 94769–94779. [Google Scholar] [CrossRef]
- Abdullah, A.K.; Mohammed, S.L.; Al-Naji, A. Computer-aided diseases diagnosis system based on tongue color analysis: A review. In AIP Conference Proceedings; AIP Publishing: Long Island, NY, USA, 2023; Volume 2804, no. 1. [Google Scholar]
- Susanto, A.; Dewantoro, Z.H.; Sari, C.A.; Setiadi, D.R.I.M.; Rachmawanto, E.H.; Mulyono, I.U.W. Shallot Quality Classification using HSV Color Models and Size Identification based on Naive Bayes Classifier Shallot Quality Classification using HSV Color Models and Size Identification based on Naive Bayes Classifier. J. Phys. Conf. Ser. 2020, 1577, 012020. [Google Scholar] [CrossRef]
- Zhang, N.; Wu, L.; Yang, J.; Guan, Y. Naive Bayes Bearing Fault Diagnosis Based on Enhanced Independence of Data. Sensors 2018, 18, 463. [Google Scholar] [CrossRef] [PubMed]
- Goswami, M.; Sebastian, N.J. Performance Analysis of Logistic Regression, KNN, SVM, Naïve Bayes Classifier for Healthcare Application During COVID-19. In Innovative Data Communication Technologies and Application; Springer Nature: Singapore, 2022; pp. 645–658. [Google Scholar]
- Jun, Z. The Development and Application of Support Vector Machine. J. Phys. Conf. Ser. 2021, 1748, 052006. [Google Scholar] [CrossRef]
- Sarker, I.H. Machine Learning for Intelligent Data Analysis and Automation in Cybersecurity: Current and Future Prospects. Ann. Data Sci. 2023, 10, 1473–1498. [Google Scholar] [CrossRef]
- Bhavani, R.R.; Jiji, G.W. Image registration for varicose ulcer classification using KNN classifier. Int. J. Comput. Appl. 2018, 40, 88–97. [Google Scholar] [CrossRef]
- Wang, A.X.; Chukova, S.S.; Nguyen, B.P. Ensemble k-nearest neighbors based on centroid displacement. Inf. Sci. 2023, 629, 313–323. [Google Scholar] [CrossRef]
- Zafra, A.; Gibaja, E. Nearest neighbor-based approaches for multi-instance multi-label classification. Expert. Syst. Appl. 2023, 232, 120876. [Google Scholar] [CrossRef]
- Villarreal-Hernández, J.Á.; Morales-Rodríguez, M.L.; Rangel-Valdez, N.; Gómez-Santillán, C. Reusability Analysis of K-Nearest Neighbors Variants for Classification Models. In Innovations in Machine and Deep Learning: Case Studies and Applications; Rivera, G., Rosete, A., Dorronsoro, B., Eds.; Springer Nature: Cham, Switzerland, 2023; pp. 63–81. [Google Scholar]
- Kesavaraj, G.; Sukumaran, S. A study on classification techniques in data mining. In Proceedings of the 2013 Fourth International Conference on Computing, Communications and Networking Technologies (ICCCNT), Tiruchengode, India, 4–6 July 2013; pp. 1–7. [Google Scholar]
- Charbuty, B.; Abdulazeez, A. Classification Based on Decision Tree Algorithm for Machine Learning. J. Appl. Sci. Technol. Trends 2021, 2, 20–28. [Google Scholar] [CrossRef]
- Damanik, I.S.; Windarto, A.P.; Wanto, A.; Poningsih; Andani, S.R.; Saputra, W. Decision Tree Optimization in C4.5 Algorithm Using Genetic Algorithm. J. Phys. Conf. Ser. 2019, 1255, 012012. [Google Scholar] [CrossRef]
- Barros, R.C.; Basgalupp, M.P.; Carvalho, A.C.P.L.F.D.; Freitas, A.A. A Survey of Evolutionary Algorithms for Decision-Tree Induction. IEEE Trans. Syst. Man. Cybern. Part. C Appl. Rev. 2012, 42, 291–312. [Google Scholar] [CrossRef]
- Anuradha; Gupta, G. A self explanatory review of decision tree classifiers. In Proceedings of the International Conference on Recent Advances and Innovations in Engineering (ICRAIE-2014), Jaipur, India, 9–11 May 2014; pp. 1–7. [Google Scholar]
- Gavankar, S.S.; Sawarkar, S.D. Eager decision tree. In Proceedings of the 2017 2nd International Conference for Convergence in Technology (I2CT), Mumbai, India, 7–9 April 2017; pp. 837–840. [Google Scholar]
- Jagtap, S.T.; Phasinam, K.; Kassanuk, T.; Jha, S.S.; Ghosh, T.; Thakar, C.M. Towards application of various machine learning techniques in agriculture. Mater. Today Proc. 2022, 51, 793–797. [Google Scholar] [CrossRef]
- Zounemat-Kermani, M.; Batelaan, O.; Fadaee, M.; Hinkelmann, R. Ensemble machine learning paradigms in hydrology: A review. J. Hydrol. 2021, 598, 126266. [Google Scholar] [CrossRef]
- Persson, I.; Grünwald, A.; Morvan, L.; Becedas, D.; Arlbrandt, M. A Machine Learning Algorithm Predicting Acute Kidney Injury in Intensive Care Unit Patients (NAVOY Acute Kidney Injury): Proof-of-Concept Study. JMIR Form. Res. 2023, 7, e45979. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Lu, P.; Zheng, Z.; Tolliver, D.; Keramati, A. Accident Prediction Accuracy Assessment for Highway-Rail Grade Crossings Using Random Forest Algorithm Compared with Decision Tree. Reliab. Eng. Syst. Saf. 2020, 200, 106931. [Google Scholar] [CrossRef]
- Chen, R.-C.; Dewi, C.; Huang, S.-W.; Caraka, R.E. Selecting critical features for data classification based on machine learning methods. J. Big Data 2020, 7, 52. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016. [Google Scholar] [CrossRef]
- Tigga, N.P.; Garg, S. Prediction of Type 2 Diabetes using Machine Learning Classification Methods. Procedia Comput. Sci. 2020, 167, 706–716. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).