Abstract
In recent years, the proliferation of health data sources due to computer technologies has prompted the use of visualization techniques to tackle epidemiological challenges. However, existing reviews lack a specific focus on the spatial and temporal analysis of epidemiological data using visualization tools. This study aims to address this gap by conducting a scoping review following the PRISMA-ScR guidelines, examining the literature from 2000 to 2024 on spatial–temporal visualization techniques when applied to epidemics, across five databases: PubMed, IEEE Xplore, Scopus, Google Scholar, and ACM Digital Library until 24 January 2024. Among 1312 papers reviewed, 114 were selected, emphasizing aggregate measures, web platform tools, and geospatial data representation, particularly favoring choropleth maps and extended charts. Visualization techniques were predominantly utilized for real-time data presentation, trend analysis, and predictions. Evaluation methods, categorized into standard methodology, user experience, task efficiency, and accuracy, were observed. Although various open-access datasets were available, only a few were commonly used, mainly those related to COVID-19. This study sheds light on the current trends in visualizing epidemiological data over the past 24 years, highlighting the gaps in standardized evaluation methodologies and the limited exploration of individual epidemiological data and diseases acquired in hospitals during epidemics.
1. Introduction
Epidemiology is the study of the distribution and determinants of health-related events, along with the application of this study to disease control [1]. Epidemiological research can be divided into two branches: descriptive studies, which include surveillance carried out to analyze the distribution of a disease, and analytical studies, which are carried out to confirm the determining factors of that disease. However, the application of modern technologies to the field of health has resulted in the production of massive amounts of data originating from various sources with different types and qualities. In addition to this, it is well known that human beings’ perceptual and cognitive capacities limit the use and exploitation of all available data. Computational applications must provide solutions to present and interact with said data, thus enabling experts in the field to analyze and evaluate them appropriately [2]. One example is the case of the coronavirus disease of 2019 (COVID-19) outbreak. In this unprecedented scenario with several large databases, various approaches have been used for data processing and visualization, though we still lack a clear and widespread use of them.
The objective of the information visualization area is to reduce the complexity of research and facilitate the understanding of information for humans through the design of visual representations of data [3]. In the field of health, information visualization and visual analytics aim to support exploration; monitoring; insight discovery; collaboration; and explanation to patients, clinicians, policymakers, and the general public [4]. Health datasets are inherently spatial and temporal; thus, the exploration of these data and the correlation between datasets to derive insights remains a challenge [4]. This has consequently led to the development of tools for synthesizing data from different and heterogeneous sources, where space and time play an important role, particularly in the field of epidemiology. Both geospatial and temporal dimensions, as well as network information, are primordial when analyzing the distribution of a disease and devising control strategies, as is evident in the pandemic resulting from COVID-19. Visualization systems coupled with models of disease spread can help predict the future course of an epidemic and evaluate strategies to control it [4].
The use of visual representations to study data regarding epidemics helps to better understand the current state of public health at both population (e.g., the number of cases or case counts, incidence, and prevalence) and individual (e.g., contacts, movement, and start of disease) levels [5]. It is therefore necessary to communicate the spatial and temporal aspects of these data through visualizations to both experts, to enable them to take measures, and the population, to raise awareness.
The relevant literature contains several reviews regarding visualization in the human healthcare domain. Bucalon et al. [6] conducted a scoping review to analyze the literature on dashboards that support the reflective task of clinicians based on routinely collected clinical indicator data. Carroll et al. [7] carried out an analysis of visualization tools for representing infectious diseases published between 1980 and mid-2013, and this focused principally on GIS, molecular epidemiology, and social network analysis. Crisan et al. [8] conducted a general categorization study of the geographic visualization techniques used to visualize outbreak data and risk patterns for public health professionals. Chishtie et al. [9] synthesized the literature concerning the use of visual analysis tools, techniques, and frameworks regarding population health and health services research, focusing on the years between 2005 and early 2019. Although these reviews provide helpful information on visualization research, very little attention is paid to the analysis of the spatial–temporal visualization of epidemiological data. In this work, we address this gap, providing a specific analysis of the techniques and tools available in the most recent scientific literature.
Our main objective is to answer the following research question: how has epidemiological data been represented through the use of spatial–temporal visualization? We seek to examine the scientific literature on visualization techniques applied to epidemics, focusing on the epidemiological measures adopted and how they are represented, the geospatial and temporal visualization techniques used and how they are combined, the most common target population, and the identification of the principal tools adopted to implement visualization.
2. Methods
2.1. Research Questions
This scoping review was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analysis extension for Scoping Reviews (PRISMA-ScR) guidelines [10]. We further used the PRISMA-ScR checklist to guide this review, which is shown in Table S1. This scoping review aims to present the state of the art of the most recent spatial–temporal visualization techniques employed to represent measures of epidemiological data. The data extracted from the articles will give insight into 3 main fields of interest: (A) the use of medical and epidemiological data, (B) the application of visualization techniques, and (C) applicable methodologies. Thus, we defined 9 research questions to guide this study:
- Q1. What are the most commonly visually represented measures of an epidemic, and what are the visualization purposes? We analyzed the epidemic data represented in each paper to identify general trends in the representation of the measures, the information involved, and the final use of the visualization techniques.
- Q2. Which techniques are most frequently used to visualize epidemiological spatial information? We classified the papers based on what type of visualization they used to represent the spatial component of epidemiological data. The classification types were (1) maps, if they used a geographical representation; (2) buildings, if the visualization represented the inside of a building structure; and (3) none, if the visualization technique was conceptual.
- Q3. Which techniques are most frequently used to visualize epidemiological temporal data? We studied the current trend in the visualizations used to represent the temporal component of epidemiological information and whether this took place through the use of time series, temporal granularity (e.g., week, month, and year), or interactive filters.
- Q4. Which combinations of spatial–temporal visualizations are most frequently used to represent epidemiological data? We studied how the papers represented both the spatial and temporal information concerning an epidemic, and whether they used just one technique or a set of several techniques.
- Q5. Do the existing visualizations use individual-level data or aggregated data? We analyzed whether it is common practice to work with individual data or with population-level data.
- Q6. Are there any visualizations of patients who became infected in a hospital? We analyzed how many papers carried out a study of hospital-acquired infections.
- Q7. What software tools are used? We explored the software used in the implementation of the study (e.g., programming languages and data management tools).
- Q8. What datasets and evaluation methods are used? We analyzed which datasets were most frequently used for the development and evaluation of each study, and what methods were used to evaluate them.
- Q9. Is the dataset open-access? It was of interest to determine how many of the papers presented open-access datasets.
These questions correspond to the mentioned fields of study as follows: (A) is answered using questions Q1 and Q6; (B) is investigated through questions Q2 to Q5; and (C) is looked into through questions Q7 to Q9.
2.2. Data Sources
As this review focused on visualization techniques and epidemiological data, we carried out the search in databases that would allow us to find one computational application over another. We therefore based our search criteria on preliminary research and recommendations obtained from existing systematic reviews [11,12] contained in the literature, and, consequently, we selected IEEE Xplore, PubMed, Scopus, Google Scholar, and ACM Digital Library. We restricted our search to journal articles from the last twenty-four years (from January 2000 to January 2024) and only those written in English. We performed 6 rounds of searching, and the last one was performed on 24 January 2024.
2.3. Search Strategy
We first searched for titles and abstracts using a selected catalog of search strings, after which we screened the titles and abstracts by employing inclusion and exclusion criteria. Finally, we screened the full text of each paper to ensure that it met the inclusion criteria.
2.3.1. Search Strings
After some rounds of trialing and refining the search terms, we chose the following terms in order to make the searches as wide-ranging as possible within the scope of our theme: “epidemic”, “epidemiological”, or “epidemiology”, with “visual analysis”, “temporal visualization”, “spatial visualization”, or “geographic visualization”. We also searched for works that might include the term “inpatients”, but only one database (i.e., IEEE Xplore) showed different essential results when adding this term. We then performed a search of medical subject heading (MeSH) terms for PubMed, which returned the following keywords: “epidemics”, “disease outbreaks”, and “pandemics”.
These search terms had to be adjusted for each database in order to attain the best possible results. Details of the search strings employed for each database are shown in Table 1.

Table 1.
Search strings for each database.
2.3.2. Search Process
We adopted a two-step search strategy, carrying out an exhaustive and systematic search in the digital libraries, followed by a manual search, where we checked the references of the included papers, and we also included other papers that were of interest for our scoping review but that were not retrieved by the automatic search. For example, we searched in the Computer Graphics Forum journal due to its importance in the field of visualization. This measure was adopted to reduce the bias of the automatic search.
2.4. Study Selection
All 1312 results were imported to a reference management tool (Zotero library) to automatically delete duplicates. This strategy allowed us to identify 415 duplicated papers. The remaining papers were exported to a spreadsheet containing essential information for screening: database key, item type (journal article, conference paper, etc.), publication year, list of authors, title, DOI, and URL.
We then screened the titles and abstracts of the remaining papers. The studies were screened by two reviewers. When in doubt as to whether to include a work, a discussion took place between the four reviewers until an agreement was reached. The exclusion criteria employed were as follows: (1) papers that were not written in English; (2) studies whose approach was the modeling of epidemic data but did not make use of spatial–temporal visualization techniques; (3) studies whose target group was not people (e.g., animal or plant infections); (4) papers for which only the abstract was available; and (5) papers not categorized as original journal articles, as we consider the latter to be the most complete and reliable sources.
The inclusion criteria were as follows: (IC1) studies carried out between 2000 and 2024 presenting a spatial or temporal visualization of epidemic data; (IC2) studies whose target group was individual persons or populations; (IC3) studies that indicated the tool or tools used for visualization implementation; and (IC4) in the case of duplication, the most recent and complete paper. For a paper to be included, it was mandatory for it to meet criterion IC1 and at least one of the other inclusion criteria, and it could not meet any exclusion criterion.
This screening led to the removal of 690 papers, and 246 were eventually chosen for the analysis of the body of the manuscript. We classified these studies based on the terms of interest for our scoping review, since this classification would help us decide the level of importance of each paper in answering our research questions. We arranged the papers that were chosen for reading in a table that would allow us to see and compare the terms of interest that they addressed at a glance. These terms were spatial visualization, temporal visualization, visual analysis, interactive simulation, 3D/AR, epidemic, GIS techniques, geographic visualization, contacts, outbreaks, individual patients, population, infectious disease, bacterial disease, incidence, prevalence, and indoor.
The next step consisted of screening the full text of the papers chosen. We excluded 132 more papers because they met at least one of the aforementioned exclusion criteria or because they did not meet the mandatory inclusion criterion (IC1). Including the papers that we found manually, a total of 114 papers remained. This process is outlined in Figure 1.
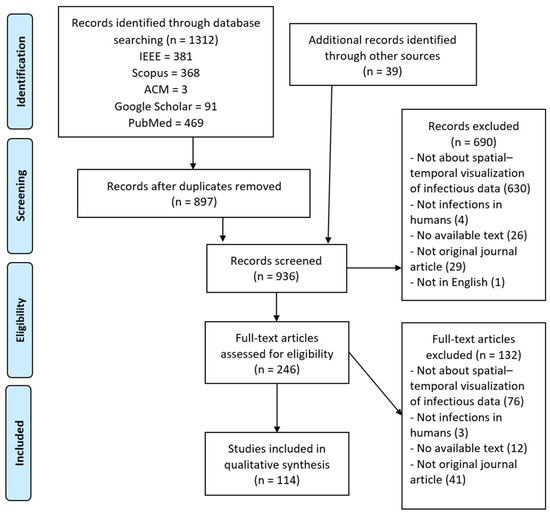
Figure 1.
PRISMA flow diagram.
2.5. Charting the Data
We developed a data-charting form to focus on the information that we wanted to extract from each article. This data-charting form contained the following variables: (1) meta-information, including title, year of publication, database, and type of publication (journal article, conference paper, etc.); (2) visualization techniques; (3) analyzed epidemiological data; (4) studied population; (5) target place and population; (6) data accessibility; (7) evaluation methods; and (8) tools used. The form was iteratively updated to meet the scope of the research questions.
In the section on visualization techniques, we analyzed which visualization techniques were applied to represent the spatial aspect, the temporal aspect, and, where appropriate, combinations of the spatial–temporal aspects. Within the spatial aspect, we classified the employed techniques as to whether they were geographic maps, representations of building structures, or statistical methods. In addition to this, we classified each article depending on the objective of the tool, whether it was real-time data presentation, real-time data detection, post-analysis, or trend prediction.
