Abstract
The intricate neuroinflammatory diseases multiple sclerosis (MS) and neuromyelitis optica (NMO) often present similar clinical symptoms, creating challenges in their precise detection via magnetic resonance imaging (MRI). This challenge is further compounded when detecting the active and inactive states of MS. To address this diagnostic problem, we introduce an innovative framework that incorporates state-of-the-art machine learning algorithms applied to features culled from MRI scans by pre-trained deep learning models, VGG-NET and InceptionV3. To develop and test this methodology, we utilized a robust dataset obtained from the King Abdullah University Hospital in Jordan, encompassing cases diagnosed with both MS and NMO. We benchmarked thirteen distinct machine learning algorithms and discovered that support vector machine (SVM) and K-nearest neighbor (KNN) algorithms performed superiorly in our context. Our results demonstrated KNN’s exceptional performance in differentiating between MS and NMO, with precision, recall, F1-score, and accuracy values of 0.98, 0.99, 0.99, and 0.99, respectively, using leveraging features extracted from VGG16. In contrast, SVM excelled in classifying active versus inactive states of MS, achieving precision, recall, F1-score, and accuracy values of 0.99, 0.97, 0.98, and 0.98, respectively, using leveraging features extracted from VGG16 and VGG19. Our advanced methodology outshines previous studies, providing clinicians with a highly accurate, efficient tool for diagnosing these diseases. The immediate implication of our research is the potential to streamline treatment processes, thereby delivering timely, appropriate care to patients suffering from these complex diseases.
1. Introduction
Multiple sclerosis is an inflammatory and autoimmune disorder that attacks the myelinated axons in the central nervous system (CNS) [1]. As a result, it destroys the myelin in multiple severities and results in multiple clinical features [2]. A combination of signs and symptoms, magnetic resonance imaging findings, and laboratory findings lead to the diagnosis of MS [3,4]. However, many diseases mimic the clinical and imaging findings of MS, such as neuromyelitis optica (NMO), neoplasm, and acute disseminated encephalomyelitis. Therefore, this mimicking can lead to misdiagnosis [5]. Most MS over-diagnosed cases are associated with nonspecific white matter abnormalities on brain MRIs and misinterpreting radiographic diagnostic criteria [6]. For example, inexperienced neurologists may easily give a false-positive result of MS for people with nonspecific neurological symptoms such as diplopia weakness or dizziness combined with white spots that are suggested related to MS [7].
One of the demyelination diseases closely related to MS is NMO; some authors have considered NMO to be a particular form of MS [8]. NMO mainly affects the optic nerve and spinal cord. Also, within five years, half of patients cannot walk independently or lose their functional vision in one eye at least [8]. Differentiating between these two demyelinating disorders was challenging in several studies [9]. J de Seze compared NMO patients with MS patients presenting with optic neuritis or myelopathy. They compared 30 consecutive cases of MS with 50 consecutive cases of optic neuritis or acute myelopathy to determine the place of NMO in the MS spectrum. Patients with MS were only included if they had a relapse, showing time and space dissemination. A comparison of clinical presentation, laboratory findings (MRI, CSF), and clinical outcomes was conducted. Neuroimaging investigations showed abnormal brain MRIs in 10% of NMOs and 66% of MSs, as well as a large spinal cord lesion in 67% of NMOs and 7.4% of MSs. Finally, this study illustrated that MS and NMO should be considered as different disorders [10].
Moreover, it is reported that NMO sometimes reveals asymptomatic lesions on brain MRI at onset, when when the patient presents with symptomatic brain involvement. To correctly diagnose MS, NMO should be excluded before diagnosis because of the imperfect sensitivity of AQP4-ab assays and the presence of brain lesions in NMO [7]. Therefore, a study was conducted and supposed that it is difficult to differentiate MS from NMO by considering the MRI criteria for MS on brain MRI at onset [11].
Many patients suffering from initial symptoms of NMO are misdiagnosed with multiple sclerosis [6]. It is essential to differentiate between these two disorders to obtain optimum treatments since both treatments completely differ. Moreover, this is useful to prevent the increased mortality and morbidity rates for these disorders (i.e., MS and NMO). Immunosuppressive drugs are the best treatment for NMO [8]. However, immunomodulatory treatments are highly recommended for the optimum treatment of multiple sclerosis [12]. On one hand, it is significant to obtain a precise diagnosis of MS and NMO; on the other hand, it is challenging to achieve a precise diagnosis. Therefore, it is necessary to improve the para-clinical method to make the diagnosis more accurate. Moreover, due to the influence of laboratory tests by assay methodology and the patient’s clinical status, we should shed light on MRI, considered the gold standard test for CNS demyelination disease, depending on morphological characteristics, and improve this method by developing an algorithm. This growing method processes the imaging data so that neurologists can differentiate the demyelinating disorder [10].
An active lesion (defined as an acute inflammatory process or the demyelination of nerve cells) appears as a white spot on MRI imaging, while an inactive lesion is the remyelination that appears on the MR image as plaque shadow [13]; detection in MS is crucial to inspect the spatial and temporal distribution of this white spot that represents the active lesion. As a result, the disease can be diagnosed after the onset of symptoms [14]. As part of the criteria for the international panel, dissemination is included [11]. In order to use the active lesion as an indicator of outcome, it is vital to follow it closely. As a result, we can test the effectiveness of the current treatment and explore several new therapeutic agents as well as the prognosis of the condition [15]; the number of MRI errors, time-consuming manual inspections, and physical effort have led to the development of artificial-intelligence-based imaging [16]. The method relies on morphological and textural characteristics. Therefore, we should improve the robust mathematical models for a more precise diagnosis and new lesions.
2. Related Work
Prior research has demonstrated the capacity of machine learning (ML) and deep learning (DL) techniques to accurately classify multiple sclerosis (MS) and identify active lesions in MS patients using both MRI images and clinical data. Buyukturkoglu et al. formulated a predictive framework for evaluating the white matter status in early onset multiple sclerosis [16]. The study employed 183 magnetic resonance imaging (MRI) images and stratified patients into two distinct groups based on their demographic and clinical traits, lesion size and localization, local and global tissue volume, local and global diffusion tensor imaging, and whole-brain resting-state functional connectivity measures. Utilizing random forest with pertinent features yielded the highest area under the curve (AUC) value of 90%, signifying remarkable predictive accuracy. Ramanujam et al. [17] gathered clinical data from 14387 individuals with multiple sclerosis in Sweden, encompassing vital demographic and clinical information. Employing a decision tree algorithm, the team attained exceptional accuracies averaged at 89.5% (89.1–90.1).
The outcomes demonstrated favorable results compared to medical professionals’ precision, and three neurologists validated the results with an accuracy of 84.3%. In the context of multiple sclerosis, Loizou et al. [18] applied a machine learning framework using a support vector machine (SVM) algorithm to identify white matter regions in brain T2-weighted MRI images of patients who underwent brain MRI at different intervals. Employing segmentation, the algorithm extracted crucial features of MS lesions in the images and subsequently trained and tested using SVM to distinguish between NAWM0 vs. ROISC0, NAWM0 vs NAWM6-12, NAWM0 vs. L0, NAWM6-12 vs. L6-12, ROIS0 vs. L0, ROIS0 vs L6-12, and ROIS0 vs ROISC0, achieving corresponding precise classification scores of 89%, 95%, 98%, 92%, 85%, 90%, and 65%, respectively, by using clinical data based on magnetic resonance images. Saccà et al. [19], employing five distinct machine learning algorithms, namely, artificial neural network, support vector machine, K-nearest-neighbor, naïve Bayes, and random forest, endeavored to distinguish between patients with multiple sclerosis and those under the control.
The random forest and SVM algorithms performed most effectively, yielding an accuracy of 85.7%. Samah et al. [20] proposed a novel method for identifying multiple sclerosis lesions in the brain through MRI images using decimal descriptor patterns (DDP). DDP leverages a 3D Brainweb dataset to analyze MRI images, and SVM is employed to classify them. With T1, DDP achieved a success rate of 97.12%, while with T2, it was 94.21%. To evaluate white lesions in MS by utilizing information from MRI images, Maggi et al. [21] devised a 3D convolutional neural network (CVSnet) based on the central vein sign (CSV), an imaging-active biomarker for MS identification. Trained on 47 datasets and 33 tests, CVSnet demonstrated a 91% accuracy in identifying lesion white matter in the brain through MR images. Roy et al. [22] proposed an advanced method utilizing a convolutional neural network (CNN) architecture comprising two distinct routes. The first route incorporated parallel convolutional filter banks that supported magnetic resonance techniques. Meanwhile, the second route eventually employed additional convolutional filter outputs to produce binary segmentation. A dataset consisting of 100 patients who had multiple sclerosis was utilized to perform segmentation and isolate lesions suggestive of MS illness.
