Extraction and Characterization of β-Viginin Protein Hydrolysates from Cowpea Flour as a New Manufacturing Active Ingredient
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Cowpea Flour
2.3. Isolation of Cowpea β-Vignin
2.4. Gel Electrophoresis
2.5. Enzymatic Hydrolysis and Fractionation
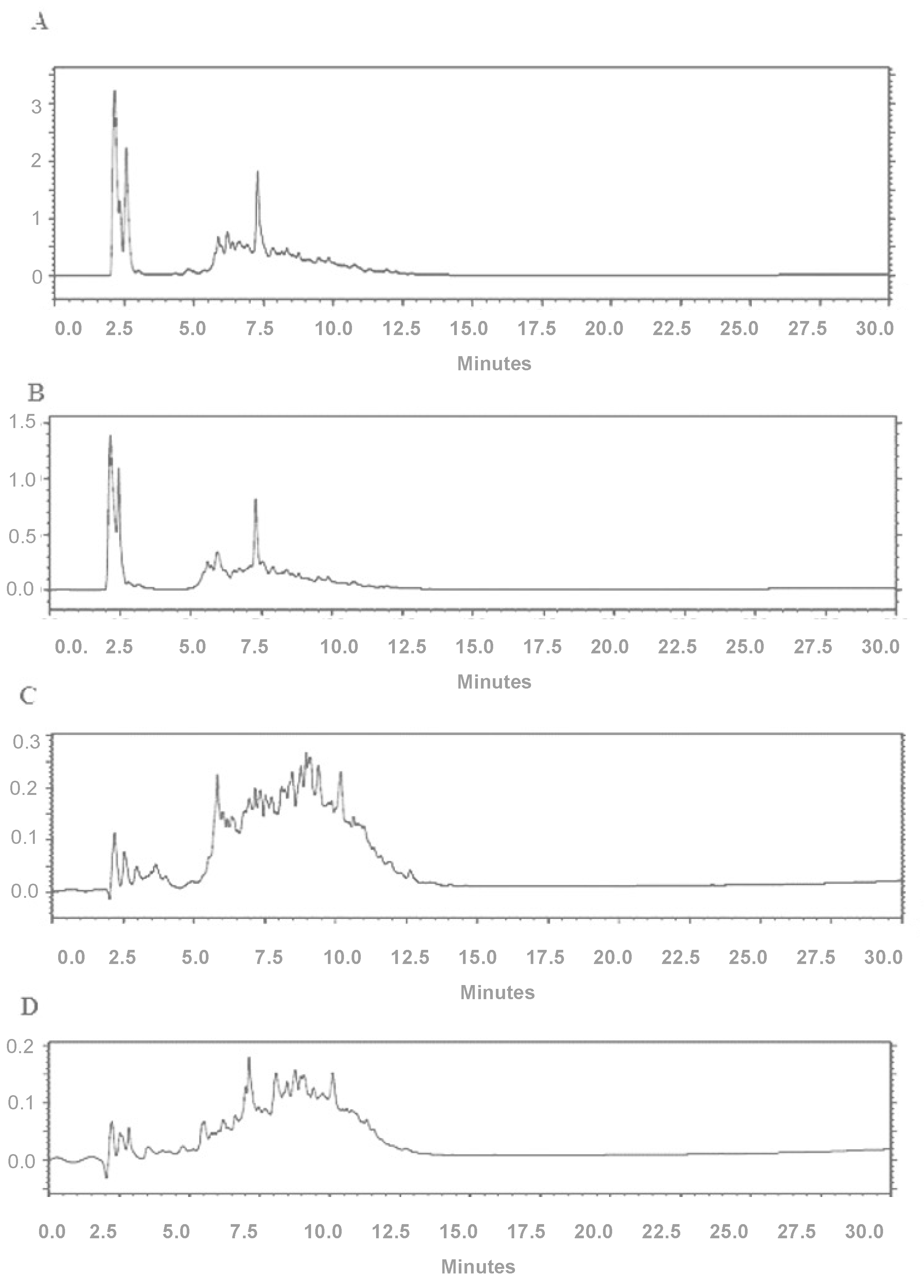
2.6. High-Performance Liquid Chromatography
2.7. In Silico Screening of Peptides with Antimicrobial Properties
2.8. Minimum Inhibitory Concentration