In the section on analyzed epidemiological data, we examined which epidemiological indicators were preferred for the analysis of the population and at what level of temporal and spatial granularity they were studied. For example, this may include the number of cases by country, contacts, mortality, and incidence, among others.
In the section on studied population, we wanted to know at what level the study was conducted, that is, whether it was at the individual or population level. In the section on target place and population, we analyzed whether the study was carried out in a hospital structure and whether the studied population was patients.
In the section on data accessibility, we analyzed what datasets were used and whether they are accessible. In evaluation methods, we listed the methods used in each article to validate the visualization techniques. And, in the section on tools, we enumerated the software tools used for the implementation or application of said visualizations.
2.6. Synthesis
We grouped the studies focusing on answering the research questions. This resulted in (1) an overview of the epidemic measures studied; (2) an overview of the types of temporal and spatial visualizations applied to represent those measures; (3) the classification of the target population in the studies; and (4) an overview of the tools and evaluation methods applied.
3. Results
From a scientific production point of view, all 114 of the studies eventually obtained were conducted between 2000 and 2023. Figure 2 illustrates the evolution of the number of publications. The number remained consistently low (0–5) until 2020, after which there was a notable increase in the number of papers (22). This is probably due to the need to study the spread of COVID-19 on a global scale. Most studies originated from the USA (33.33%, 38/114), China (14.91%, 17/114), Europe (20.18%, 23/114), and other countries in Asia (17.54%, 20/114). The results of research questions Q1 to Q9 are explained below.
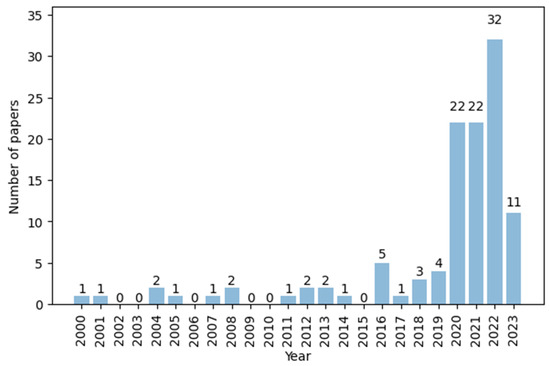
Figure 2.
Number of papers per year.
3.1. Q1: What Are the Most Commonly Visually Represented Measures of an Epidemic, and What Are the Visualization Purposes?
In this section, we analyze the type of epidemic data used in each study to identify general trends in their representation and the final use of the visualization techniques.
In this scoping review, we adopted “epidemic measures” as an umbrella term to include (1) key data parameters (such as the number of cases, number of deaths, symptoms, and number of medical resources), (2) the principal measures of risk indicated by the CDC [13] (morbidity, incidence, prevalence, etc.), and (3) contact and outbreak information. These pre-established statistical measures are usually applied to describe the current situation of an epidemic and predict its progression with the purpose of making decisions to alleviate it. Among all these measures, the most common ones, according to the literature [13], include the number of cases, the number of deaths, incidence, prevalence, contacts, and outbreaks.
We investigated the measures that were most often visually represented in the studies and found that a great number of papers graphically displayed the number of cases (75/114) and deaths (31/114). Other parameters were represented to a lesser extent, such as incidence (25/114), contacts (23/114), and accumulative cases (13/114), among others. The most repeated measures are shown in Figure 3, in which the category “Others” refers to the measures that appeared in only one study, which are connectedness between regions, the total and available capacity of a hospital, and the number of ventilators. Figure 4 shows the evolution of their use over time.
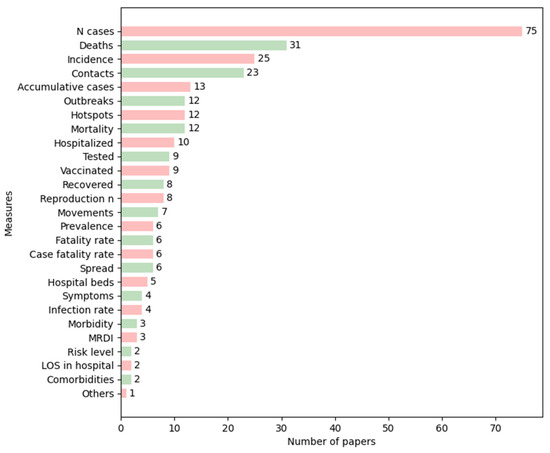
Figure 3.
Number of papers by epidemic measure. Colors are used so that each measure is easily followed. LOS = length of stay, MRDI = medical resource deficiency index.
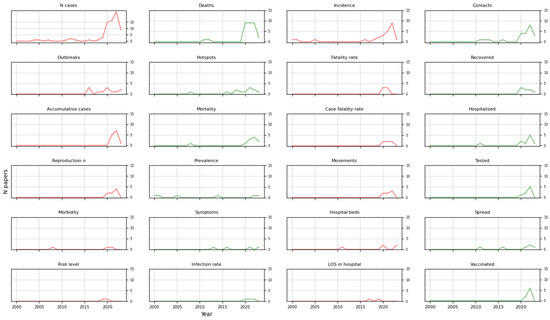
Figure 4.
Trends of principal measures over time. Colors coordinate the measures with Figure 3. LOS = length of stay.
In addition to the most represented epidemic measures, we analyzed the purpose of the visualization techniques designed in each paper. Table 2 summarizes the main uses and what measures were applied to achieve each target use, and the size of the dots represents the number of papers in each category.

Table 2.
Main uses of visualization techniques.
3.2. Q2: Which Techniques Are Most Frequently Used to Visualize Epidemiological Spatial Information?
In this section, we analyze the type of visualization techniques that the studies used to represent the spatial component of epidemiological data.
Of the 114 studies, 90.06% (79/114) employed maps as a basis on which to represent the data spatially; 36.48% (32/114) adopted a conceptual visualization approach (e.g., bar charts and line charts) to represent spatial information, and only 3.42% (3/114) represented the epidemiological data in an architectural format (i.e., building structure or building plans).
The studies that used a map-style representation (79/114) adopted different strategies: static geographic 2D or 3D maps [14,15,19,20,22,31,33,36,38,42,43,44,50,75,81,83,88,93,102,104,107,109,117,118,123], dynamic 2D maps [20,21,22,25,61,65,80,85,89,90,94,97], or choropleth maps [17,23,24,27,30,32,34,35,37,40,46,48,49,52,55,56,58,60,62,64,68,71,76,77,78,79,82,84,87,99,103,105,111,113,114,115,116,119,120,121,124,127]. Choropleth maps are widely used to visualize the state of a variable across a geographic area. In the papers, this technique was mostly used to represent the number of cases or cumulative cases in an area [17,22,24,27,46,48,52,56,58,60,61,62,63,65,67,71,76,79,84,87,99,103,105,113,114,115,116,119,121,127], along with the incidence of a disease [30,32,56,62,63,105,119,120,127]. It was also used to a lesser extent to visualize other measures, such as prevalence [35,64,76] and morbidity [111].
Moreover, 24 of the studies that used a map-style representation combined a map with one or more types of charts, most of which were bar charts, pie charts, and networks [17,21,30,43,44,49,50,76,80,82,83,85,93,102,107,111,115,117,123]. An example of the application of bar charts over maps to create symbol maps is the representation of confirmed, death, or recovery cases [102,107].
Networks were used to represent movement, flows, or contacts: the authors of [83] represented population flows between countries on a map, while those of [123] showed a food chain network to trace outbreaks of food-borne disease. Networks can also be used to depict correlations, as in [43], in which they were placed on a map to show the correlation of changes in the number of confirmed cases between two geographic areas. In other examples that combined maps with charts, it was common to employ circles of different radii to depict the number of cases or incidence by region [27,115]. These combinations allow for the study of the spread of a disease on a large scale and the comparison of the same measure in different locations, among other things.
Of the 32 studies that used only conceptual representations of spatial information regarding an epidemic, the dominant visualization methods were line charts and networks [28,54,57,67,69,72,73,86,100,108,110,112,122,125,126]. Line charts were used principally to represent the number of cases and fatality rates. For example, in one study, they were used to represent the number of confirmed deaths, recovered cases, and active cases with regard to the globe, the United States, and Spain in a particular period [28]. In another study, they were used to represent daily incident cases and the CFR in a period by country [100]. In a third, they were used to represent the attack rate, population fatality rate, CFR, and basic reproduction number by country [108]. Networks, however, were primarily used to represent contacts and to explain how an epidemic could spread: Ref. [110] visualized the people epidemiologically linked to an outbreak by employing person–division and person–person networks; Ref. [122] showed contact networks in a matrix view; and Ref. [112] represented the movement of patients carrying drug-resistant bacteria across three hospitals.
Finally, regarding the representation within buildings, it is worth mentioning that only three papers represented a building structure: Ref. [26] used a 2D plane of each floor, differentiating the areas of greatest interest to the user; Ref. [18] created a 3D model to represent aerosol transmission in a room; and Ref. [91] created a situation awareness dashboard with the bed layout and information on staff and equipment for clinicians working in ICUs.
The main techniques studied are represented in Figure 5, together with the evolution of their use over time. In recent years, there has been an increasing use of choropleth maps compared to the rest, and building representations are starting to emerge in the study of epidemiological data, although the number is still low.
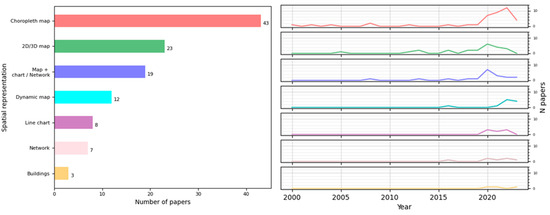
Figure 5.
Number of papers that applied a visual representation to depict space and trends of principal techniques over time.
3.3. Q3: Which Techniques Are the Most Frequently Used to Visualize Epidemiological Temporal Data?
In this section, we analyze the type of visualization techniques that the studies used to represent the temporal component of epidemiological data.
According to our review, more than half of the 114 studies (67/114) used 2D or 3D line charts as depictions of time series to represent temporal data related to a pandemic. They were not only used to depict the number of cases per week, month, or year [16,21,22,24,25,30,36,50,53,54,56,59,60,62,65,67,68,71,72,73,75,79,84,85,86,87,97,98,100,103,104,106,109,113,114,120,121,127] or cumulative cases [22,27,52,55,65,70,71,98,99,100,105,115] but were also used to depict other disease measures, such as the basic reproduction number [19,23,55,75,87,100,104,108,115], fatality rate [85,100,101,108], or incidence [32,39,57,59,62,67,97,98,104,105,120,127], among others [31,33,44,56,66,77,82,88,90,94,95,102,107,112,118,125,126]. Other papers (24/114) used bar charts based on different time granularities (days, months, and years) to illustrate the progression of the number of cases [16,28,32,41,43,48,51,63,67,68,71,90,92,96,97,104,106,120], deaths [33,68,70,96], and recovered cases [42], among others [60,88,117]. All the techniques employed to represent time in epidemiology are depicted in Figure 6, where “Others” comprises pie charts, Gantt charts, quilt plots, timelines, and specific line charts such as epidemic curves. The trends of the main techniques over the years are also depicted in Figure 6, color-coded similarly to the bar chart. It is notable that there is an increasing use of techniques such as line charts and heatmaps.
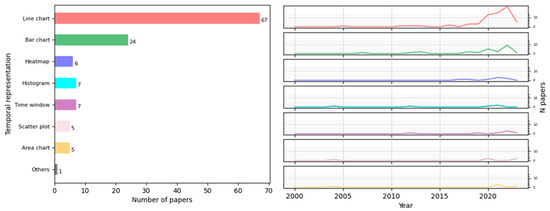
Figure 6.
Number of papers that applied a visual representation to depict time and trends of principal techniques over time.
Some of the studies used more than one technique to depict information concerning time. The most frequent combination was bar charts with line charts [16,30,33,60,67,68,70,71,88,90,97,104,106,120], which comprised 14 out of 35 combinations. Bar charts are particularly useful for comparing certain measures at different levels of temporal granularity to detect any breakdown of time that could influence the appearance of diseases. They were usually used to represent the number of cases [30,106,120] or other measures. In [88], the authors showed the daily progress of patients classified by a specific group of symptoms, and, in [16], the authors compared the weekly incidence rate by genre. Conversely, line charts depicted the continuous evolution of a particular measure: the incidence rate [120], number of patients [88], deaths [33], or cases of infection [16].