In addition, the proposed method was evaluated using the ISBI 2015 challenge data, which involved 14 patients, and attained an impressive score of 90.48. Sepahvand et al. [23] used the CNN model to classify multiple sclerosis and neuromyelitis spectrum disorders. Due to their limited numbers, we used SqueezeNet to extract features from the data of 35 patients with MS and 18 patients with NMOSD to prevent overfitting. The model’s accuracy was 81.1%, and its sensitivities to NMOSD and MS were 83.3% and 80.0%, respectively. Eshaghi et al. [24] used a random forest algorithm to classify between MS and NMO through brain gray matter imaging. The random forest model was evaluated on ninety patients on clinical data, wherein the model was classified between MS vs. HCs and NMO vs. HCs, where the accuracy between MS vs. HCs was 92% and 88% between NMO vs. HCs. Zhang et al. [25] devised a neural network architecture, QSMRim-Net, to accurately identify active lesions in patients with multiple sclerosis by analyzing MRI images. The model was trained on a dataset of 172 MRI images, and the relevant features were extracted from the images after two trained experts identified the active lesions. The model’s performance was evaluated using the pROC AUC metric, and it achieved an impressive score of 0.760. Coronado et al. [26] utilized a state-of-the-art CNN architecture, the U5 model, to detect active lesions in patients with multiple sclerosis by analyzing MRI images. The model was trained on a dataset of 1006 MRI images, and it achieved remarkable DSC/TPR/FPR scores of 0.77/0.90/0.23, correspondingly, for various improved lesions.
Gaj et al. [27] introduced an automated 2D-UNet augmented lesion segmentation method to accurately identify active lesions in patients with multiple sclerosis by analyzing MRI images. The model was trained on a dataset of 600 MRI images consisting of T1 post-contrast, T1 pre-contrast, FLAIR, T2, and proton-density images. The model’s performance was evaluated using the random forest classifier, and it achieved an impressive accuracy of 87.7%. Narayana et al. [28] proposed a convolutional neural network architecture, based on the pre-trained VGG16 model, to accurately distinguish between enhanced and unenhanced lesions in patients with multiple sclerosis by analyzing MRI images. The model was trained on a dataset of 1008 MRI images, and it achieved high accuracy scores of AUCs 0.75 ± 0.03 and 0.82 ± 0.02 for participant-wise enhancement prediction and slice-wise analysis, respectively. Sepahvand et al. [23] developed an advanced 3D U-Net model to accurately detect T2w active lesions in patients with multiple sclerosis by analyzing MRI images. The model was trained on a large dataset of 1677 MRI scans and achieved outstanding performance metrics, including an AUC of 0.95, a sensitivity of 0.69, and a specificity of 0.97 for classifying lesions as active or inactive. Table 1 shows a summary of previous studies. Seok proposed a deep learning model for classification between multiple sclerosis (MS) and neuromyelitis optica spectrum disorder (NMOSD), wherein modified ResNet18 was used, and Grad-CAM was used to clarify white matter lesions in MRI images; in terms of results, the following results were achieved: accuracy of 76.1%, sensitivity of 77.3%, and specificity of 74.8%. One of the limitations is that it was satisfied with only one model and did not compare it with modern models, and this is not sufficient to verify the results [29]. Ciftci worked on detecting multiple sclerosis through optical coherence tomography (OCT) features in children through machine learning algorithms, wherein the study was evaluated on a dataset of 512 eyes from 187 children, where the random forest model outperformed other models with a detection accuracy of 80% for MS and 75% for demyelinating diseases [30]. Kenny did so on separating optic neuritis and people with multiple sclerosis. The data was divided into 2/3 for training, and 1/3 for testing, and the model was evaluated using SVM. Among 1568 PwMS and 552 controls, variable selection models identified GCIPL IED, average GCIPL thickness (both eyes), and binocular 2.5% LCLA as most important for classifying PwMS vs. controls [31]. This composite score performed best: the area under the curve (AUC) = 0.89 (95% CI 0.85–0.93), sensitivity = 81%, and specificity = 80%. Carcia included 79 patients recently diagnosed with RRMS with average features, and the difference between the eye diff was obtained in four retinal layers. Avg features plus recursive feature elimination with cross-validation (SVM-RFE-LOOCV) technique were used to identify OCT features for early diagnosis. Sensitivity = 0.86 and specificity = 0.90 were obtained [32].

Table 1.
Summary of previous studies related to MS and NMO.
It is imperative to distinguish between MS and NMO in order to give the patient the appropriate medication. Otherwise, there could be catastrophic complications [8]. Detection of active lesions helps avoid using contrast media before the MRI, which is sometimes considered poisonous or allergic. Because MS and NMO lesions have very similar shapes and are in the same brain region, the exciting approach to diagnosis relies on the interpretation of MRIs by a highly skilled physician. Also, a lab test is needed to support the diagnosis, which sometimes retrieves normal results. Existing methods are time consuming and require much effort and knowledge. Therefore, finding an objective method to leverage in deciding the precise diagnosis is crucial, such as an algorithmic method that provides an instant result. The previous cutting-edge methods of deep learning and machine learning retrieved satisfactory results. However, we suppose a hybrid model in order to provide more accuracy, which can be highly recommended in health care institutions.
Also, based on previous studies, we noticed a lack of studies on the classification between active and inactive in multiple sclerosis. Moreover, there is a weakness in the performance of the approaches used in previous studies. Our work aims to achieve the best possible performance in addition to conducting many experiments with different models. This research will be a reference for many researchers to benefit from these experiences. We note that the special dataset used in this work is real patient data, which can contribute to the assistance of many upcoming studies on the same topic.
3. Methodology
In this study, we strived to develop an accurate framework for classifying MS and MS_NMO datasets and classifying MS into active and inactive states. This is accomplished by extracting and preparing features from the dataset for classification. Three pre-trained deep learning models were used to extract features from the images: the Visual Geometry Group (VGG16, VGG19) and GoogLeNet (Inception V3). These models (given in Appendix A) are deep-learning convolution neural networks that use small convolutional filters to classify and recognize images accurately. Afterward, principal component analysis was used to reduce the dimensionality of the features to a reasonable number. Finally, the data were normalized into a specified range to be ready for classification.
For the classification of MS and MS_NMO datasets as well as active and inactive MS, we applied different machine learning algorithms, namely, logistic regression, KNN, SVM, naïve Bayes, DT, perceptron, stochastic gradient descent, RF, extra trees classifier, gradient boost, Ada boost, Ridge regression, and LGBM. KNN achieved the best result among all the other ML algorithms when applied to MS and MS_NMO datasets. It also achieved the best results when applied to MS active and inactive datasets using the Inception V3 feature extraction model. At the same time, SVM provides better results than all other algorithms for this type of classification using VGG16 and VGG19. Furthermore, we developed and evaluated a model without common feature selection to demonstrate the effects of common feature selection. As a result, we found that the feature selection methods we adopted positively affected the classification of the datasets with the highest performance. KNN with the value of k = 3 was considered the best ML algorithm for classifying MS and MS_NMO datasets, wherein SVM can distinguish between active and inactive MS under the VGG feature selection model. Figure 1 shows the proposed model for our work.
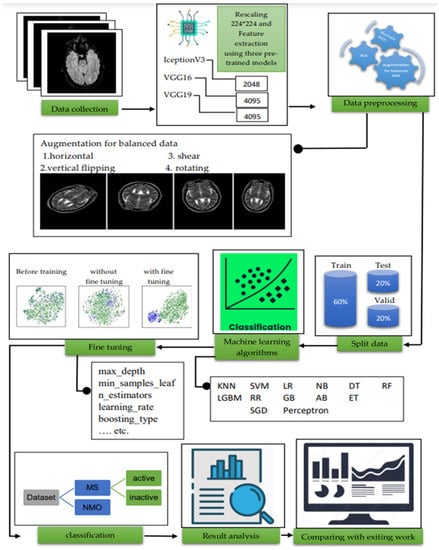
Figure 1.
The proposed methodology.
3.1. Data Acquisition
3.1.1. Participants
Our study involved retrospectively reviewing the medical records of MS and NMO patients who visited King Abdullah University Hospital from January 2020 to February 2023. Patients with MS who met the 2010 McDonald criteria [34] and those with NMO who had AQP4 immunoglobulin G (AQP4-IgG) seropositivity, according to the 2015 International Consensus on NMO [35], were included. We excluded patients whose brain MRI data were not available, as well as patients with clinically isolated syndromes suspected of having MS. Patients with MS ranged in age from 16 to 65, with an average age of 45. Conversely, the ages of patients with NMO ranged from 14 to 49, with AVG reaching 30. For all MRI images included, the number of females was twice that of males, and for NMO, males made up one-fifth, without a significant difference between the two diseases. Nevertheless, the mean age for patients with MS was 45, and for those with NMO was 30. There was no significant difference between the two groups. This was like the duration of a disease. For those with NMO, the average disease duration was six years, compared to nine years for those with MS. The following Table 2 illustrates the demographic data.

Table 2.
Demographics of patients.
3.1.2. MRI Acquisition
Pre- and post-contrast sequences were acquired using Philips 1.5 T or Philips 3.0 T scanners at KAUH in 2D FLAIR, T2W, and T1W (axial and sagittal). An MRI protocol with inversion recovery was used here, with the following scanning parameters: inversion time = 2500 MS; slice thickness = 3 mm; slice gap = 1 mm; and several acquisitions = 2, field of view (FOV) = 220 × 200 mm, a 336 × 231 acquisition matrix, time of repetition = 11,000 MS, time of echo = 125 MS, and voxel size = 0.65 mm × 0.82 mm. As was aforementioned, the enhanced lesion is a significant marker of disease progression and checking treatment efficacy. However, small active lesions are very challenging. Hence, the formulas in Figure 2 show the label-flip map generated using Paint 3D for each active lesion.
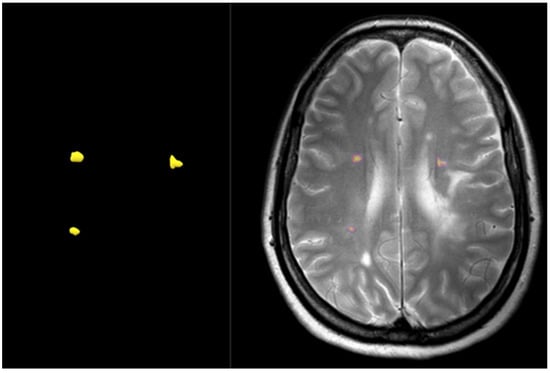
Figure 2.
Example of detecting active lesions in multiple sclerosis based on a map of T2W images.
3.1.3. MS vs. NMO Diagnosis
KAUH has a team of neurologists and neuroradiologists. Additionally, a physiotherapist will test the patient’s physical activity to determine their functional level. All patients are independent in their activities of daily living to achieve homogeneity in all aspects among patients. Using FLAIR and T2W images with and without contrast, they diagnosed MS and NMO with the following clinical information of each patient: age at disease onset, age at MRI, sex, disease duration, number of episodes, the behavior of episodes, duration from last relapse, lumber puncture, and laboratory testing. After that, a classification of these two conditions is conducted. Finally, the total images for MS were 424, and active and inactive MS were 200 and 224 in number, respectively; moreover, the final included images for NMO were 261 in number. Figure 3 shows the distribution of the data used in this research.
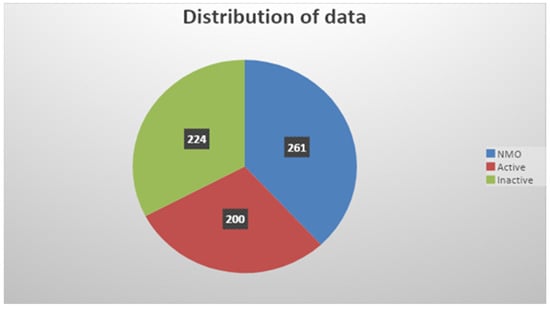
Figure 3.
Distribution of the data.
3.2. Pre-Processing
The following section outlines the crucial pre-processing steps to prepare our dataset for the machine learning algorithms. Pre-processing is essential in any data analysis process, particularly in machine learning, as the data’s quality and suitability directly influence our models’ performance. These pre-processing measures aim to transform the raw, complex data into a structured and understandable format, ensuring our machine learning algorithms can effectively interpret and learn from it. This involves dealing with missing or noisy data, standardizing or normalizing values, reducing dimensionality, and much more. Refining our dataset through pre-processing, we pave the way for more accurate, reliable, and efficient outcomes from our machine learning processes.
3.2.1. Augmentation
Deep learning models necessitate an extensive volume of high-quality data, and our dataset consisting of MS and NMO contains 424 and 261 MRI images, respectively, with MS segregated into 200 active and 224 inactive images. The three deep learning models extracted notable features and employed a suite of machine learning algorithms for classification purposes. Inadequate data can lead to suboptimal results, compounded by the problem of overfitting. In this regard, augmentation enhanced the number of data instances while achieving balance. Augmentation enabled the expansion of the MS images to 2120, comprising 1000 active and 1120 inactive images, with NMO accounting for 2088 images. Various types of augmentation, including image rescaling, horizontal and vertical flipping, rotating, and shearing, were implemented. Overall, the augmentation approach was proven to be effective for our dataset, enhancing accuracy and promising outcomes. Figure 4 shows some examples of the augmented data of one case.
Figure 4.
Original dataset after applying data augmentation.
3.2.2. Feature Extraction with Deep Learning Models
In this study, the feature extraction step was one of the necessary tasks in image processing and feature extraction, and it is considered the process of extracting information from raw data such as images, videos, or sounds.
Reference [36] is considered one of the most important studies to have presented a survey of techniques, one of the most important studies that has provided a survey of feature extraction techniques extracted recently, which encouraged us to proceed with our approach, which relies mainly on feature extraction.
Orange3 is a popular special tool for data science applications and machine learning algorithms called “Orange Data Mining”. Three pre-trained deep learning models (VGG16, VGG19, InceptionV3) were employed to generate three distinct datasets, extracting features through convolutional and pooling layers. When an image is fed into the network of the proposed models, it undergoes a series of convolutional operations. These convolutions act as filters that slide over the image’s pixels, extracting different patterns, textures, and edges at different scales. The network gradually learns more complex and abstract features as one goes deeper into the layers. The pooling layers subtract the extracted features, reducing spatial dimensions and preserving the underlying information. This hierarchical feature extraction process captures low-level features such as edges and textures and higher-level features such as shapes and object parts, enabling models to represent and differentiate objects within an image effectively. The extracted features are then normalized and passed through fully connected layers for classification. This process allows models to understand and classify images based on the hierarchical features they learned during training. The VGG16 and VGG19 architectures were designed by the Visual Geometry Group in 2014, comprising 16 and 19 layers, respectively, including convolutional and max pooling layers [37]. Due to their pre-training on the ImageNet dataset containing 1000 classes, VGG16 and VGG19 are frequently used for feature extraction in image processing applications [38]. InceptionV3 [39], on the other hand, is a convolutional neural network developed by Google in 2015, with 42 layers and a similar feature extraction mechanism as VGG, but with the addition of a special global average pooling layer that aids in preventing overfitting and reducing the number of parameters. The features extracted from vgg16, vgg19, and inceptionv3 were 4095, 4095, and 2048, respectively.
3.2.3. Principal Component Analysis
In our approach, we harnessed the power of principal component analysis (PCA) [40], a widely adopted statistical method, to simplify and interpret our data while maintaining a significant proportion of the original variance. PCA enabled us to reduce the dimensions in our dataset—extracted from image data—while retaining the essence of the original information. By implementing this technique, we effectively streamlined our dataset, compacting it to a more manageable size of 75 principal components. This dimensionality reduction strategy enhances computational efficiency while preserving the core structure of our data.
3.2.4. Normalization
Data normalization represents a cornerstone technique within the realm of data pre-processing. It scales numerical data into a broader, uniform range, typically between −1 and 1, ensuring that all data points contribute proportionally to the result. This technique is of paramount importance when deploying machine learning algorithms, as it mitigates the disproportionate influence of outliers, thus enhancing the performance and stability of the models. Adopting normalization ensures a balanced data distribution, allowing for a more robust and reliable machine-learning process [41].
3.3. Machine Learning Algorithms
This study used various advanced machine-learning techniques and algorithms to classify MS and MS_NMO datasets, wherein we used twelve machine-learning algorithms, namely, RF, SVM, NB, KNN, LR, DT, extra tress classifier, LGBM, Ada boost, gradient boost, SGD, and RR, as well as a set of hyperparameters including max_depth, n_estimators, learning rate, min_child_weight, boosting type, and min_samples_leaf that have a significant impact on the behavior and performance of the models, wherein through optimization and experimentation, the optimal values of the hyperparameters that provide the best performance are chosen.
4. Result and Discussion
4.1. Performance Measures and Experiments
In this section, the results obtained from thirteen machine learning algorithms that were trained on the features extracted were divided into 60% for training, 20% validation, and 20% testing using collab using a Python tool on a computer with an i5-10H CPU, NVIDIA GeForce GTX 1660Ti 4 GB, 8 GB RAM, 250 GB SSD, and 1 TB HDD from MRI images, presented by the three pre-trained models in order to build a strategy to extract the most effective features and algorithms that are suitable for these features and achieve the best performance. The performance of the algorithms for classifying the MS_NMO dataset is compared in Table 2, and the algorithms are compared for the MS dataset, which contains active and inactive classes, in Table 3. In this research, we relied on the evaluation of our model on the accuracy, precession, recall, and F1-score with a preference for F1-score and accuracy.

Table 3.
Performance for multi-machine learning algorithms using feature extraction based on pre-trained MS and NMO classification models.