We also identified other noteworthy combinations, such as in the case of [115], where the authors not only used line charts, as mentioned earlier, but also epidemic curves based on daily COVID-19 confirmed cases to monitor the effectiveness of control measures.
3.4. Q4: Which Combinations of Spatial–Temporal Visualizations Are Most Frequently Used to Represent Epidemiological Data?
In this section, we analyze how the studies represented both the spatial and temporal information concerning an epidemic, and what technique or combination of techniques was employed.
According to our research, 79.83% (91/114) of the studies produced a spatial–temporal representation of epidemiological data. Three main cases will be discussed below: (1) maps applied with other techniques found in 34 papers, (2) maps applied alone found in 16 papers, and (3) the use of other techniques to depict time and space.
First, of all the combinations, that most frequently used was maps with animations [22,25,38,42,61,65,87,89,111,113,115,117,121], which represented 14.29% (13/91). Some of them also used line charts to compare the progression of the epidemic in different regions or countries [25,30,52,65,121]. We also found other combinations, such as maps with color-coding: [107] used three colors to show areas within the map with variations in the number of cases, and [31] used disc areas with a degree of colors to show the areas in which the earliest and latest outbreaks had occurred. There were also maps with filters of time by day [21,61,79,89,102,105,115,116,117,118,119], with a time range slider [113], or with timelines to show, for example, the daily progress of the death rate, rate of infection, and medical resource deficiency indices (MRDIs), along with their representation on a map [82]. To a lesser extent, some studies combined maps with bar charts [43,60,78,102] to compare different regions over time.
Second, the rendering of only maps occurred in the form of snapshots of choropleth maps to show and compare the differences between a single characteristic of an epidemic in an area from one point in time to another [19,37,49,55,62,64,71,76,84,120,124]. There were also maps that changed and showed the target measure at different moments [14,90]. Maps were applied for the achievement of the four main uses that we categorized in question Q1. Combinations of techniques with animations were used to a greater extent for the post-analysis of data and trend prediction and estimation and to a lesser extent for the real-time detection of data.
Third, 26 of the 91 studies applied line charts [16,21,23,24,25,28,30,36,48,57,65,68,85,86,95,97,99,100,103,106,108,109,121,125,126,127], followed by bar charts, which were applied in 16 combinations of space and time [16,33,41,43,48,51,60,67,68,70,77,78,96,102,106,109]. These studies used bar charts to depict the spread of an epidemic through districts with a time filter [106], color code by month [33], or classification by year [41]. They were also applied to represent the incidence of comorbidity by week in a selected state [16] or to represent periods linked to a map [43,109]. Table 3 shows all the references according to the visualization technique combinations used for spatial–temporal representation, where the category “Others” includes techniques only used once and not in any combination. In Table S2, the main topics or uses of each group of visualization techniques are also represented. This is encoded with the size attribute depending on the target use of the visualization: the size of each circle in the table indicates whether more or fewer papers performed each target use.

Table 3.
Articles by combinations of techniques used for spatial–temporal representation. The X marks the visualization methods employed in the articles.
3.5. Q5: Do the Existing Visualizations Use Individual-Level Data or Aggregated Data?
In this section, we analyze which studies represented individual-level data and which represented population-level data.
The target groups of most of the papers analyzed were populations (97/114). Only 4 papers studied data regarding individual persons [18,26,91,110], all of them between 2020 and 2021, while 13 papers observed both populations and individual people [14,32,34,48,51,69,89,90,92,104,109,112,117].
3.6. Q6: Are There Any Visualizations of Patients Who Became Infected in a Hospital?
In this section, we analyze how many of these studies were conducted in hospitals and whether they surveyed patients who had become infected inside the buildings.
We found that only 4 papers out of 114 focused on patients infected inside hospitals [29,51,91,112]. Ref. [29] developed a heatmap to study the relation between the length of stay of patients in intensive care units and the occurrence of hospital-acquired infections (HAIs) during their stay. Ref. [112] investigated the movement of patients carrying drug-resistant bacteria across a hospital to detect disease transmissions. Ref. [51] analyzed the transmission of COVID-19 in an emergency childcare center. Additionally, Ref. [91] created a dashboard with bed layouts and information on staff and equipment for clinicians working in ICUs.
3.7. Q7: What Software Tools Are Used?
In this section, we explore and break down the software used in the implementation of each study.
In this scoping review, the term tools refers to any piece of software used in the implementation of the study. This also includes programming languages, the packages most frequently used in the studies, and data management. The workflows, functionality, and algorithms of the programs developed are not within the scope of this study.
According to our results, there is a great variety of specific tools with which to visualize and work with epidemic data. We classified these tools according to their purpose: web development, data management, the visualization of maps or GIS, the statistic processing of data and generation of charts, and the development of network and graph representations. The papers that indicated the tools used are listed in Table 4, which also provides a breakdown of the tool categories.

Table 4.
Software tools utilized by papers.
3.8. Q8: What Datasets and Visualization Evaluation Methods Are Used?
In this section, we analyze the methods used to evaluate each study and the datasets most frequently employed for development and evaluation purposes.
The aim of visualization programs is to assist experts in their work in some respect, and, as such, there is no single way in which to evaluate them. In Table 5, we classified the papers according to whether they applied a standard evaluation methodology and the type of evaluation conducted. In the latter, we identified three main groups: user experience, task efficiency, and task accuracy. Papers where no evaluation method was specified were classified as “N/A”.

Table 5.
Evaluation method applied. The X marks the method applied in each paper.
In our research, 57% (65/114) of the studied papers did not indicate whether they used a validation method. Of the methods that we observed, the most used was the analysis of task accuracy, applied in 24 studies. Most evaluation methods were applied in works that developed an interactive tool [15,17,19,20,21,23,25,26,62,63,74,80,85,86,90,91,93,94,95,96,97,99,103,104,105,107,109,111,114,115,116,119,121,122,123,125]. Out of those that used visualization techniques to communicate data with some sort of interactivity (7/114), only 1 [92] validated its techniques by obtaining users’ feedback. Out of those that did not use any type of interaction, a few evaluated their implementations with task accuracy [40,50,51,60,66,71,87,118,126], task efficiency [31,36], and users’ experience [48,67].
With regard to datasets, 72.81% (83/114) of the studies indicated which ones they used. However, owing to the extent and variety of the studies reviewed, there were only eight duplicate sources: Korea Centers for Disease Control & Prevention (KCDC) [30,44,71,106,107], Johns Hopkins University datasets [15,23,28,42,46,47,53,57,75,81,84,85,86,95,97,98,125], WHO COVID-19 Daily Report [30,42,43,76,102], European Centre of Disease Prevention and Control (ECDC) [57,86,100,108,118], Centre of Disease Prevention and Control (CDC) on COVID-19 [24,30,31,55,95,102], the National Health Commission [42,43,52,55,87], the US Census Bureau [31,46,49,58,75,89,97], and the New York Times Github repository of COVID-19 [49,99].
3.9. Q9: Is the Dataset Open-Access?
In this section, we explore the open-access datasets that were used by the studies. Data accessibility is an increasingly important issue with regard to the reliability of a paper. Upon observing the proportions of studies that presented accessible data and those that did not, 72.81% (83/114) of the studies gave access to their data, and 38.6% (44/114) did not. In Figure 7, we can see the progression of papers that gave access, and it appears that there is currently an increasing trend regarding how data are presented. All the open-access datasets are presented in Table A1.
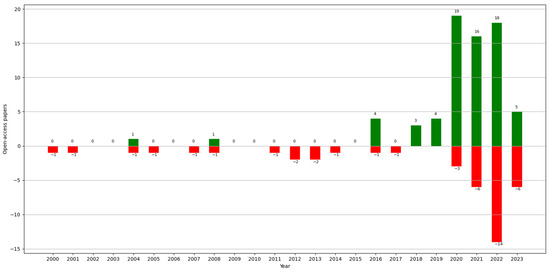
Figure 7.
Number of papers with open-access datasets (green) vs. without open-access datasets (red) by year.
4. Discussion
This scoping review aimed to summarize and analyze the current state of the literature on the development and research of spatial–temporal visualization techniques applied to epidemiological data. This scoping review, along with its results, contributes to the literature in several ways. First, it provides a mapping between the study of epidemiological events, the visualization techniques used, and their applications. Second, it analyzes the trends in the development and use of these techniques and their purposes in recent years. Third, it explores studies conducted at local and individual scales, with a focus on the search for studies within hospitals. The latter is particularly relevant in the context of the pandemic situation due to COVID-19. Finally, it summarizes the methodologies used in this field, including the tools, evaluation methods for visualizations, and datasets employed. In this section, we discuss the results obtained for the main research questions.
The most relevant tasks in epidemiology for visual representation have, to date, been the visualization of the number of health-related events (e.g., the number of cases of a disease in a population) and the recognition of the occurrences of these events concerning time and space to detect both temporal (at the level of year, season, week, etc.) and spatial patterns (geographic variations, urban and rural differences, etc.) [13]. Most of the studies reviewed focused on representing aggregate data, such as the number of cases, deaths, and incidence (Figure 3), which are investigations at a population level. We could see that the most frequently used measures peaked in recent years, except for prevalence, which was applied more in the early 2000s than recently. Regarding the other measures, we could not identify any trend in the papers that met the inclusion criteria defined.
Regarding the spatial visualization of the epidemiological events, the techniques most frequently employed were maps or GIS technologies often combined with widely used charts, such as bar charts, pie charts, and networks. These allow for the representation of movement and the comparison of the same measure in different locations, among other functions. We highlighted the increasing use of choropleth maps in recent years. This kind of representation is a great option to provide a comprehensive view of a situation and facilitates pattern recognition at a glance. However, choropleth maps have some features that need to be taken care of: they are more effective with relative data rather than absolute values when comparing regions. For instance, comparing the number of cases in different places using a choropleth map without considering population size might not provide useful insights; for these cases, alternative approaches may be more suitable, such as symbol maps. It is also important to consider the size of regions when making the comparison, as it can alter the perception of the colored areas. This issue can be addressed by incorporating interactions and text or by replacing the choropleth map with a symbol map or a cartogram, though the latter depends on the user’s knowledge of the geography.
We analyzed the main practical objectives of each presented application and identified four big groups: data presentation, real-time data detection, post-analysis, and trend prediction. Some papers only focused on one objective, while others covered many of them (Table 2). In the first place, the most common purpose was analytical, primarily for the subsequent study of epidemic situations. The next one was the informative purpose, achieved through data presentation. From Table 2 and Table 3, we could infer that there is a trend towards the creation of more complex tools, since, in recent years, there has been an increase in works covering many or all of these objectives. Besides this, there has been a rise in real-time presentation and real-time detection applications, reflecting the growing need for better monitoring and the development of new technologies.
Following this, we found that many papers focused on the analysis and prediction of trends, which is logical given the large number of studies that have emerged to help explain and understand the global situation during the COVID-19 pandemic. In this context, network-based methods can be very useful, as they can be integrated with powerful analytics, such as graph algorithms or arc prediction using Deep Learning. Through an effective layout, networks enable insights into data and facilitate the discovery of trends, outliers, and behavior patterns in complex datasets.
The detection of temporal patterns is essential for studying an epidemic. The most widely used visualization techniques for this were line charts and bar charts, which, along with heatmaps, have seen an increased use recently. Line charts are mainly employed to represent the evolution of a given measure, while bar charts enable the comparison of a measure across different levels of temporal granularity to detect any temporal breakdown that could influence disease appearance. The intuitive and extended nature of these visual representations makes them the preferred options over more specialized techniques [128].
With regard to spatial–temporal visualizations, most of the papers reviewed sought to capture the epidemic situation of a population or area. The most commonly employed techniques were line charts, maps (either alone or in combination with other techniques), and combinations of charts, such as line, bar, and pie charts, followed by graphs. For real-time data detection, studies mainly used maps with other techniques and heatmaps with graphs. In the post-analysis of data, maps (alone or with other techniques), combinations of charts, line charts with heatmaps, and bar charts with several techniques were utilized. Finally, for trend predictions and estimations, line charts were predominantly used, followed by maps, combinations of statistical charts, and the application of heatmaps and graphs. Thus, the current state of the art involves the combination of maps with animations, which, being dynamic representations, are suitable means to express the general development and the main trends in the data analyzed [128]. Additionally, a modern application should include, along with maps, interactive time filters or range sliders, allowing the user to choose a temporal granularity or a range of temporal granules (days or weeks, depending on the application), with the map displaying the situation at that moment.