- Accuracy: Refers to the correctly expected cases from the sum of all cases as shown in Formula (1):Accuracy = (TP + TN)/(TP + FP + FN + TN) SEQ Equation\* ARABIC
- Precision: Refers to the positive cases that were correctly predicted for all positive cases as shown in Formula (2):Precision = TP/(TP + FP) SEQ Equation\* ARABIC
- Recall: The proportion of predicted true positive cases that were correctly predicted relative to all actual positive cases stated in Formula (3):Recall = TP/(TP + FN) SEQ Equation\* ARABIC
- F1-Score: Balances precision and recall to evaluate overall performance stated in Formula (4):F1-score = 2 × (precision×recall)/(precision + recall) SEQ Equation\* ARABIC
4.2. Result Analysis
The K-nearest neighbor (KNN) algorithm demonstrated superior performance on the MS_NMO dataset when applied to the features extracted from three deep learning models, namely, vgg16, vgg19, and inceptionV3. The results were exceptional, with precision, recall, F1-score, and accuracy of 0.98, 0.99, 0.99, and 0.99 for the vgg16 features, respectively. The vgg19 features also yielded impressive results with precision, recall, F1-score, and accuracy of 0.96, 0.98, 0.97, and 0.97, respectively. However, the inceptionV3 features produced slightly lower results with precision, recall, F1-score, and accuracy of 0.92, 0.95, 0.93, and 0.93, respectively.
In contrast, the support vector machine (SVM) algorithm achieved the best results on the MS dataset with the features extracted from vgg16 and vgg19. For the vgg16 features, the precision, recall, F1-score, and accuracy were 0.99, 0.97, 0.98, and 0.98, respectively, while the vgg19 features resulted in the precision, recall, F1-score, and accuracy of 0.99, 0.98, 0.98, and 0.98, respectively. The KNN algorithm achieved the best results on the inceptionV3 features with precision, recall, F1-score, and accuracy of 0.95, 1.00, 0.98, and 0.98, respectively. Table 2 and Table 3 identified several promising algorithms that produced results above 90, such as random forest (RF), gradient boosting (GB), and light gradient boosting machine (LGBM) on both datasets, and extra tree classifier (ETC) on the MS dataset. However, some algorithms, such as naïve Bayes and Ada boost, consistently produced lower performance across all CNN models, as evidenced by their lower precision, recall, and F1-scores. Therefore, these algorithms may not be well-suited for this particular task.
As shown in Table 3, we conducted a rigorous evaluation of multiple machine learning algorithms using feature extraction based on pre-trained CNN models, and notable patterns and key findings emerged. When features were extracted using VGG16 for MS and NMO classification, the K-nearest neighbor (KNN) and support vector machine (SVM) algorithms demonstrated the most promising performance, with precision, recall, F1-score, and accuracy values consistently exceeding 0.94. This remarkable result suggests the robustness and reliability of KNN and SVM when used with features derived from VGG16.
However, some algorithms did not perform as well. For instance, logistic regression and Naïve bayes achieved lower performance metrics, with their precision, recall, F1-score, and accuracy values barely exceeding 0.70. Despite this, these algorithms still demonstrated reasonable accuracy, highlighting the overall effectiveness of using VGG16 for feature extraction. When transitioning to features extracted using VGG19, we found a similar pattern. The KNN, SVM, random forest (RF), and light gradient boosting machine (LGBM) algorithms displayed the most impressive results, with all performance metrics exceeding 0.90. Yet again, the results indicated the high efficacy of these machine-learning models with features extracted from VGG19.
Finally, for the features extracted using InceptionV3, the top performers were SVM, RF, gradient boost, and LGBM, all of which achieved precision, recall, F1-score, and accuracy values close to or above 0.90. The consistency of these results underscores the versatility and performance of these machine-learning models when used with features derived from different pre-trained CNN models. Our findings revealed a consistently strong performance by KNN, SVM, RF, and LGBM across all feature sets, highlighting their potential for accurate and reliable classification in this context. These insights lay a strong foundation for future research and the development of machine-learning models for classifying complex neuroinflammatory diseases such as MS and NMO.
Table 4 showcases the performance of various machine learning algorithms for classifying active and inactive states of multiple sclerosis (MS), with features extracted from pre-trained CNN models.

Table 4.
Performance for multi-machine learning algorithms using feature extraction based on pre-trained models for (active and inactive) MS classification.
When analyzing the VGG16 model, it becomes evident that the K-nearest neighbor (KNN) and support vector machine (SVM) algorithms demonstrated superior performance, with all performance metrics exceeding 0.96. This confirms the efficacy of these models in discerning the active and inactive stages of MS using VGG16-derived features. The random forest (RF), extra trees classifier, gradient boost, and light gradient boosting machine (LGBM) algorithms also exhibit strong results, with precision, recall, F1-score, and accuracy values consistently above 0.95.
In examining the VGG19 model, the KNN and SVM algorithms continued to excel, boasting impressive precision, recall, F1-score, and accuracy values above 0.97. Additional algorithms, including RF, extra trees classifier, and LGBM, likewise maintained strong performance across all metrics, highlighting the robustness and reliability of these machine learning algorithms when used with VGG19-derived features.
Finally, when features were extracted using the InceptionV3 model, the KNN and SVM algorithms persisted in an outstanding performance, each achieving precision, recall, F1-score, and accuracy values nearing or reaching 0.98. Similarly, the RF and extra trees classifier delivered high-quality results, with all performance metrics near 0.94.
We saw a clear distinction between KNN and SVM in light of the results obtained. The reason was that these models had superior performance on the features extracted from pre-trained models, as the following studies showed, as they achieved promising results using the classifier SVM and KNN on the extracted features of pre-trained models, including VGG16, ResNet50, and DenseNet, which proved the effectiveness and superiority of these classifiers in our study over the proposed approach [29,42,43,44]. Overall, these findings underscored the remarkable performance of KNN, SVM, RF, and LGBM across different feature sets derived from pre-trained models in classifying the active and inactive states of MS. The consistent high performance of these machine learning algorithms demonstrated their potential as valuable tools in aiding the accurate diagnosis and subsequent management of MS.
Figure 5, Figure 6, Figure 7, Figure 8, Figure 9 and Figure 10 provide valuable insights into the performance of our diagnostic tests, highlighting various strengths and potential areas for improvement. In Figure 5, we observe a substantial quantity of true positives (TP), demonstrating the test’s efficacy in correctly diagnosing disease presence. The minimal false negatives (FN) indicated few disease cases were overlooked. However, an elevated count of false positives (FP) raised concerns over potentially unwarranted further testing or treatment. Figure 6 depicts a decrease in TP and an increase in FP compared to the previous figure, implying diminished overall accuracy.
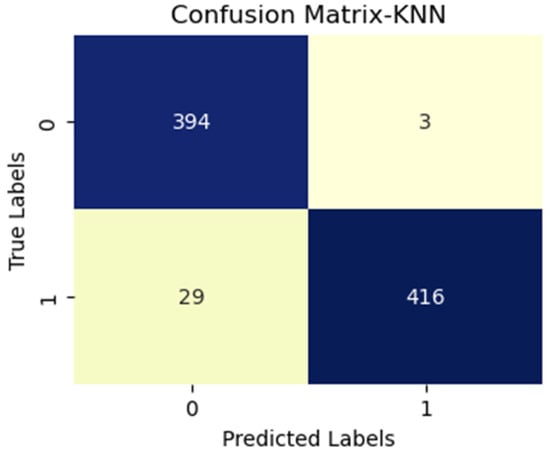
Figure 5.
KNN with vgg19 (MS).
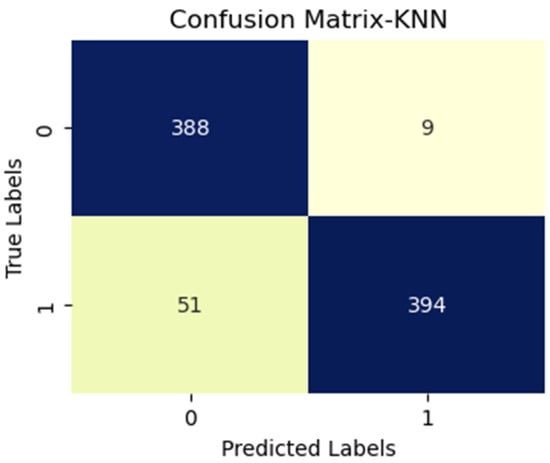
Figure 6.
KNN with InceptionV3 (MS).
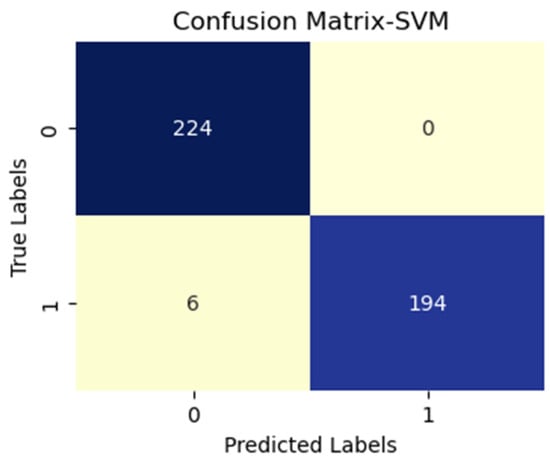
Figure 7.
SVM with vgg19 (active and inactive).