There are currently many tools, programming language packages, and other resources that increasingly facilitate the development of maps and geographic displays. A wide range of tools were used for this purpose, and there appears to be a trend towards the development of web applications. This trend is likely because most of the programs are for informational purposes: the objective is to offer the population easy access to updated information at any time. This also justifies the choice of datasets for these programs, as most of the repeated datasets consist of open sources with information on COVID-19.
Contrary to this, we verified that there has been very little development in the scientific literature regarding programs aimed at facilitating decision-making at the hospital level to mitigate the spread of infectious diseases or epidemics. Only a small number of papers studied the transmission of diseases among patients in hospitals, with just three papers focusing on the spread of an epidemic within a building. Despite the low number of these, all of them were published in recent years, suggesting a potentially emerging trend to represent these data at a more local scale, and we expect more publications of this kind. Although this type of representation is not very extensive, it has several advantages that are more valued nowadays, such as enabling cost-effective studies of the efficacy of mitigation strategies in controlled environments or facilitating monitoring at a local scale, which can reveal patterns and aid in the decision-making process.
Another field of interest for us was the methodology employed in these studies. To this end, we analyzed the developed tools, the chosen evaluation methods, and the datasets. One of the main principles of information visualization is that the tool must be interactive, as it enhances cognition. This means that it must help us better understand the information or solve a problem more effectively. Furthermore, validation is a crucial step in the development of a visualization. Not all visual encodings can solve all tasks, so assessing the quality of a visualization for its specific objectives is essential. Despite this, our study shows that many tools were not interactive but rather static statistical presentations of data, and not all the tools were validated in the papers. However, most of the tools that were validated belong to recent years. This, together with the increasing accessibility of data, suggests a move towards greater transparency and reproducibility in development methodologies.
The papers analyzed did not adopt a single methodology to validate the visualization techniques and tools, but rather different approaches were employed. This coincides with the observation made by Chittaro [3], who noted the lack of established guidelines and disciplined methodologies for creating information visualizations. Moreover, in some fields such as healthcare, time constraints and other factors (such as work overload and a lack of resources for investigation) do not play in favor of the development of new technologies, including the fulfillment of a valid and formal evaluation. Evaluating visualizations requires a specific analysis of how well they achieve their intended objectives. However, we lack a more rigorous and formal method to validate visualization work in general, as was also pointed out in [129], since the evaluation of visualization techniques is a key issue in human interaction research.
Finally, regarding data accessibility, this is an increasingly important issue concerning the reliability of a paper. Since the “reproducibility crisis”, studied by a survey in 2016 [130], more and more works in the area of Computer Science are allowing access to the data that they use (unless it compromises privacy) or making the code available for reproducibility. For easier access, the open-access datasets found in the analyzed papers are listed and referenced in Table A1. Most of these datasets contain information published by governments on websites. In some cases, we found data collected by universities also available on websites or public repositories (e.g., GitHub). Even though there is a growing trend towards open-access data, these continue to be aggregated information, and, sometimes, the computations used are not indicated. Despite the importance at the health and control levels of studying contact between individuals to detect potential risk points or to analyze the transmission of a disease, along with the availability of hospital resources at critical moments of an epidemic, their study is not as extensive as with aggregate data. One potential solution to this issue could be the use of synthetic clinical data, which do not compromise patients’ privacy.
This scoping review has some limitations. First, it is possible that we overlooked relevant works during the search process despite our efforts to make it as extensive as possible through several rounds of searching. Second, we included only papers written in English, so there is a possibility that we missed out interesting works written in other languages, even though the search was conducted at a worldwide scale. Third, this study considered only journal articles to guarantee the study of solid research results. However, this might be considered a potential source of bias in some exceptional cases.
The literature contains several reviews involving data visualization and the human healthcare domain. Here, we analyze and compare our scoping review using two reviews that are the closest to ours in terms of subject matter: a systematic review conducted by Lauren Carroll et al. in 2014 [7] and a scoping review conducted by Jawad A Chishtie et al. in 2020 [9].
In the first case, the study carried out by Carroll had several objectives: (1) to identify public health user needs and preferences for infectious disease visualization tools; (2) to identify existing infectious disease information visualization tools; (3) to identify commonalities among approaches applied to different data types; and (4) to describe tool usability evaluation efforts and barriers to the adoption of such tools. It consisted of a systematic review that also followed the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidelines, choosing papers written in English between 1980 and mid-2013, of which the authors eventually studied 88. Of all the results attained, it is worth highlighting the analysis of the architecture of existing tools: this term refers to how the systems were created in the sense of software design. The authors sought to study the structure of each system as a whole, whereas we focused on what software tool was used for each part (for the management of data, web implementation, the incorporation of maps, etc.). They then focused on three types of tools: GIS, molecular epidemiology, and social network analysis. Concerning GIS, they detailed what GIS functionalities they found, whereas we indicated only whether or not the programs used GIS technology. In addition to this, they indicated that the systems that combined GIS with time did so using animations and time windows, which coincides with our results. Within molecular epidemiology, they sought to understand the clustering distribution of different molecular groupings of pathogens. In the social network analysis, they studied networks of social contact or exposure to disease that they found in the literature and concluded that there were no visualization methods that would help the user understand network structures at a more aggregate level. Regarding the usability study and evaluation, they discussed the latter at the level of utility and usability and, like us, analyzed the evaluation methods that were repeated in the literature. Finally, unlike our inferences, their conclusions were focused more on the user and the implementation and adoption of visualization tools.
In the second case, the objective of the scoping review conducted by Chishtie [9] was to summarize the use of visual analytics tools, techniques, and frameworks in interrelated areas of population health and health services research. This review was based on the PRISMA-ScR methodology [10], and the authors chose articles published in English between 2005 and early 2019 and obtained a final total of 55 articles. With regard to the results, it should be noted that, although they analyzed the software programs used for the programming of applications, as we did, they focused on their analytic capacities. Regarding the characteristics of data and datasets, they distinguished whether the studies used single dataset sources, multiple, or both, and they categorized said sources depending on whether they were simulation data, social network or web data, EMR or EHR data, and national or administrative survey data, unlike us, as we were more interested in discovering the data sources that were used in the papers and that were accessible. In the results concerning analytics and visualization engines, they, like us, indicated the main development tools, but they dedicated only a small paragraph to the visualization techniques most frequently employed. Finally, regarding the availability of tools, they studied the percentage of investigations that were open-source or available, and they indicated the main evaluation methods found, attaining similar conclusions to ours in this respect.
5. Conclusions
The purpose of our scoping review is to provide a global vision of the current state of visualization techniques applied to epidemiological data published in the scientific literature. In this work, we put the efforts made during the last two decades into perspective, paying particular attention to how epidemic measures, and temporal and spatial information are displayed. There is a trend towards the generation of geographic programs and web-based programs, along with the implementation of common and extended visualizations with little complexity to ensure their rapid comprehension. Our results suggest that the vast majority of the works have a more informative purpose for a general understanding. There is also a growing trend in recent years in the representation of data at an individual level, as well as the representation of scenarios inside buildings to study the spread of diseases at a more local scale. This, combined with the study of epidemic situations within hospitals, is a move forward in the analysis of severe diseases, such as COVID-19. However, these studies are still very few.
With this work, we have helped identify some gaps in the development of visualizations for epidemic data processes:
- There is a need for the development of good practices for the generation of more effective visualizations: we are faced with a situation where, though the need to apply visualizations in the field of health is more recognized (proven by the increase in publications in this field in recent years), we lack a clear methodology for developing and evaluating visualization techniques.
- There is still a lack of studies of epidemiological data at an individual level.
- There is a lack of representations of buildings in both 2D and 3D: this is key to helping identify contacts and the spread of diseases at a local level.
- There is little development of programs that would help decision-making at a hospital level to prevent the spread of infectious diseases or epidemics.
- A very small number of papers studied the acquisition of diseases by patients inside a hospital as the result of any epidemic.
The last point is an issue that is gaining more weight and is increasingly necessary. From a technological perspective, the COVID-19 pandemic has evidenced the need to address new challenges regarding the management of complex multi-source information: the generation of new methods for spatial–temporal patient contact visualization and the potential role of Artificial Intelligence techniques to support adaptive and personalized methods to display visual information. With all this, future visualization methods could help control and restrict rapid spread more effectively. In this field, the development of 3D visualization methods of buildings and the gamification of contact between patients, healthcare staff, and visitors could help clinicians control the spread of infections inside hospitals. Although these developments face the lack of availability of quality clinical data, the use of generated synthetic datasets could help address this challenge.
With this review, we provide a new perspective for the study of visualization techniques applied or developed to capture the spatial–temporal component of epidemiological data. We shed some light on what has been implemented so far and what we still lack. With this, we can help search for new developments that can be very useful for the study of epidemics that have not yet been assessed or in which much effort has not yet been put.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/technologies12030031/s1, Table S1: PRISMA-ScR checklist; Table S2: Papers by combinations of techniques for spatial–temporal representation with main uses for each paper, encoded through the size of the circles. The size indicates whether more or fewer papers from each group performed the respective target use.
Author Contributions
Conceptualization, M.C. and J.M.J.; methodology, D.K.; database search D.K.; manual search, D.K. and M.C.; screening, D.K. and M.C.; discussion, D.K., M.C., J.M.J. and B.C.-S.; writing—original draft preparation, D.K.; writing—review and editing, D.K., M.C., J.M.J. and B.C.-S.; visualization, D.K. and J.M.J. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially funded by the CONFAINCE project (Ref: PID2021-122194OB-I00), the Spanish Ministry of Science and Innovation, the Spanish Agency for Research (MCIN/AEI/10.13039/501100011033), the European Regional Development Fund (ERDF) A way of making Europe, the GRALENIA project (Ref: 2021/C005/00150055) supported by the Spanish Ministry of Economic Affairs and Digital Transformation, the Spanish Secretariat of State for Digitization and Artificial Intelligence, Red.es, and NextGenerationEU funding. This research was also partially funded by the FPI program grant (Ref: PRE2019-089806).
Data Availability Statement
All the data can be found in the body of the paper and the Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A

Table A1.
Open-access datasets used by the reviewed papers.
Table A1.
Open-access datasets used by the reviewed papers.
| Datasets | Papers |
|---|---|
| KCDC [131] | [25,39,106,107] |
| COVID-19 analysis tools at Instituto de Informática of the Federal University of Rio Grande do Sul (Brazil) [132] | [22] |
| COVID-19 Data Repository by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University [133] | [15,23,37,41,42,48,52,84,95,125] |
| NSF Spatiotemporal Innovation Center (STC) dataset on COVID-19 in the US [134] | [82] |
| Arrivals of non-resident tourists at Brazilian national borders by country of residence [135] | [83] |
| Trips abroad by Brazilian resident visitors to countries of destination [136] | [84] |
| Flutrack [137] | [14] |
| Notification on the pneumonia epidemic situation from the China National Health Commission [138] | [25] |
| Press statement by Hubei province Health Commission [139] | [25] |
| Press statement of novel coronavirus (in Japanese) by the Ministry of Health, Labour and Welfare, Japan [140] | [25] |
| CDC [141] | [25,26,50,95,102] |
| Data on school start dates in the US [142] | [26] |
| US Census Bureau [143] | [26,41,44] |
| Statistics Canada and Census datasets [144] | [27] |
| District Health Information System (DHIS) [145] | [31] |
| Taiwan Open Data Platform [146] | [20] |
| Taiwan Centers for Disease Control (TCDC) [147] | [20] |
| Interactions in a French primary school [148] | [122] |
| FoodRisk-Labs [149] | [123] |
| Epidemic Situation by the Ministry of Civil Affairs of Bosnia and Herzegovina [150] | [115] |
| The New York Times Coronavirus in the United States dataset [151] | [44,99] |
| Open Data of Epidemiology by the National Governments Health Secretary [152] | [16] |
| National Epidemiological Surveillance of Infectious Diseases [153] | [32] |
| European Centre for Disease Prevention and Control (ECDC) [154] | [52,100,108,118] |
| COVID-19 situation reports by the World Health Organization (WHO) [155] | [25,37,38,102] |
| Chinese Center for Disease Control and Prevention [156] | [37] |
| National Health Commission of the PCR [157] | [37,38,47,50] |
| COVID-19 from Datadista [158] | [101] |
| US coronavirus cases by county from USAFacts [159] | [17] |
| COVID-19 data from the Italian National Institute of Health [160] | [118] |
| Compendium of US Health Systems [161] | [41] |
| Directorate of Health Services, Government of Kerala, India [162] | [90] |
| NSF Spatiotemporal Innovation Center (STC) dataset on COVID-19 [163] | [113] |
| genEpi dataset [164] | [43] |
| The COVID-19 Tracking Project [165] | [84] |
| COVID-19 dataset from the Presidency of the Council of Ministers- Department of Civil Protection [166] | [84] |
| Tencent COVID-19 dataset [167] | [124] |
| Infectious Disease App by the International Civil Aviation Organization [168] | [45] |
| esCOVID-19 data [169] | [103] |
| Information about different aspects of the Spanish reality from Spanish National Institute of Statistics [170] | [103] |
| Mortality monitoring system from the Spanish Health Institute Carlos III [171] | [103] |
| Information about meteorological data stemming from Spanish State Agency of Meteorology [172] | [103] |
| HIV-1 Transmission Dynamics in Germany [173] | [93] |
| Oxford COVID-19 Government Response Tracker [174] | [125] |
| COVID-19 Data from the NSW Government [175] | [109] |
| Victorian Coronavirus (COVID-19) Data [176] | [109] |
| Locations Visited by Confirmed COVID-19 Cases in Western Australia [177] | [109] |
| Australian Statistical Geography Standard (ASGS) [177] | [109] |
References
- Last, J.M.; International Epidemiological Association (Eds.) A Dictionary of Epidemiology, 4th ed.; Oxford University Press: New York, NY, USA, 2001; ISBN 978-0-19-514168-9. [Google Scholar]
- Combi, C.; Keravnou-Papailiou, E.; Shahar, Y. Temporal Information Systems in Medicine, 1st ed.; Springer Publishing Company, Incorporated: Berlin/Heidelberg, Germany, 2010; ISBN 978-1-4419-6542-4. [Google Scholar]
- Chittaro, L. Information visualization and its application to medicine. Artif. Intell. Med. 2001, 22, 81–88. [Google Scholar] [CrossRef]
- Shneiderman, B.; Plaisant, C.; Hesse, B.W. Improving Healthcare with Interactive Visualization. Computer 2013, 46, 58–66. [Google Scholar] [CrossRef]
- Chen, H.; Zeng, D.; Yan, P. Data Visualization, Information Dissemination, and Alerting. In Infectious Disease Informatics: Syndromic Surveillance for Public Health and BioDefense; Chen, H., Zeng, D., Yan, P., Eds.; Springer: New York, NY, USA, 2010; pp. 73–87. [Google Scholar] [CrossRef]
- Bucalon, B.; Shaw, T.; Brown, K.; Kay, J. State-of-the-art Dashboards on Clinical Indicator Data to Support Reflection on Practice: Scoping Review. JMIR Med. Inform. 2022, 10, e32695. [Google Scholar] [CrossRef]
- Carroll, L.N.; Au, A.P.; Detwiler, L.T.; Fu, T.-C.; Painter, I.S.; Abernethy, N.F. Visualization and analytics tools for infectious disease epidemiology: A systematic review. J. Biomed. Inform. 2014, 51, 287–298. [Google Scholar] [CrossRef]
- Crisan, A.; Gardy, J.L.; Munzner, T. A systematic method for surveying data visualizations and a resulting genomic epidemiology visualization typology: GEViT. Bioinformatics 2018, 35, 1668–1676. [Google Scholar] [CrossRef]
- Chishtie, J.A.; Marchand, J.-S.; A Turcotte, L.; Bielska, I.A.; Babineau, J.; Cepoiu-Martin, M.; Irvine, M.; Munce, S.; Abudiab, S.; Bjelica, M.; et al. Visual Analytic Tools and Techniques in Population Health and Health Services Research: Scoping Review. J. Med. Intern. Res. 2020, 22, e17892. [Google Scholar] [CrossRef]
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.K.; Colquhoun, H.; Levac, D.; Moher, D.; Peters, M.D.J.; Horsley, T.; Weeks, L.; et al. PRISMA Extension for Scoping Reviews (PRIS-MA-ScR): Checklist and Explanation. Ann. Intern. Med. 2018, 169, 467–473. [Google Scholar] [CrossRef]
- Tawfik, G.M.; Dila, K.A.S.; Mohamed, M.Y.F.; Tam, D.N.H.; Kien, N.D.; Ahmed, A.M.; Huy, N.T. A step by step guide for conducting a systematic review and meta-analysis with simulation data. Trop. Med. Health 2019, 47, 46. [Google Scholar] [CrossRef]
- Xie, B.; Tao, C.; Li, J.; Hilsabeck, R.C.; Aguirre, A. Artificial Intelligence for Caregivers of Persons With Alzheimer’s Disease and Related Dementias: Systematic Literature Review. JMIR Med. Inf. 2020, 8, e18189. [Google Scholar] [CrossRef]
- Dicker, R.; Coronado, F.; Koo, D.; Gibson Parrish, R. Principles of Epidemiology in Public Health Practice, 3rd ed.; An Introduction; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2006; p. 511.
- Chorianopoulos, K.; Talvis, K. Flutrack.org: Open-source and linked data for epidemiology. Health Inform. J. Engl. 2016, 22, 962–974. [Google Scholar] [CrossRef]
- Jen, T.-H.; Chien, T.-W.; Yeh, Y.-T.; Lin, J.-C.J.; Kuo, S.-C.; Chou, W. Geographic risk assessment of COVID-19 transmission using recent data: An observational study. Medicine 2020, 99, e20774. [Google Scholar] [CrossRef]
- Galván-Tejada, C.E.; Zanella-Calzada, L.A.; Villagrana-Bañuelos, K.E.; Moreno-Báez, A.; Luna-García, H.; Celaya-Padilla, J.M.; Galván-Tejada, J.I.; Gamboa-Rosales, H. Demographic and Comorbidities Data Description of Population in Mexico with SARS-CoV-2 Infected Patients(COVID19): An Online Tool Analysis. Int. J. Env. Res. Public. Health 2020, 17, 5173. [Google Scholar] [CrossRef]
- Kaul, S.; Coleman, C.; Gotz, D. A rapidly deployed, interactive, online visualization system to support fatality management during the coronavirus disease 2019 (COVID-19) pandemic. J. Am. Med. Inform. Assoc. 2020, 27, 1943–1948. [Google Scholar] [CrossRef]
- Makhsous, S.; Segovia, J.M.; He, J.; Chan, D.; Lee, L.; Novosselov, I.V.; Mamishev, A.V. Methodology for Addressing Infectious Aerosol Persistence in Real-Time Using Sensor Network. Sensors 2021, 21, 3928. [Google Scholar] [CrossRef]
- Su, W.; Fu, W.; Kato, K.; Wong, Z.S.-Y. “Japan LIVE Dashboard” for COVID-19: A Scalable Solution to Monitor Real-Time and Regional-Level Epidemic Case Data. Stud. Health Technol. Inf. Neth. 2021, 286, 21–25. [Google Scholar]
- Cintron, R.; Whitmer, S.L.M.; Moscoso, E.; Campbell, E.M.; Kelly, R.; Talundzic, E.; Mobley, M.; Chiu, K.W.; Shedroff, E.; Shankar, A.; et al. HantaNet: A New MicrobeTrace Application for Hantavirus Classification, Genomic Surveillance, Epidemiology and Outbreak Investigations. Viruses 2023, 15, 2208. [Google Scholar] [CrossRef]
- Neto, O.L.; Paolotti, D.; Dalton, C.; Carlson, S.; Susumpow, P.; Parker, M.; Phetra, P.; Lau, E.H.Y.; Colizza, V.; van Hoek, A.J.; et al. Enabling Multicentric Participatory Disease Surveillance for Global Health Enhancement: Viewpoint on Global Flu View. JMIR Public Health Surveill. 2023, 9, e46644. [Google Scholar] [CrossRef]
- Meneses, M.V.; Riva, A.; Salemi, M.; Mavian, C. ARCA: The interactive database for arbovirus reported cases in the Americas. BMC Bioinform. 2023, 24, 312. [Google Scholar] [CrossRef]
- Shi, A.; Gaynor, S.M.; Dey, R.; Zhang, H.; Quick, C.; Lin, X. COVID-19 Spread Mapper: A multi-resolution, unified framework and open-source tool. Bioinform. Engl. 2022, 38, 2661–2663. [Google Scholar] [CrossRef]
- Mason, L.; Hicks, B.; Almeida, J.S. EpiVECS: Exploring spatiotemporal epidemiological data using cluster embedding and interactive visualization. Sci. Rep. 2023, 13, 21193. [Google Scholar] [CrossRef]
- Chen, C.-C.; Teng, Y.-C.; Lin, B.-C.; Fan, I.-C.; Chan, T.-C. Online platform for applying space–time scan statistics for prospectively detecting emerging hot spots of dengue fever. Int. J. Heal. Geogr. 2016, 15, 43. [Google Scholar] [CrossRef]
- Van Woensel, W.; Roy, P.C.; Abidi, S.S.R.; Abidi, S.R. Indoor location identification of patients for directing virtual care: An AI ap-proach using machine learning and knowledge-based methods. Artif. Intell. Med. 2020, 108, 101931. [Google Scholar] [CrossRef]
- Comba, J.L.D. Data Visualization for the Understanding of COVID-19. Comput. Sci. Eng. 2020, 22, 81–86. [Google Scholar] [CrossRef]
- Muthusami, R.; Saritha, K. Statistical analysis and visualization of the potential cases of pandemic coronavirus. Virusdisease 2020, 31, 204–208. [Google Scholar] [CrossRef]
- Ohannessian, R.; Bénet, T.; Argaud, L.; Guérin, C.; Guichon, C.; Piriou, V.; Rimmelé, T.; Girard, R.; Gerbier-Colomban, S.; Vanhems, P. Heat map for data visualization in infection control epidemiology: An application describing the relationship between hospi-tal-acquired infections, Simplified Acute Physiological Score II, and length of stay in adult intensive care units. Am. J. Infect. Control. 2017, 45, 746–749. [Google Scholar] [CrossRef]
- Xu, X.; Zeng, J.; Liu, R.; Liu, Y.; Zhou, X.; Zhou, L.; Dong, T.; Cha, Y.; Wang, Z.; Deng, Y.; et al. Should we remain hopeful? The key 8 weeks: Spatiotemporal epidemic characteristics of COVID-19 in Sichuan Province and its compar-ative analysis with other provinces in China and global epidemic trends. BMC Infect. Dis. 2020, 20, 807. [Google Scholar] [CrossRef]