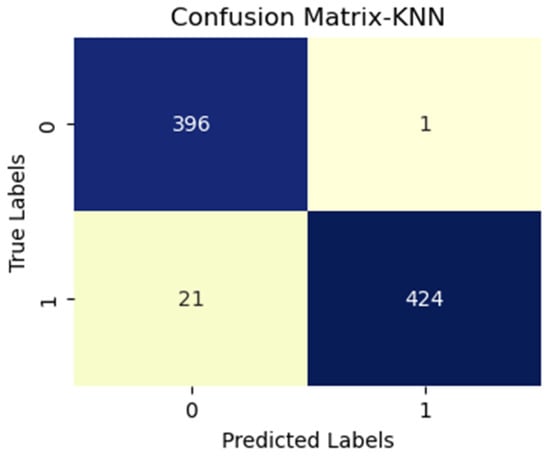
Figure 8.
KNN with vgg16 (MS).
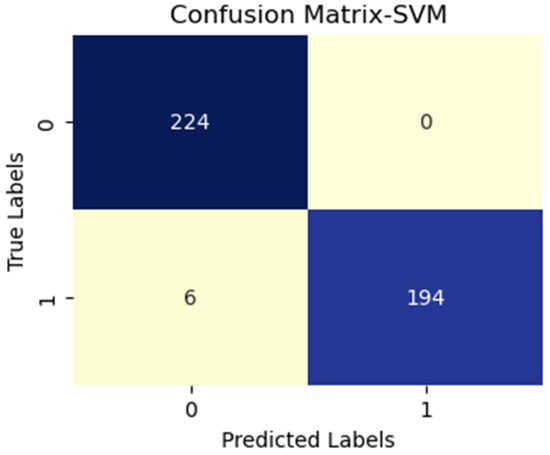
Figure 9.
SVM with vgg16 (active and inactive).
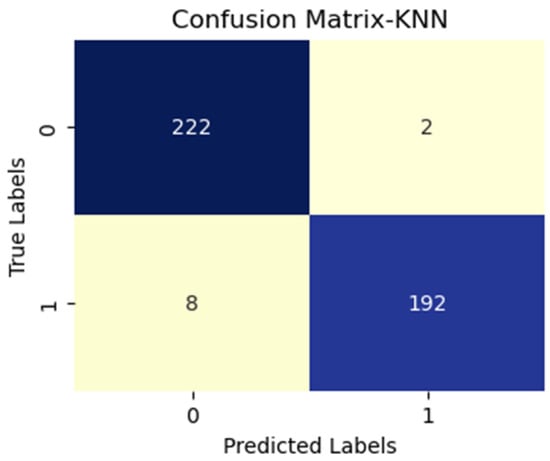
Figure 10.
KNN with InceptionV3 (active and inactive).
Additionally, FN figures were higher, suggesting that the test may overlook some disease instances. Figure 7 presents low TP and FN numbers, implying that the test might lack sensitivity in detecting the disease. However, the scarcity of FP implied a high degree of specificity in correctly identifying disease absence. Figure 8 illustrates high TP figures, supporting the test’s robustness in accurately detecting disease presence. Low FN and FP counts imply that the test rarely misses disease instances and maintains overall accuracy. In Figure 9, similar to Figure 7, we see low TP and FN numbers, suggesting limited disease sensitivity. Still, the low FP numbers indicate a high specificity in recognizing individuals without the disease. Figure 10 presents low TP and FN numbers akin to Figure 7 and Figure 9, implying suboptimal sensitivity. A moderately elevated FP number could lead to unnecessary subsequent tests or treatments. However, a high count of true negatives (TN) reflects good overall test accuracy.
In summary, these figures indicate varying test sensitivity and specificity levels, with different TP, FN, FP, and TN rates observed across distinct experiments. Further in-depth analysis is warranted to ascertain the test’s accuracy and reliability.
In this paper, we thoroughly examined how two common machine learning methods, k-nearest neighbor (KNN) and support vector machine (SVM), performed when paired with three different convolutional neural network (CNN) architectures: VGG16, VGG19, and InceptionV3, as shown in Figure 11 and Figure 12. Our results revealed interesting insights into how the choice of classifier and CNN architecture interact. Specifically, KNN_MS_VGG16 and SVM_Active_VGG19 achieved outstanding AUC scores of 99.9%, indicating their strong ability to tell classes apart. This suggests a good match between these architectures’ and the classifiers’ extracted features. Furthermore, SVM classifiers consistently showed impressive AUC performance across all CNN architectures, highlighting their strength in capturing complex decision boundaries. These findings underscore the delicate balance between classifier sensitivity and CNN architecture characteristics, offering a clearer understanding of how image classification accuracy is influenced in deep learning contexts. Figure 13, Figure 14, Figure 15, Figure 16, Figure 17 and Figure 18 indicates the ROC curve of our highest performance.
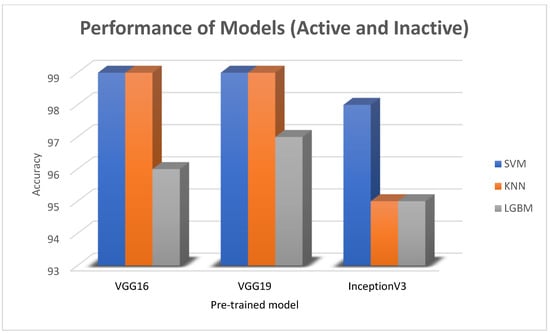
Figure 11.
The bar charts for the best three machine learning results for the active and inactive datasets.
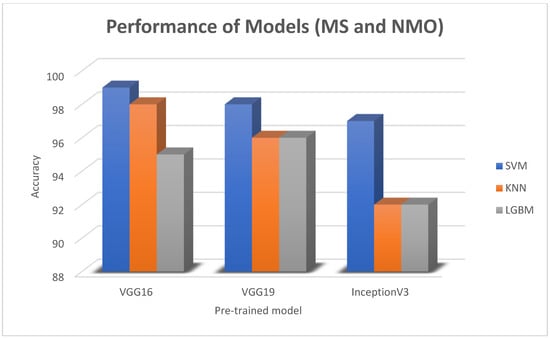
Figure 12.
The bar charts for the best three machine learning results for MS and NMO datasets.
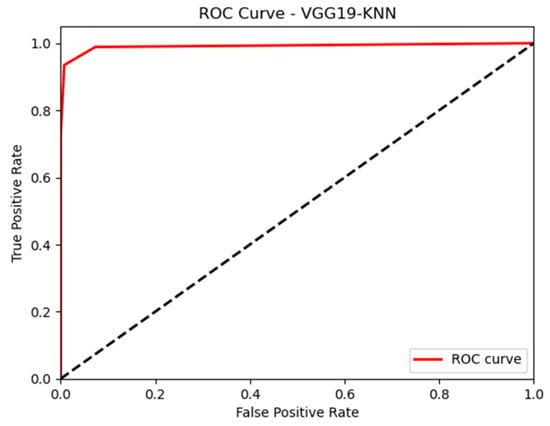
Figure 13.
KNN with vgg19 (MS).
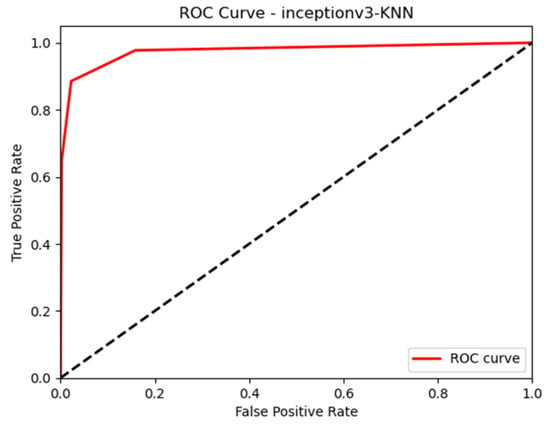
Figure 14.
KNN with InceptionV3 (MS).
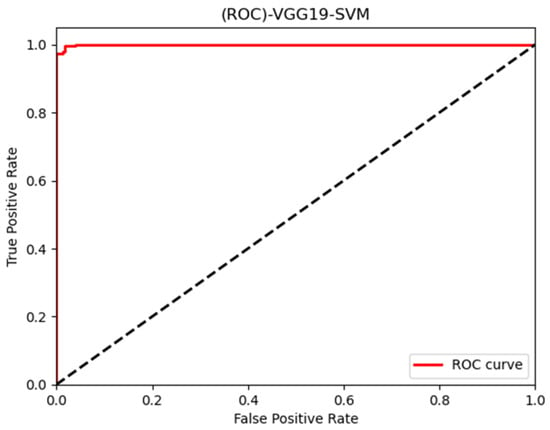
Figure 15.
SVM with vgg19 (active and inactive).
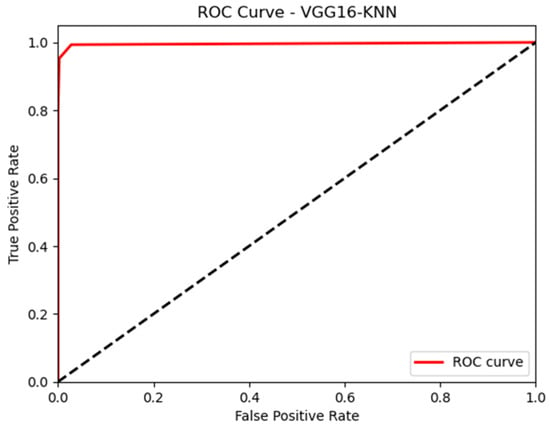
Figure 16.