- Kissler, S.M.; Gog, J.R.; Viboud, C.; Charu, V.; Bjørnstad, O.N.; Simonsen, L.; Grenfell, B.T. Geographic transmission hubs of the 2009 influenza pandemic in the United States. Epidemics 2019, 26, 86–94. [Google Scholar] [CrossRef]
- Brown, E.M.; McTaggart, L.R.; Dunn, D.; Pszczolko, E.; Tsui, K.G.; Morris, S.K.; Stephens, D.; Kus, J.V.; Richardson, S.E. Epidemiology and Geographic Distribution of Blastomycosis, Histoplasmosis, and Coccidioidomycosis, Ontario, Canada, 1990–2015. Emerg. Infect. Dis. 2018, 24, 1257–1266. [Google Scholar] [CrossRef]
- Séguy, I.; Bernigaud, N.; Bringé, A.; Signoli, M.; Tzortzis, S. A geographic information system for the study of past epidemics: The 1705 epidemic in Martigues (Bouches-du-Rhône, France). Can. Stud. Popul. 2012, 39, 107–122. [Google Scholar] [CrossRef]
- Curtis, A.J. Three-dimensional visualization of cultural clusters in the 1878 yellow fever epidemic of New Orleans. Int. J. Health Geogr. 2008, 7, 47. [Google Scholar] [CrossRef][Green Version]
- Wasley, A.; Alter, M.J. Epidemiology of Hepatitis C: Geographic Differences and Temporal Trends. Semin. Liver Dis. 2000, 20, 1–16. [Google Scholar] [CrossRef]
- Ullah, S.; Daud, H.; Dass, S.C.; Fanaee-T, H.; Khalil, A. An Eigenspace approach for detecting multiple space-time disease clusters: Application to measles hotspots detection in Khyber-Pakhtunkhwa, Pakistan. PLoS ONE 2018, 13, e0199176. [Google Scholar] [CrossRef]
- Sakai, T.; Suzuki, H.; Sasaki, A.; Saito, R.; Tanabe, N.; Taniguchi, K. Geographic and Temporal Trends in Influenzalike Illness, Japan, 1992–1999. Emerg. Infect. Dis. 2004, 10, 1822–1826. [Google Scholar] [CrossRef]
- Meliker, J.R.; Slotnick, M.J.; AvRuskin, G.A.; Kaufmann, A.; Jacquez, G.M.; Nriagu, J.O. Improving exposure assessment in environmental epidemiology: Application of spatio-temporal visualization tools. J. Geogr. Syst. 2005, 7, 49–66. [Google Scholar] [CrossRef]
- Massart, F.; Seppia, P.; Pardi, D.; Lucchesi, S.; Meossi, C.; Gagliardi, L.; Liguori, R.; Fiore, L.; Federico, G.; Saggese, G. High incidence of central precocious puberty in a bounded geographic area of northwest Tuscany: An estrogen disrupter epidemic? Gynecol. Endocrinol. 2005, 20, 92–98. [Google Scholar] [CrossRef]
- Zenilman, J.M.; Glass, G.; Shields, T.; Jenkins, P.R.; Gaydos, J.C.; McKee, K.T., Jr. Geographic epidemiology of gonorrhoea and chlamydia on a large military installation: Application of a GIS system. Sex. Transm. Infect. 2002, 78, 40–44. [Google Scholar] [CrossRef][Green Version]
- Blanc, D.S.; Petignat, C.; Wenger, A.; Kuhn, G.; Vallet, Y.; Fracheboud, D.; Trachsel, S.; Reymond, M.; Troillet, N.; Siegrist, H.H.; et al. Changing Molecular Epidemiology of Methicillin-Resistant Staphylococcus aureus in a Small Geographic Area over an Eight-Year Period. J. Clin. Microbiol. 2007, 45, 3729–3736. [Google Scholar] [CrossRef]
- Dey, S.K.; Rahman, M.; Siddiqi, U.R.; Howlader, A. Analyzing the epidemiological outbreak of COVID-19: A visual exploratory data analysis approach. J. Med. Virol. 2020, 92, 632–638. [Google Scholar] [CrossRef]
- So, M.K.; Tiwari, A.; Chu, A.M.; Tsang, J.T.; Chan, J.N. Visualizing COVID-19 pandemic risk through network connectedness. Int. J. Infect. Dis. 2020, 96, 558–561. [Google Scholar] [CrossRef]
- Park, J.Y. Spatial Visualization of Cluster-Specific COVID-19 Transmission Network in South Korea During the Early Epidemic Phase. medRxiv 2020. [Google Scholar] [CrossRef]
- Wand, H.; Iversen, J.; Law, M.; Maher, L. Quilt Plots: A Simple Tool for the Visualisation of Large Epidemiological Data. PLoS ONE 2014, 9, e85047. [Google Scholar] [CrossRef]
- Ronquillo, J.G.; Lester, W.T.; Zuckerman, D.M. Using informatics to guide public health policy during the COVID-19 pandemic in the USA. J. Public Health 2020, 42, 660–664. [Google Scholar] [CrossRef]
- Pang, M.-F.; Liang, Z.-R.; Cheng, Z.-D.; Yang, X.-P.; Wu, J.-W.; Lyu, K.; Xi, J.-J.; Li, Z.-J.; Shi, G.-Q.; Zhang, Y.-P.; et al. Spatiotemporal visualization for the global COVID-19 surveillance by balloon chart. Infect. Dis. Poverty 2021, 10, 21. [Google Scholar] [CrossRef]
- Crisan, A.; Fisher, S.E.; Gardy, J.L.; Munzner, T. GEViTRec: Data Reconnaissance Through Recommendation Using a Do-main-Specific Visualization Prevalence Design Space. IEEE Trans. Vis. Comput. Graph. 2021, 28, 4855–4872. [Google Scholar] [CrossRef]
- Cahill, G.; Kutac, C.; Rider, N.L. Visualizing and assessing US county-level COVID19 vulnerability. Am. J. Infect. Control 2021, 49, 678–684. [Google Scholar] [CrossRef]
- Chu, A.M.; Chan, J.N.; Tsang, J.T.; Tiwari, A.; So, M.K. Analyzing Cross-country Pandemic Connectedness During COVID-19 Using a Spatial-Temporal Database: Network Analysis. JMIR Public Health Surveill. 2021, 7, e27317. [Google Scholar] [CrossRef]
- Soto, J.C.; Barakat, M.; Hutter, J.A.; Kiely, M.; Moreira, S.; Shapiro, B.J.; Murall, C.L.; Parenteau, N.; Désilets, J.; Lessard, R. Outbreak inves-tigation of SARS-CoV-2 transmission in an emergency childcare centre. Can. J. Public Health 2021, 112, 566–575. [Google Scholar] [CrossRef]
- Ma, Q.; Gao, J.; Zhang, W.; Wang, L.; Li, M.; Shi, J.; Zhai, Y.; Sun, D.; Wang, L.; Chen, B.; et al. Spatio-temporal distribution characteristics of COVID-19 in China: A city-level modeling study. BMC Infect. Dis. 2021, 21, 816. [Google Scholar] [CrossRef] [PubMed]
- Corso, K.A.; Kielbasa, K.; Calkin, A.B. Using standard celeration makes COVID-19 data more meaningful. Fam. Syst. Health 2021, 39, 101–111. [Google Scholar] [CrossRef]
- Johansson, B. Challenges and Controversies in COVID-19: Masking the General Population may Attenuate This Pandemic’s Outbreak. Front. Public Health 2021, 9, 643991. [Google Scholar] [CrossRef]
- Hu, Y.; Kong, L.; Yao, T.; Chen, X.; Du, W. Does lock-down of Wuhan effectively restrict early geographic spread of novel coronavirus epidemic during chunyun in China? A spatial model study. BMC Public Health 2021, 21, 825. [Google Scholar] [CrossRef] [PubMed]
- Belay, E.D.; Abrams, J.; Oster, M.E.; Giovanni, J.; Pierce, T.; Meng, L.; Prezzato, E.; Balachandran, N.; Openshaw, J.J.; Rosen, H.E.; et al. Trends in Geographic and Temporal Distribution of US Children with Multisystem Inflammatory Syndrome during the COVID-19 Pandemic. JAMA Pediatr. 2021, 175, 837–845. [Google Scholar] [CrossRef]
- Manz, K.M.; Mansmann, U. Inequality indices to monitor geographic differences in incidence, mortality and fatality rates over time during the COVID-19 pandemic. PLoS ONE 2021, 16, e0251366. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, S.; Bauer, C.X.; Hendricks, B.; Stopka, T.J. Spatial epidemiology: An empirical framework for syndemics research. Soc. Sci. Med. 2020, 295, 113352. [Google Scholar] [CrossRef]
- de Sousa, P.A.; Rodrigues, C.A.; Nascimento Sobrinho, C.L.; da Cruz, L.A.; Santos Junior, E.G.D.; Nunes, P.C.; Costa, M.G.R.; da Costa Rocha, M.O. COVID-19 epidemic curve in Brazil: A sum of multiple epidemics, whose inequality and population density in the states are correlated with growth rate and daily acceleration. An ecological study. Rev. Soc. Bras. Med. Trop. Braz. 2022, 55, e0118. [Google Scholar]
- Haileselassie, W.; Getnet, A.; Solomon, H.; Deressa, W.; Yan, G.; Parker, D.M. Mobile phone handover data for measuring and ana-lysing human population mobility in Western Ethiopia: Implication for malaria disease epidemiology and elimination efforts. Malar. J. Engl. 2022, 21, 323. [Google Scholar] [CrossRef]
- Simpson, R.B.; Babool, S.; Tarnas, M.C.; Kaminski, P.M.; Hartwick, M.A.; Naumova, E.N. Dynamic mapping of cholera outbreak during the Yemeni Civil War, 2016–2019. J. Public Health Policy 2022, 43, 185–202. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, P.S.; Woodyatt, C.R.; Kouzouian, O.; Parrish, K.J.; Taussig, J.; Conlan, C.; Phillips, H. America’s HIV Epidemic Analysis Dashboard: Protocol for a Data Resource to Support Ending the HIV Epidemic in the United States. JMIR Public Health Surveill. 2022, 8, e33522. [Google Scholar] [CrossRef]
- Ma, W.; Shi, L.; Li, M. A fast and accurate method for SARS-CoV-2 genomic tracing. Brief. Bioinform. 2023, 24, bbad339. [Google Scholar] [CrossRef]
- Su, Y.-J.; Ma, Z.-D.; Qiao, X.; Wang, P.-T.; Kang, Y.-T.; Yang, N.-A.; Jia, W.; Zhao, Z.-J. Geospatial epidemiology of Toxoplasma gondii infection in livestock, pets, and humans in China, 1984–2020. Parasitol. Res. 2022, 121, 743–750. [Google Scholar] [CrossRef]
- Ho, S.Y.-C.; Chien, T.-W.M.; Shao, Y.; Hsieh, J.-H. Visualizing the features of inflection point shown on a temporal bar graph using the data of COVID-19 pandemic. Medicine 2022, 101, e28749. [Google Scholar] [CrossRef]
- Garcia-Carretero, R.; Vazquez-Gomez, O.; Ordoñez-Garcia, M.; Garrido-Peño, N.; Gil-Prieto, R.; Gil-De-Miguel, A. Differences in Trends in Admissions and Outcomes among Patients from a Secondary Hospital in Madrid during the COVID-19 Pandemic: A Hospital-Based Epidemiological Analysis (2020–2022). Viruses 2023, 15, 1616. [Google Scholar] [CrossRef]
- Missaghi, B.; Malik, M.W.; Shaukat, W.; Ranjha, M.A.; Ikram, A.; Barkema, H.W. Associations of the COVID-19 pandemic with the reported incidence of important endemic infectious disease agents and syndromes in Pakistan. BMC Infect. Dis. 2022, 22, 887. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.K.; Rahman, M.; Siddiqi, U.R.; Howlader, A.; Tushar, A.; Qazi, A. Global landscape of COVID-19 vaccination progress: Insight from an exploratory data analysis. Hum. Vaccines Immunother. 2022, 18, 2025009. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Phinney, S.; Angell, K.; Grimm, B.; Tegomoh, B.; Figliomeni, J.; Abdalhamid, B.; Khan, A.S.; Donahue, M.; Brett-Major, D.M.; et al. Outbreak of SARS-CoV-2 B.1.617.2 (Delta Variant) in a Youth Camp Associated With Community Spread, Nebraska, June–July 2021. Public Health Rep. 2022, 138, 157–163. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, H.; Wen, Z.; Gu, M.; Liu, L.; Li, X. Asymptomatic Transmissibility Calls for Implementing a Zero-COVID Strategy to End the Current Global Crisis. Front. Cell. Infect. Microbiol. 2022, 12, 836409. [Google Scholar] [CrossRef]
- Kang, D.; Choi, J.; Kim, Y.; Kwon, D. An analysis of the dynamic spatial spread of COVID-19 across South Korea. Sci. Rep. 2022, 12, 9364. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, J.; Gao, S.; Wang, H. Complex Contact Network of Patients at the Beginning of an Epidemic Outbreak: An Analysis Based on 1218 COVID-19 Cases in China. Int. J. Environ. Res. Public Health 2022, 19, 689. [Google Scholar] [CrossRef]
- Zhangbo, Y.; Zheng, C.; Hui, W. Contact network analysis of COVID-19 Delta variant outbreak in urban China -based on 2,050 confirmed cases in Xi’an, China. BMC Public Health Engl. 2022, 22, 2408. [Google Scholar] [CrossRef]