KNN with vgg16 (MS).
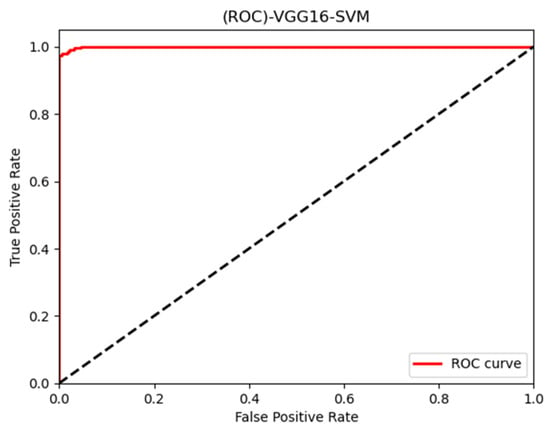
Figure 17.
SVM with vgg16 (active and inactive).
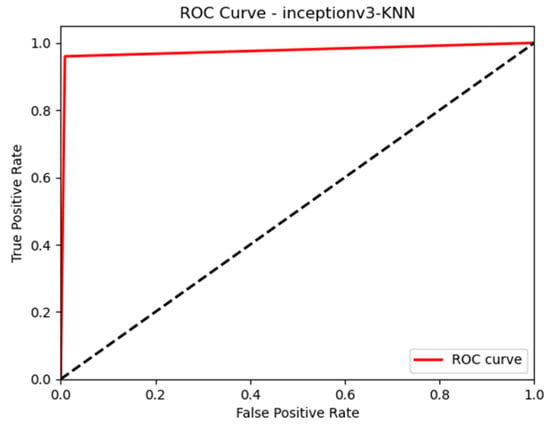
Figure 18.
KNN with InceptionV3 (active and inactive).
In our fine-tuning process, we optimized machine learning algorithms via a rigorous parameter optimization technique to identify the ideal parameters for each model that would yield optimal results. This selection process was data driven, relying on comprehensive experimentation.
We initially focused on fine-tuning the max_depth between [2, 12, dtype = int] parameter, determining a model’s maximum depth. Its selection depends on the problem’s complexity and the dataset’s size. Choosing an excessively high depth value risks overfitting, hence the need to identify an optimal value through experimentation. Next, we examined the n_estimators parameter between [20, 300], a hyperparameter common to algorithms such as RF, ET, GB, Ada boost, and LGBM. The choice of n_estimators depends on the data size, and while larger values may enhance performance, this must be balanced against the computational complexity and the time required for training. The learning rate between [0, 0.05, 0.0001] parameters, which significantly impacts training accuracy, was another pivotal hyperparameter we adjusted. This rate dictated the step size for each iteration during algorithm optimization and was determined experimentally. We also fine-tuned the min_child_weight between [1, 9, 0.025] parameters used in the LGBM algorithm. This parameter helps determine the minimum sum of instance weight in the child nodes. Likewise, sub, another parameter employed in the decision tree algorithm, prevents overfitting and sets the tree depth. Higher min_child_weight values generally result in simpler trees and help prevent overfitting. The ideal min_child_weight value was experimentally derived.
4.3. Comparison with Other Studies
Table 5 and Table 6 show a comparison between the results of our study and the results of previous studies related to the discovery of the same diseases, wherein a clear superiority is evident in our results. We achieved a promising performance in the classification between MS and NMO, as the highest accuracy was achieved through the hybrid model VGG16 + KNN, with an accuracy of 99%. And 98% for the model VGG16 + SVM shows that the proposed approach is more effective for detecting these diseases than the other works. In this study, our goal was to create a machine learning methodology capable of distinguishing between MS and NMO and identifying active and inactive states in MS. Further, the absence of active and inactive state classification in some studies underscores an additional advantage of our study.

Table 5.
Comparison of existing studies detecting MS.

Table 6.
Comparison of existing studies detecting active states.
The reason is that the methods they used in previous studies did not work effectively. The MRI images need high-efficiency pre-processing in addition to extracting the important features that represent the disease well and training these extracted features with appropriate algorithms.
Regarding limitations, the problem begins with obtaining the real dataset, so we needed time to collect it, in addition to the approval of the hospital and patients. Despite the success of obtaining it, it was relatively small, and we needed to increase it through the augmentation step. Among the problems we encountered when extracting features, extracting features through pre-trained models requires a large amount of time, in addition to a device with high specifications.
5. Conclusions and Recommendations
We present an innovative, data-driven framework that successfully leverages cutting-edge machine learning algorithms and leading-edge feature selection methodologies to differentiate between multiple sclerosis (MS) and neuromyelitis optica (NMO) and to further identify active and inactive stages of MS. Through rigorous and comprehensive experimentation, we demonstrated our approach’s prowess, combining the capabilities of support vector machine (SVMs) and K-nearest neighbor (KNN) classifiers. These classifiers, applied to feature vectors derived through utilizing the VGG16 model, yielded highly encouraging results, effectively making strides in disease classification. The data for these feature vectors were meticulously collected and curated from King’s Abdullah University Hospital in Jordan, ensuring the reliability and thoroughness of the information.
Our approach goes beyond these specific disorders. We envision a future where our framework can be extended to classify a wide spectrum of neuroinflammatory diseases. This will be realized by integrating diverse, comprehensive datasets and broadening the applicability of our methodology. Furthermore, our proposed framework stands ready to integrate segmentation strategies, enhancing disease detection capabilities. By doing so, we anticipate a substantial improvement in diagnostic accuracy, inevitably leading to more effective treatment strategies for neurological disorders. This will ultimately pave the way for improved patient care and a better understanding of these conditions. Our work symbolizes an important step forward in the application of machine learning in medical diagnostics, demonstrating the potential for more accurate, rapid, and extensive classification of complex neurological disorders. In the future, we will work hard to obtain a larger dataset, add different optimization methods to improve the results, and work in this way to include other diseases. In addition, we will work on segmentation methods to extract features more accurately.
Author Contributions
Conceptualization, M.G., W.A., N.A.A., A.N., H.G., M.E.-H., M.A., A.F. and L.A.; methodology, A.N., N.A.A., H.G. and W.A.; software, A.N. and H.G.; validation, A.N., H.G. and N.A.A.; formal analysis, L.A.; investigation, A.N., H.G., W.A. and N.A.A.; resources, W.A., A.N. and H.G.; data curation, W.A.; writing—original draft preparation, M.G., W.A., N.A.A., A.N., H.G., M.E.-H., A.F. and L.A.; writing—review and editing, M.G., W.A., N.A.A., H.G., A.N., M.E.-H., A.F. and L.A.; visualization, A.N., H.G. and W.A. All authors have read and agreed to the published version of the manuscript.
Funding
The researchers wish to thank the Deanship of Scientific Research at Taif University for funding this research.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data will be available upon reasonable request from the corresponding authors.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
This study employs diverse advanced machine learning techniques and algorithms to classify MS and MS_NMO datasets.
- Random Forest (RF)
It is an algorithm that is used in binary and multi-classification problems. Random forest is a set of trees that predict classification, which is a group learning method for classification, regression, and other tasks, working by creating many decision trees; in this work, (n_estimater = [20–300]) was tested to obtain a mode of the classes (classification) or mean prediction (regression) [33].
- Support Vector Machine (SVM)
SVM is a machine learning algorithm that separates categories by setting boundaries, which sorts data into more than one category, and these boundaries are called the hyperplane. There are three types of hyperplanes, namely, optimal, positive, and negative [39].
- Naïve Bayes (NB)
NB classifiers are a set of simple probability classifiers based on Bayes’ theory. The classifier is a function that maps the output of the input features to obtain the class labels. The following equation shows the relationship between the output and the features, which is a highly scalable classifier but requires several linear parameters in the number of features in the learning problem [45].
- k-Nearest Neighbor (KNN)
KNN is an algorithm developed due to the need to conduct a discriminatory analysis when the parametric estimates are difficult to determine. The principle of its work is to choose the neighbors closest to the sample to be classified, and based on the distance between it and its neighbors, the sample class is chosen. The number of neighbors was chosen as K = [2, 400], wherein the best number of neighbors gives the best result [41].
- Logistic Regression
A supervised learning approach called logistic regression estimates the likelihood of a sample patch depending on how closely it resembles the label. Changes to the input values may impact the final prediction of this algorithm, and the size of the input vector must be modest because it influences the cost of training. When a problem requires various variables that must be merged into a binary classification task and requires little complexity, logistic regression can be a useful model [46].
- Decision Tree (DT)
An approach for machine learning called DT produces coherent classification rules. It creates a tree structure by iteratively dividing the data based on principles promoting data separation. The basic goal of this method is to determine the key characteristics based on which the dataset is separated into more manageable groupings. To forecast the class starting from the root, the process is continued until it reaches the leaf node and assigns it a predicted label by comparing the test sample with the values of the root features [47].