- Sondag, M.; Turkay, C.; Xu, K.; Matthews, L.; Mohr, S.; Archambault, D. Visual Analytics of Contact Tracing Policy Simulations During an Emergency Response. Comput. Graph. Forum 2022, 41, 29–41. [Google Scholar] [CrossRef]
- Dallas, T.A.; Foster, G.; Richards, R.L.; Elderd, B.D. Epidemic time series similarity is related to geographic distance and age structure. Infect. Dis. Model. 2022, 7, 690–697. [Google Scholar] [CrossRef] [PubMed]
- Taran, A.; Alfanatseh, A.; Rawashdeh, S.; Almayouf, F. The Temporal and Spatial Analysis of Corona Pandemic in Jordan using the Geographic Information System: An Applied Geographical Study. Indones. J. Geogr. 2022, 55, 155–166. [Google Scholar] [CrossRef]
- Rangachev, A.; Marinov, G.K.; Mladenov, M. The demographic and geographic impact of the COVID pandemic in Bulgaria and Eastern Europe in 2020. Sci. Rep. 2022, 12, 6333. [Google Scholar] [CrossRef]
- Ngom, R.; Gueye, A.S.; Lassieur, S.; Oloo, S.; Shahid, R.; Mize, V.; Okot, C.L.; Waogodo, J.C.; Fall, I.S. Five decades of infectious diseases outbreaks in the African region (1970–2018) a geographic snapshot. Soc. Sci. Humanit. Open 2023, 8, 100625. [Google Scholar] [CrossRef]
- Li, M. Epidemic Data Analysis and Visualization System based on Big Data. Int. Core J. Eng. 2023, 9, 206–220. [Google Scholar]
- Yang, C.; Zhang, Z.; Fan, Z.; Jiang, R.; Chen, Q.; Song, X.; Shibasaki, R. EpiMob: Interactive Visual Analytics of Citywide Human Mobility Restrictions for Epidemic Control. IEEE Trans. Vis. Comput. Graph. 2023, 29, 3586–3601. [Google Scholar] [CrossRef]
- Zheng, T.; Zhang, C.; Shi, Y.; Chen, D.; Liu, S. Influencing Factors and Clustering Characteristics of COVID-19: A Global Analysis. Big Data Min. Anal. 2022, 5, 318–338. [Google Scholar] [CrossRef]
- Sha, D.; Miao, X.; Lan, H.; Stewart, K.; Ruan, S.; Tian, Y.; Tian, Y.; Yang, C. Spatiotemporal analysis of medical resource deficiencies in the U.S. under COVID-19 pandemic. PLoS ONE 2020, 15, e0240348. [Google Scholar] [CrossRef]
- Moraga, P.; Dorigatti, I.; Kamvar, Z.N.; Piatkowski, P.; Toikkanen, S.E.; Nagraj, V.P.; Donnelly, C.A.; Jombart, T. epiflows: An R package for risk assessment of travel-related spread of disease. F1000Research 2018, 7, 1374. [Google Scholar] [CrossRef]
- Zhou, S.; Braca, P.; Marano, S.; Willett, P.; Millefiori, L.M.; Gaglione, D.; Pattipati, K.R. Application of Hidden Markov Models to Analyze, Group and Visualize Spatio-Temporal COVID-19 Data. IEEE Access 2021, 9, 134384–134401. [Google Scholar] [CrossRef]
- Healey, C.G.; Simmons, S.J.; Manivannan, C.; Ro, Y. Visual Analytics for the Coronavirus COVID-19 Pandemic. Big Data 2022, 10, 95–114. [Google Scholar] [CrossRef]
- Balogh, A.; Harman, A.; Kreuter, F. Real-Time Analysis of Predictors of COVID-19 Infection Spread in Countries in the European Union Through a New Tool. Int. J. Public Health 2022, 67, 1604974. [Google Scholar] [CrossRef]
- Zhong, H.; Wang, K.; Wang, W. Spatiotemporal pattern recognition and dynamical analysis of COVID-19 in Shanghai, China. J. Theor. Biol. 2022, 554, 111279. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Palm, L.; Lu, X.; Nie, S.; Xu, B.; Zhao, Q.; Tao, T.; Cheng, L.; Tan, L.; Dong, H.; et al. ISS-An Electronic Syndromic Surveillance System for Infectious Disease in Rural China. PLoS ONE 2013, 8, e62749. [Google Scholar] [CrossRef] [PubMed]
- Foraker, R.; Landman, J.; Lackey, I.; Haslam, M.D.; Antes, A.L.; Goldfarb, D. Enabling Hotspot Detection and Public Health Response to the COVID-19 Pandemic. Prev. Chronic Dis. 2022, 19, E35. [Google Scholar] [CrossRef]
- Akpan, G.U.; Bello, I.M.; Touray, K.; Ngofa, R.; Oyaole, D.R.; Maleghemi, S.; Babona, M.; Chikwanda, C.; Poy, A.; Mboussou, F.; et al. Leveraging Polio Geographic Information System Platforms in the African Region for Miti-gating COVID-19 Contact Tracing and Surveillance Challenges: Viewpoint. JMIR Mhealth Uhealth Can. 2022, 10, e22544. [Google Scholar] [CrossRef]
- Boudreault, L.; Hebert-Lavoie, M.; Ung, K.; Mahmoudhi, C.; Vu, Q.P.; Jouvet, P.; Doyon-Poulin, P. Situation Awareness-Oriented Dashboard in ICUs in Support of Resource Management in Time of Pandemics. IEEE J. Transl. Eng. Health Med. 2023, 11, 151–160. [Google Scholar] [CrossRef]
- Ulahannan, J.P.; Narayanan, N.; Thalhath, N.; Prabhakaran, P.; Chaliyeduth, S.; Suresh, S.P.; Mohammed, M.; Rajeevan, E.; Joseph, S.; Balakrishnan, A.; et al. A citizen science initiative for open data and visualization of COVID-19 outbreak in Kerala, India. J. Am. Med. Inform. Assoc. 2020, 27, 1913–1920. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.M.; Boyles, A.; Shankar, A.; Kim, J.; Knyazev, S.; Cintron, R.; Switzer, W.M. MicrobeTrace: Retooling molecular epidemiology for rapid public health response. PLoS Comput. Biol. 2021, 17, e1009300. [Google Scholar] [CrossRef]
- Burkom, H.; Loschen, W.; Wojcik, R.; Holtry, R.; Punjabi, M.; Siwek, M.; Lewis, S. Electronic Surveillance System for the Early Notification of Community-Based Epidemics (ESSENCE): Overview, Components, and Public Health Applications. JMIR Public Health Surveill. 2021, 7, e26303. [Google Scholar] [CrossRef]
- Almeida, J.S.; Shiels, M.; Bhawsar, P.; Patel, B.; Nemeth, E.; Moffitt, R.; Closas, M.G.; Freedman, N.; Berrington, A. Mortality Tracker: The COVID-19 case for real time web APIs as epidemiology commons. Bioinformatics 2021, 37, 2073–2074. [Google Scholar] [CrossRef] [PubMed]
- Mirza, M.; Grant-Greene, Y.; Valles, M.P.; Joseph, P.; Juin, S.; Brice, S.; Dely, P.; Clement, M.G.; Kumar, M.; Silver, M.; et al. Leveraging PEPFAR-Supported Health Information Systems for COVID-19 Pandemic Response. Emerg. Infect. Dis. 2022, 28, S49–S58. [Google Scholar] [CrossRef]
- Baxter, L.; Baynes, J.; Weaver, A.; Neale, A.; Wade, T.; Mehaffey, M.; Lobdell, D.; Widener, K.; Cascio, W. Development of the United States Environmental Protection Agency’s Facilities Status Dashboard for the COVID-19 Pandemic: Approach and Challenges. Int. J. Public Health 2022, 67, 1604761. [Google Scholar] [CrossRef]
- Massri, M.B.; Costa, J.P.; Bauer, A.; Grobelnik, M.; Brank, J.; Stopar, L. A global COVID-19 observatory, monitoring the pandemics through text mining and visualization. Informatica 2022, 46, 49–55. [Google Scholar] [CrossRef]
- Arneson, D.; Elliott, M.; Mosenia, A.; Oskotsky, B.; Solodar, S.; Vashisht, R.; Zack, T.; Bleicher, P.; Butte, A.J.; Rudrapatna, V.A. COVID-Counties is an interactive real time tracker of the COVID19 pandemic at the level of US counties. Sci. Data 2020, 7, 405. [Google Scholar] [CrossRef]
- Tebé, C.; Valls, J.; Satorra, P.; Tobías, A. COVID19-world: A shiny application to perform comprehensive country-specific data visualization for SARS-CoV-2 epidemic. BMC Med. Res. Methodol. 2020, 20, 235. [Google Scholar] [CrossRef] [PubMed]
- Valls, J.; Tobías, A.; Satorra, P.; Tebé, C. COVID19-Tracker: una aplicación Shiny para analizar datos de la epidemia de SARS-CoV-2 en España. Gac. Sanit. 2021, 35, 99–101. [Google Scholar] [CrossRef]
- Salehi, M.; Arashi, M.; Bekker, A.; Ferreira, J.; Chen, D.-G.; Esmaeili, F.; Frances, M. A Synergetic R-Shiny Portal for Modeling and Tracking of COVID-19 Data. Front. Public Health 2021, 8, 623624. [Google Scholar] [CrossRef]
- Martínez Beltrán, E.T.; Quiles Pérez, M.; Pastor-Galindo, J.; Nespoli, P.; García Clemente, F.J.; Gómez Mármol, F. COnVIDa: COVID-19 multidisciplinary data collection and dashboard. J. Biomed. Inf. 2021, 117, 103760. [Google Scholar] [CrossRef]
- Silenou, B.C.; Verset, C.; Kaburi, B.B.; Leuci, O.; Ghozzi, S.; Duboudin, C.; Krause, G. A Novel Tool for Real-time Estimation of Epide-miological Parameters of Communicable Diseases Using Contact-Tracing Data: Development and Deployment. JMIR Public Health Surveill. Can. 2022, 8, e34438. [Google Scholar] [CrossRef]
- Chaudhuri, S.; Giménez-Adsuar, G.; Saez, M.; Barceló, M.A. PandemonCAT: Monitoring the COVID-19 Pandemic in Catalonia, Spain. Int. J. Env. Res. Public Health Switz. 2022, 19, 4783. [Google Scholar] [CrossRef]
- Lee, M.; Kim, J.W.; Jang, B. DOVE: An Infectious Disease Outbreak Statistics Visualization System. IEEE Access 2018, 6, 47206–47216. [Google Scholar] [CrossRef]
- Jang, B.; Lee, M.; Kim, J.W. PEACOCK: A Map-Based Multitype Infectious Disease Outbreak Information System. IEEE Access 2019, 7, 82956–82969. [Google Scholar] [CrossRef]
- Tobías, A.; Satorra, P.; Valls, J.; Tebé, C. COVID19-Global: A Shiny Application to Perform a Global Comparative Data Visualization for the SARS-CoV-2 Epidemic; medRxiv Cold Spring Harbor Laboratory Press: New York, NY, USA, 2020. [Google Scholar] [CrossRef]
- Field, E.; Dyda, A.; Hewett, M.; Weng, H.; Shi, J.; Curtis, S.; Law, C.; McHugh, L.; Sheel, M.; Moore, J.; et al. Development of the COVID-19 Real-Time Information System for Preparedness and Epidemic Response (CRISPER), Australia. Front. Public Health 2021, 9, 753493. [Google Scholar] [CrossRef]
- Kırbıyık, U.; Binder, A.M.; Ghinai, I.; Zawitz, C.; Levin, R.; Samala, U.; Smith, M.B.; Gubser, J.; Jones, B.; Varela, K.; et al. Network Characteristics and Visualization of COVID-19 Outbreak in a Large Detention Facility in the United States—Cook County, Illinois, 2020. MMWR. Morb. Mortal. Wkly. Rep. 2020, 69, 1625–1630. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Mioc, D.; Anton, F.; Yi, X.; Coleman, D.J. Online GIS services for mapping and sharing disease information. Int. J. Health Geogr. 2008, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Myall, A.C.; Peach, R.L.; Weiße, A.Y.; Davies, F.; Mookerjee, S.; Holmes, A.; Barahona, M. Network memory in the movement of hospital patients carrying drug-resistant bacteria. arXiv 2020, arXiv:2009.14480. [Google Scholar]
- Chintala, S.; Dutta, R.; Tadmor, D. COVID-19 spatiotemporal research with workflow-based data analysis. Infect. Genet. Evol. 2021, 88, 104701. [Google Scholar] [CrossRef]
- Ali, M.; Ahsan, Z.; Amin, M.; Latif, S.; Ayyaz, A.; Ayyaz, M. ID-Viewer: A visual analytics architecture for infectious diseases surveillance and response management in Pakistan. Public Health 2016, 134, 72–85. [Google Scholar] [CrossRef]
- Ponjavić, M.; Karabegović, A.; Ferhatbegović, E.; Tahirović, E.; Uzunović, S.; Travar, M.; Pilav, A.; Mulić, M.; Karakaš, S.; Avdić, N.; et al. Spatio-temporal data visualization for monitoring of control measures in the prevention of the spread of COVID-19 in Bosnia and Herzegovina. Med. Glas. (Zenica) Bosnia Herzeg. 2020, 17, 265–274. [Google Scholar]
- Livnat, Y.; Rhyne, T.; Samore, M. Epinome: A Visual-Analytics Workbench for Epidemiology Data. IEEE Comput. Graph. Appl. 2012, 32, 89–95. [Google Scholar] [CrossRef]
- Ramírez-Ramírez, L.L.; Gel, Y.R.; Thompson, M.; de Villa, E.; McPherson, M. A new surveillance and spatio-temporal visualization tool SIMID: SIMulation of Infectious Diseases using random networks and GIS. Comput. Methods Programs Biomed. 2013, 110, 455–470. [Google Scholar] [CrossRef]