- Extra Trees Classifier
Extra tree classifier is a kind of ensemble learning algorithm that combines the ideas of decision trees with packaging to improve the performance of decision trees. Additional random trees are generated by adding randomness to the tree induction process. This helps reduce model variance and improve its performance on noisy datasets. Additional randomness is introduced by randomly selecting split points at each node in the tree from a uniform distribution across the range of feature values. This contrasts with the traditional decision tree approach, where split points are chosen based on a specific criterion, such as maximizing information acquisition or minimizing Gini impurities. In short, the extra tree classifier is a powerful ensemble learning algorithm that can improve the performance of decision trees by adding more randomness to the tree induction process [48].
- LightGBM (LGBM)
LGBM is designed based on gradient boosting decision tree, a popular algorithm for machine learning, and has many effective applications such as XGboost and pGBRT, developed to obtain satisfactory efficiency and scalability when the feature dimension is high and the data volume is large. LGBM is built on two new technologies: gradient-based one-side sampling (GOSS) and exclusive feature bundling (EFB). Its goal was to develop a solution to the challenge of defining the best possible combination of properties so that features can be reduced efficiently without significantly affecting the accuracy of split point determination [48].
- Ada Boost
Adaptive boosting, sometimes called Ada boost, is one of the most widely used methods for classifier boosting. It seeks to improve the model’s performance by combining many weak learners into strong learners. Ada boost trains the samples and chooses the weights for underperforming classifiers per each iteration. The process is continued until the greatest amount of data is correctly classified. In the event of misclassification, the weights are increased and decreased for each classifier, and then the samples are trained again to generate a classifier for the new data [49].
- Gradient Boosting
A new technique for addressing classification and regression problems is gradient boosting, an enhancement of the traditional gradient boosting algorithm. This technique aims to construct predictive models by amalgamating numerous simple models, such as decision trees, while striving to minimize residuals. This is achieved by initially fitting a model to the data, then fitting a new model to the residuals of the previous model and repeating the process. As its name suggests, random gradient boosting incorporates random data samples into the algorithm at each iteration of the boosting process. This feature permits the algorithm to handle noisy input more efficiently and avoid overfitting, a frequent concern in machine learning. Several parameters must be optimized to attain the best results, including the model’s learning rate and the number of iterations. Stochastic gradient boosting has emerged as a prevalent and remarkably effective machine learning technique, particularly in critical fields such as finance and healthcare, where accuracy is paramount [50].
- Stochastic Gradient Descent (SGD)
Stochastic gradient descent (SGD) is a popular optimization algorithm for training classification models in machine learning. Unlike batch gradient descent, which computes the gradient of the cost function using the entire dataset, SGD updates the model parameters for each example in the training set, making it a more efficient and scalable method for large datasets. SGD works by randomly selecting a single training example at each iteration and updating the model parameters based on the gradient of the cost function concerning that example. This random sampling of the training data introduces noise into the updates, which can help the model avoid local optima and improve generalization performance [51].
- Ridge Regression
Ridge regression is a linear regression technique introducing a penalty term to the standard least squares loss function. This penalty component, added to the objective function to limit the values of the coefficients, is proportional to the square of the size of the regression coefficients. The ridge penalty causes the regression coefficients to shrink in size towards zero, which can assist in lowering the estimates’ variance and increase the model’s stability. A tuning parameter that determines the trade-off between bias and variance controls the degree of shrinkage [52].
References
- Calabresi, P.A. Diagnosis and management of multiple sclerosis. Am. Fam. Physician 2004, 70, 1935–1944. [Google Scholar]
- Goldenberg, M.M. Multiple sclerosis review. Pharm. Ther. 2012, 37, 175. [Google Scholar]
- Brownlee, W.J.; Hardy, T.A.; Fazekas, F.; Miller, D.H. Diagnosis of multiple sclerosis: Progress and challenges. Lancet 2017, 389, 1336–1346. [Google Scholar] [CrossRef]
- Arshad, A.; Jabeen, M.; Ubaid, S.; Raza, A.; Abualigah, L.; Aldiabat, K.; Jia, H. A novel ensemble method for enhancing Internet of Things device security against botnet attacks. Decis. Anal. J. 2023, 8, 100307. [Google Scholar] [CrossRef]
- Ömerhoca, S.; Akkaş, S.Y.; İçen, N.K. Multiple sclerosis: Diagnosis and differential diagnosis. Arch. Neuropsychiatry 2018, 55, S1–S9. [Google Scholar] [CrossRef] [PubMed]
- Lennon, V.A.; Wingerchuk, D.M.; Kryzer, T.J.; Pittock, S.J.; Lucchinetti, C.F.; Fujihara, K.; Weinshenker, B.G. A serum autoantibody marker of neuromyelitis optica: Distinction from multiple sclerosis. Lancet 2004, 364, 2106–2112. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Kim, S.H.; Lee, S.H.; Li, X.F.; Kim, H.J. Brain abnormalities as an initial manifestation of neuromyelitis optica spectrum disorder. Mult. Scler. J. 2011, 17, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Mandler, R.N.; Ahmed, W.; Dencoff, J.E. Devic’s neuromyelitis optica: A prospective study of seven patients treated with prednisone and azathioprine. Neurology 1998, 51, 1219–1220. [Google Scholar] [CrossRef] [PubMed]
- Kawachi, I.; Lassmann, H. Neurodegeneration in multiple sclerosis and neuromyelitis optica. J. Neurol. Neurosurg. Psychiatry 2017, 88, 137–145. [Google Scholar] [CrossRef]
- Kim, H.; Lee, Y.; Kim, Y.H.; Lim, Y.M.; Lee, J.S.; Woo, J.; Kim, K.K. Deep learning-based method to differentiate neuromyelitis optica spectrum disorder from multiple sclerosis. Front. Neurol. 2020, 11, 599042. [Google Scholar] [CrossRef]
- Miller, D.H.; Albert, P.S.; Barkhof, F.; Francis, G.; Frank, J.A.; Hodgkinson, S.; Lublin, F.D.; Paty, D.W.; Reingold, S.C.; Simon, J. Guidelines for the use of magnetic resonance techniques in monitoring the treatment of multiple sclerosis. Ann. Neurol. 1996, 39, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Goodin, D.S.; Frohman, E.M.; Garmany, G.P.; Halper, J.; Likosky, W.H.; Lublin, F.D. Disease modifying therapies in multiple sclerosis. Neurology 2002, 58, 169–178. [Google Scholar] [PubMed]
- Lassmann, H. Multiple sclerosis pathology. Cold Spring Harb. Perspect. Med. 2018, 8, a028936. [Google Scholar] [CrossRef] [PubMed]
- Moraal, B.; Wattjes, M.P.; Geurts, J.J.G.; Dirk, L.K.; van Schijndel, R.A.; Pouwels, P.J.W.; Vrenken, H.; Barkhof, F. Improved detection of active multiple sclerosis lesions: 3D subtraction imaging. Radiology 2010, 255, 154–163. [Google Scholar] [CrossRef]
- Lladó, X.; Ganiler, O.; Oliver, A.; Martí, R.; Freixenet, J.; Valls, L.; Vilanova, J.C.; Ramió-Torrentà, L.; Rovira, À. Automated detection of multiple sclerosis lesions in serial brain MRI. Neuroradiology 2012, 54, 787–807. [Google Scholar] [CrossRef]
- Buyukturkoglu, K.; Zeng, D.; Bharadwaj, S.; Tozlu, C.; Mormina, E.; Igwe, K.C.; Lee, S.; Habeck, C.; Brickman, A.M.; Riley, C.S.; et al. Classifying multiple sclerosis patients on the basis of SDMT performance using machine learning. Mult. Scler. J. 2021, 27, 107–116. [Google Scholar] [CrossRef]
- Ramanujam, R.; Zhu, F.; Fink, K.; Karrenbauer, V.D.; Lorscheider, J.; Benkert, P.; Kingwell, E.; Tremlett, H.; Hillert, J.; Manouchehrinia, A. Accurate classification of secondary progression in multiple sclerosis using a decision tree. Mult. Scler. J. 2021, 27, 1240–1249. [Google Scholar] [CrossRef]
- Loizou, C.P.; Pantzaris, M.; Pattichis, C.S. Normal appearing brain white matter changes in relapsing multiple sclerosis: Texture image and classification analysis in serial MRI scans. Magn. Reson. Imaging 2020, 73, 192–202. [Google Scholar] [CrossRef]
- Saccà, V.; Sarica, A.; Novellino, F.; Barone, S.; Tallarico, T.; Filippelli, E.; Granata, A.; Chiriaco, C.; Bruno Bossio, R.; Valentino, P.; et al. Evaluation of machine learning algorithms performance for the prediction of early multiple sclerosis from resting-state FMRI connectivity data. Brain Imaging Behav. 2019, 13, 1103–1114. [Google Scholar] [CrossRef]
- Samah, Y.; Yassine, B.S.; Naceur, A.M. Multiple sclerosis lesions detection from noisy magnetic resonance brain images tissue. In Proceedings of the 2018 15th International Multi-Conference on Systems, Signals & Devices (SSD), Yasmine Hammamet, Tunisia, 19–22 March 2018. [Google Scholar]
- Maggi, P.; Fartaria, M.J.; Jorge, J.; La Rosa, F.; Absinta, M.; Sati, P.; Meuli, R.; Du Pasquier, R.; Reich, D.S.; Cuadra, M.B. CVSnet: A machine learning approach for automated central vein sign assessment in multiple sclerosis. NMR Biomed. 2020, 33, e4283. [Google Scholar] [CrossRef]
- Roy, S.; Butman, J.A.; Reich, D.S.; Calabresi, P.A.; Pham, D.L. Multiple sclerosis lesion segmentation from brain MRI via fully convolutional neural networks. arXiv 2018, arXiv:1803.09172. [Google Scholar]
- Sepahvand, N.M.; Arnold, D.L.; Arbel, T. CNN detection of new and enlarging multiple sclerosis lesions from longitudinal MRI using subtraction images. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020. [Google Scholar]
- Eshaghi, A.; Wottschel, V.; Cortese, R.; Calabrese, M.; Sahraian, M.A.; Thompson, A.J.; Alexander, D.C.; Ciccarelli, O. Gray matter MRI differentiates neuromyelitis optica from multiple sclerosis using random forest. Neurology 2016, 87, 2463–2470. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Nguyen, T.D.; Zhang, J.; Marcille, M.; Spincemaille, P.; Wang, Y.; Gauthier, S.A.; Sweeney, E.M. QSMRim-Net: Imbalance-aware learning for identification of chronic active multiple sclerosis lesions on quantitative susceptibility maps. NeuroImage Clin. 2022, 34, 102979. [Google Scholar] [CrossRef] [PubMed]
- Coronado, I.; Gabr, R.E.; Narayana, P.A. Narayana. Deep learning segmentation of gadolinium-enhancing lesions in multiple sclerosis. Mult. Scler. J. 2018, 27, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Gaj, S.; Ontaneda, D.; Nakamura, K. Automatic segmentation of gadolinium-enhancing lesions in multiple sclerosis using deep learning from clinical MRI. PLoS ONE 2021, 16, e0255939. [Google Scholar] [CrossRef]
- Narayana, P.A.; Coronado, I.; Sujit, S.J.; Wolinsky, J.S.; Lublin, F.D.; Gabr, R.E. Deep learning for predicting enhancing lesions in multiple sclerosis from noncontrast MRI. Radiology 2020, 294, 398–404. [Google Scholar] [CrossRef]
- Seok, J.M.; Cho, W.; Chung, Y.H.; Ju, H.; Kim, S.T.; Seong, J.K.; Min, J.H. Differentiation between multiple sclerosis and neuromyelitis optica spectrum disorder using a deep learning model. Sci. Rep. 2023, 13, 11625. [Google Scholar] [CrossRef]
- Kavaklioglu, B.C.; Erdman, L.; Goldenberg, A.; Kavaklioglu, C.; Alexander, C.; Oppermann, H.M.; Patel, A.; Hossain, S.; Berenbaum, T.; Yau, O. Machine learning classification of multiple sclerosis in children using optical coherence tomography. Mult. Scler. J. 2022, 28, 2253–2262. [Google Scholar] [CrossRef]
- Kenney, R.C.; Liu, M.; Hasanaj, L.; Joseph, B.; Abu Al-Hassan, A.; Balk, L.J.; Behbehani, R.; Brandt, A.; Calabresi, P.A.; Frohman, E. The role of optical coherence tomography criteria and machine learning in multiple sclerosis and optic neuritis diagnosis. Neurology 2022, 99, e1100–e1112. [Google Scholar] [CrossRef]
- Garcia-Martin, E.; Dongil-Moreno, F.; Ortiz, M.; Ciubotaru, O.; Boquete, L.; Sánchez-Morla, E.; Jimeno-Huete, D.; Miguel, J.; Barea, R.; Vilades, E. Diagnosis of Multiple Sclerosis using Optical Coherence Tomography Supported by Explainable Artificial Intelligence. 2023; Preprint. [Google Scholar] [CrossRef]
- Gharaibeh, M.; Alzu’bi, D.; Abdullah, M.; Hmeidi, I.; Al Nasar, M.R.; Abualigah, L.; Gandomi, A.H. Radiology imaging scans for early diagnosis of kidney tumors: A review of data analytics-based machine learning and deep learning approaches. Big Data Cogn. Comput. 2022, 6, 29. [Google Scholar] [CrossRef]
- Thompson, A.J.; Banwell, B.L.; Barkhof, F.; Carroll, W.M.; Coetzee, T.; Comi, G.; Correale, J.; Fazekas, F.; Filippi, M.; Freedman, M.S.; et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018, 17, 162–173. [Google Scholar] [CrossRef]
- Wingerchuk, D.M.; Banwell, B.; Bennett, J.L.; Cabre, P.; Carroll, W.; Chitnis, T.; De Seze, J.; Fujihara, K.; Greenberg, B.; Jacob, A.; et al. International consensus diagnostic criteria for neuromyelitis optica spectrum disorders. Neurology 2015, 85, 177–189. [Google Scholar] [CrossRef]
- Singer, G.; Marudi, M. Ordinal decision-tree-based ensemble approaches: The case of controlling the daily local growth rate of the COVID-19 epidemic. Entropy 2020, 22, 871. [Google Scholar] [CrossRef] [PubMed]
- Jaradat, A.S.; Al Mamlook, R.E.; Almakayeel, N.; Alharbe, N.; Almuflih, A.S.; Nasayreh, A.; Gharaibeh, H.; Gharaibeh, M.; Gharaibeh, A.; Bzizi, H. Automated Monkeypox Skin Lesion Detection Using Deep Learning and Transfer Learning Techniques. Int. J. Environ. Res. Public Health 2023, 20, 4422. [Google Scholar] [CrossRef] [PubMed]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Abualigah, L.M.Q. Feature Selection and Enhanced Krill Herd Algorithm for Text Document Clustering; Springer: Cham, Switzerland, 2019; p. 816. [Google Scholar]
- Fonville, J.M.; Carter, C.; Cloarec, O.; Nicholson, J.K.; Lindon, J.C.; Bunch, J.; Holmes, E. Robust data processing and normalization strategy for MALDI mass spectrometric imaging. Anal. Chem. 2012, 84, 1310–1319. [Google Scholar] [CrossRef]
- Otair, M.; Ibrahim, O.T.; Abualigah, L.; Altalhi, M.; Sumari, P. An enhanced grey wolf optimizer based particle swarm optimizer for intrusion detection system in wireless sensor networks. Wirel. Netw. 2022, 28, 721–744. [Google Scholar] [CrossRef]
- Khaledian, N.; Nazari, A.; Khamforoosh, K.; Abualigah, L.; Javaheri, D. TrustDL: Use of trust-based dictionary learning to facilitate recommendation in social networks. Expert Syst. Appl. 2023, 228, 120487. [Google Scholar] [CrossRef]
- Musleh, D.; Alotaibi, M.; Alhaidari, F.; Rahman, A.; Mohammad, R.M. Intrusion Detection System Using Feature Extraction with Machine Learning Algorithms in IoT. J. Sens. Actuator Netw. 2023, 12, 29. [Google Scholar] [CrossRef]
- Selvanayaki, K.S.; Somasundaram, R.; Shyamala, D.J. Detection and Recognition of Vehicle Using Principal Component Analysis. In Proceedings of the International Conference on ISMAC in Computational Vision and Bio-Engineering 2018 (ISMAC-CVB); Springer: Cham, Switzerland, 2019; p. 30. [Google Scholar]
- Al-Manaseer, H.; Abualigah, L.; Alsoud, A.R.; Zitar, R.A.; Ezugwu, A.E.; Jia, H. A novel big data classification technique for healthcare application using support vector machine, random forest and J48. In Classification Applications with Deep Learning and Machine Learning Technologies; Springer: Cham, Switzerland, 2023; Volume 1071, pp. 205–215. [Google Scholar]
- Houssein, E.H.; Dirar, M.; Abualigah, L.; Mohamed, W.M. An efficient equilibrium optimizer with support vector regression for stock market prediction. Neural Comput. Appl. 2022, 34, 3165–3200. [Google Scholar] [CrossRef]
- Leung, K.M. Naive Bayesian Classifier, Department of Computer Science/Finance and Risk Engineering, Polytechnic University. 2007. Available online: https://cse.engineering.nyu.edu/~mleung/FRE7851/f07/naiveBayesianClassifier.pdf (accessed on 2 July 2023).
- Abualigah, L. Classification Applications with Deep Learning and Machine Learning Technologies; Springer: Cham, Switzerland, 2022; p. 1071. [Google Scholar]
- Friedman, J.H. Stochastic gradient boosting. Comput. Stat. Data Anal. 2002, 38, 367–378. [Google Scholar] [CrossRef]
- LeCun, Y.; Bottou, L.; Orr, G.B.; Müller, K.R. Efficient backprop. In Neural Networks: Tricks of the Trade; Springer: Berlin/Heidelberg, Germany, 2002; pp. 9–50. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J.H.; Friedman, J.H. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer: Berlin/Heidelberg, Germany, 2009; Volume 2. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).