- Niu, B.; Liang, R.; Zhang, S.; Zhang, H.; Qu, X.; Su, Q.; Zheng, L.; Chen, Q. Epidemic analysis of COVID-19 in Italy based on spatio-temporal geographic information and Google Trends. Transbound. Emerg. Dis. 2020, 68, 2384–2400. [Google Scholar] [CrossRef]
- Dao, T.P.; Hoang, X.H.T.; Nguyen, D.N.; Huynh, N.Q.; Pham, T.T.; Nguyen, D.T.; Nguyen, H.B.; Do, N.H.; Nguyen, H.V.; Dao, C.H.; et al. A geospatial platform to support visualization, analysis, and prediction of tuberculosis notification in space and time. Front. Public Health 2022, 10, 973362. [Google Scholar] [CrossRef]
- Zambrano, L.I.; Rodriguez, E.; Espinoza-Salvado, I.A.; Fuentes-Barahona, I.C.; Lyra de Oliveira, T.; Luciano da Veiga, G.; Cláudio da Silva, J.; Valle-Reconco, J.A.; Rodríguez-Morales, A.J. Spatial distribution of dengue in Honduras during 2016–2019 using a geo-graphic information systems (GIS)–Dengue epidemic implications for public health and travel medicine. Travel Med. Infect. Dis. 2019, 32, 101517. [Google Scholar] [CrossRef]
- Maciejewski, R.; Livengood, P.; Rudolph, S.; Collins, T.F.; Ebert, D.S.; Brigantic, R.T.; Corley, C.D.; Muller, G.A.; Sanders, S.W. A pandemic influenza modeling and visualization tool. J. Vis. Lang. Comput. 2011, 22, 268–278. [Google Scholar] [CrossRef]
- Luo, W. Visual analytics of geo-social interaction patterns for epidemic control. Int. J. Health Geogr. 2016, 15, 28. [Google Scholar] [CrossRef]
- Weiser, A.A.; Thöns, C.; Filter, M.; Falenski, A.; Appel, B.; Käsbohrer, A. FoodChain-Lab: A Trace-Back and Trace-Forward Tool De-veloped and Applied during Food-Borne Disease Outbreak Investigations in Germany and Europe. PLoS ONE 2016, 11, e0151977. [Google Scholar] [CrossRef]
- Ma, W. Data Analysis and Visualization of COVID-19 Epidemic based on Python. Int. J. Innov. Sci. Res. Technol. 2021, 6, 504–509. [Google Scholar]
- Yu, C.-S.; Chang, S.-S.; Chang, T.-H.; Wu, J.L.; Lin, Y.-J.; Chien, H.-F.; Chen, R.-J. A COVID-19 Pandemic Artificial Intelligence-Based System With Deep Learning Forecasting and Automatic Statistical Data Acquisition: Development and Implementation Study. J. Med. Internet Res. 2021, 23, e27806. [Google Scholar] [CrossRef] [PubMed]
- Qiu, T.; Xiao, H.; Brusic, V. Estimating the Effects of Public Health Measures by SEIR(MH) Model of COVID-19 Epidemic in Local Geographic Areas. Front. Public Health 2022, 9, 728525. [Google Scholar] [CrossRef]
- Yadav, C.P.; Sharma, A. National Institute of Malaria Research-Malaria Dashboard (NIMR-MDB): A digital platform for analysis and visualization of epidemiological data. Lancet Reg. Health-Southeast. Asia 2022, 5, 100030. [Google Scholar] [CrossRef]
- Aigner, W.; Miksch, S.; Schumann, H.; Tominski, C. Visualization of Time-Oriented Data; Springer: London, UK, 2011; ISBN 978-0-85729-078-6. [Google Scholar] [CrossRef]
- Isenberg, T.; Isenberg, P.; Chen, J.; Sedlmair, M.; Moller, T. A Systematic Review on the Practice of Evaluating Visualization. IEEE Trans. Vis. Comput. Graph. 2013, 19, 2818–2827. [Google Scholar] [CrossRef]
- Baker, M. Is There a Reproducibility Crisis? Nature 2016, 533, 452–454. [Google Scholar] [CrossRef]
- KCDC. Available online: http://www.cdc.go.kr (accessed on 9 November 2022).
- Instituto de Informática UFRGS. COVID-19 Analysis Tools. Available online: https://covid19.ufrgs.dev/ (accessed on 9 November 2022).
- COVID-19 Data Repository by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University. 2022. Available online: https://github.com/CSSEGISandData/COVID-19 (accessed on 9 November 2022).
- NSF Spatiotemporal Innovation Center. COVID-19 in the US Dataset. Available online: https://github.com/stccenter/COVID-19-Data/tree/master/US (accessed on 9 November 2022).
- Moraga, P.; Dorigatti, I.; Kamvar, Z.N. Dataset 1 in: Epiflows: An R package for risk assessment of travel-related spread of disease. F1000Research 2018, 7, 1374. [Google Scholar] [CrossRef]
- Moraga, P.; Dorigatti, I.; Kamvar, Z.N. Dataset 2 in: Epiflows: An R package for risk assessment of travel-related spread of disease. F1000Research 2018, 7, 1374. [Google Scholar] [CrossRef]
- Flutrack. Available online: https://github.com/flutrack/Flutrack.org_webapp_source_code (accessed on 9 November 2022).
- China National Health Commission. Notification on the Pneumonia Epidemic Situation. Available online: http://www.nhc.gov.cn/xcs/yqtb/list_gzbd.shtml (accessed on 9 November 2022).
- Hubei Province Health Commission. Press Statement. Available online: http://wjw.hubei.gov.cn/bmdt/ztzl/fkxxgzbdgrfyyq/xxfb/ (accessed on 9 November 2022).
- Ministry of Health, Labour and Welfare, Japan. Press Statement of Novel Coronavirus. Available online: https://www.mhlw.go.jp/stf/newpage_08906.html (accessed on 9 November 2022). (In Japanese)
- Centers for Disease Control and Prevention (CDC). 2020. Available online: https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/index.html (accessed on 9 November 2022).
- Chao, D.L.; Halloran, M.E.; Longini, I.M., Jr. School Opening Dates Predict Pandemic Influenza A(H1N1) Outbreaks in the United States. J. Infect. Dis. 2010, 202, 877–880. [Google Scholar] [CrossRef]
- United States Census Bureau. U.S. Gazetteer Files. 2015. Available online: https://www.census.gov/geographies/reference-files/time-series/geo/gazetteer-files.html (accessed on 9 November 2022).
- Government of Canada SC. Statistics Canada and Census Datasets. 2015. Available online: https://www12.statcan.gc.ca/datasets/Index-eng.cfm (accessed on 9 November 2022).
- District Health Information System. Available online: http://www.dhiskp.gov.pk/ (accessed on 9 November 2022).
- Taiwan Open Data Platform. Available online: https://data.gov.tw/en (accessed on 9 November 2022).
- Taiwan Centers for Disease Control. Available online: https://www.cdc.gov.tw/En (accessed on 9 November 2022).
- Primary School–Cumulative Networks. Available online: http://www.sociopatterns.org/datasets/primary-school-cumulative-networks/ (accessed on 9 November 2022).
- FoodRisk-Labs. Available online: https://foodrisklabs.bfr.bund.de/foodrisk-labs/ (accessed on 9 November 2022).
- Ministry of Civil Affairs of Bosnia and Herzegovina. Epidemic Situation. Available online: http://mcp.gov.ba/publication/read/epidemioloska-slika-covid-19?pageId=3 (accessed on 9 November 2022).
- The New York Times. Coronavirus (COVID-19) Data in the United States. 2022. Available online: https://github.com/nytimes/covid-19-data (accessed on 9 November 2022).
- National Governments Health Secretary of Mexico. Datos Abiertos Dirección General de Epidemiología. 2020. Available online: https://www.gob.mx/salud/documentos/datos-abiertos-152127 (accessed on 9 November 2022).
- National Epidemiological Surveillance of Infectious Diseases. Available online: https://www.niid.go.jp/niid/en/ (accessed on 9 November 2022).
- European Centre for Disease Prevention and Control. Available online: https://www.ecdc.europa.eu/en (accessed on 9 November 2022).
- WHO. Coronavirus Disease (COVID-19) Situation Reports. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (accessed on 9 November 2022).
- Chinese Center for Disease Control and Prevention. Available online: https://www.chinacdc.cn/en/ (accessed on 9 November 2022).
- National Health Commission of the PRC. Available online: http://en.nhc.gov.cn/ (accessed on 9 November 2022).
- COVID-19. Datadista. Available online: https://github.com/datadista/datasets (accessed on 9 November 2022).
- USAFacts. US COVID-19 Cases and Deaths by State. 2020. Available online: https://usafacts.org/visualizations/coronavirus-covid-19-spread-map (accessed on 9 November 2022).
- EpiCentro. Coronavirus. Istituto Superiore di Sanità. Available online: https://www.epicentro.iss.it/coronavirus/ (accessed on 9 November 2022).
- Comparative Health System Performance (CHSP) Initiative. Agency for Healthcare Research and Quality. 2018. Available online: https://www.ahrq.gov/chsp/index.html (accessed on 9 November 2022).
- Directorate of Health Services; Government of Kerala, India. Available online: https://dhs.kerala.gov.in/ (accessed on 9 November 2022).
- NSF Spatiotemporal Innovation Center. COVID-19 Dataset. Available online: https://github.com/stccenter (accessed on 9 November 2022).
- Dudas, G.; Carvalho, L.M.; Bedford, T.; Tatem, A.J.; Baele, G.; Faria, N.R.; Park, D.J.; Ladner, J.T.; Arias, A.; Asogun, D.; et al. Virus genomes reveal factors that spread and sustained the Ebola epidemic. Nature 2017, 544, 309–315. [Google Scholar] [CrossRef]
- The COVID Tracking Project. Available online: https://covidtracking.com/data/api (accessed on 9 November 2022).
- Presidenza del Consiglio dei Ministri-Dipartimento della Protezione Civile. Dati COVID-19 Italia. 2022. Available online: https://github.com/pcm-dpc/COVID-19 (accessed on 9 November 2022).
- Tencent. Available online: https://www.tencent.com/en-us/responsibility/combat-covid-19.html (accessed on 9 November 2022).
- International Civil Aviation Organization. Infectious Disease App. Available online: http://quips.anbdata.com/project/dev/5c1c21b205c09f70bfe60eeeeb46316af89506e9.html (accessed on 9 November 2022).
- Escovid19data. 2022. Available online: https://github.com/montera34/escovid19data (accessed on 9 November 2022).
- Instituto Nacional de Estadística (INE). Available online: http://www.ine.es/ (accessed on 9 November 2022).
- Instituto de Salud Carlos III (ISCIII). Available online: https://www.isciii.es/Paginas/Inicio.aspx (accessed on 9 November 2022).
- Agencia Estatal de Meteorología (AEMET); Gobierno de España. Available online: http://www.aemet.es/es/portada (accessed on 9 November 2022).
- Yousef, K.P.; Meixenberger, K.; Smith, M.R.; Somogyi, S.; Gromöller, S.; Schmidt, D.; Gunsenheimer-Bartmeyer, B.; Hamouda, O.; Kücherer, C.; von Kleist, M. Inferring HIV-1 Transmission Dynamics in Germany From Recently Transmitted Viruses. AIDS J. Acquir. Immune Defic. Syndr. 2016, 73, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Oxford COVID-19 Government Response Tracker. Available online: https://github.com/OxCGRT (accessed on 9 November 2022).
- The NSW Government. NSW COVID-19 Data. Available online: https://data.nsw.gov.au/nsw-covid-19-data (accessed on 9 November 2022).
- Victorian COVID-19 Data|Coronavirus Victoria. Available online: https://www.coronavirus.vic.gov.au/victorian-coronavirus-covid-19-data (accessed on 9 November 2022).
- COVID-19: Locations Visited by Confirmed Cases. Available online: https://www.wa.gov.au/government/covid-19-coronavirus/covid-19-coronavirus-locations-visited-confirmed-cases (accessed on 9 November 2022).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
