2.9. Agar Disk Diffusion Method
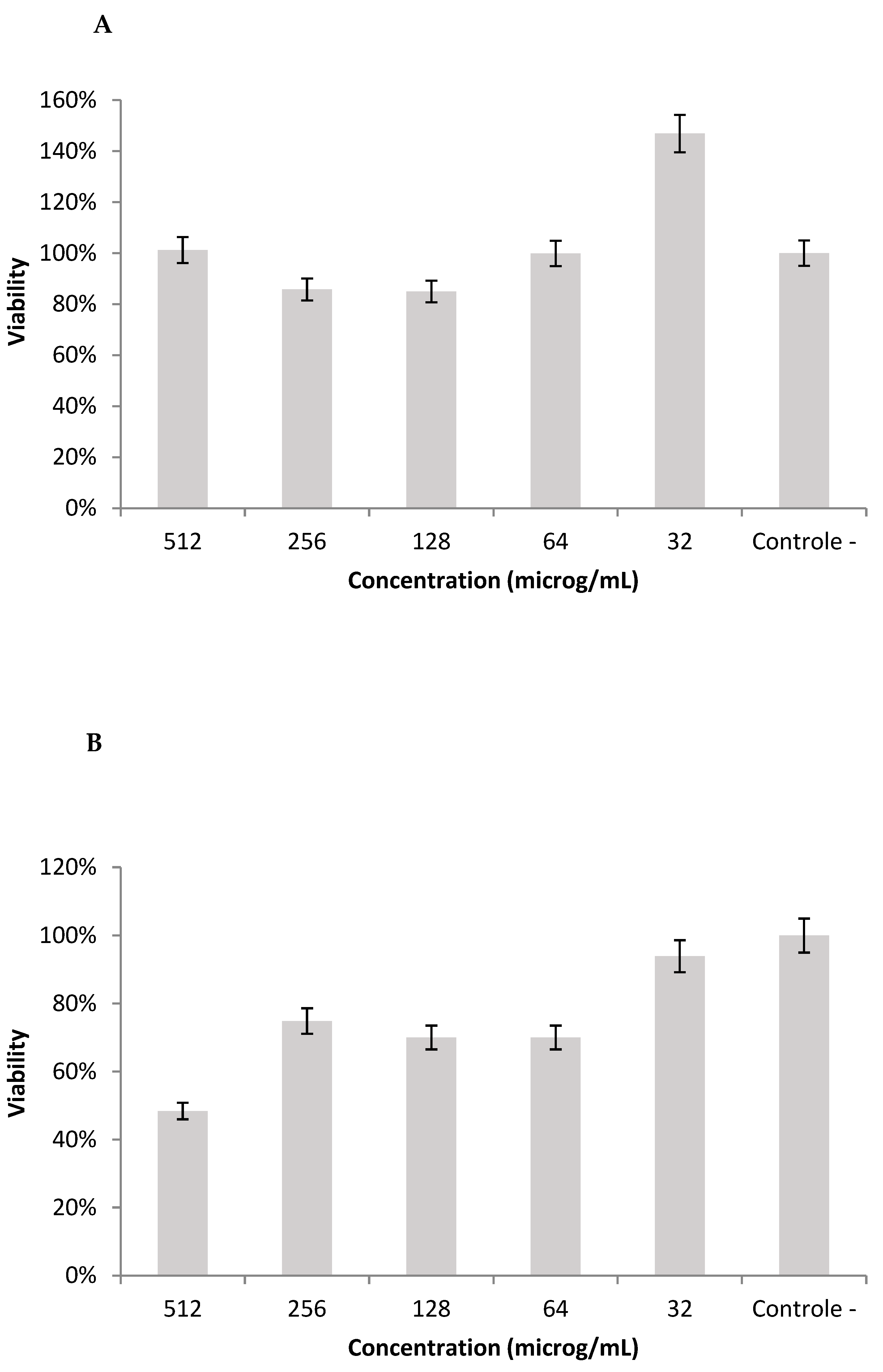
2.10. Cell Viability in L929 Cell-Line
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Brown, D. Antibiotic resistance breakers: Can repurposed drugs fill the antibiotic discovery void? Nat. Rev. Drug Discov. 2015, 14, 821–832. [Google Scholar] [CrossRef] [PubMed]
- Okoye, C.O.; Ezeorba, T.P.C.; Okeke, E.S.; Okagu, I.U. Recent Findings on the Isolation, Identification and Quantification of Bioactive Peptides. Appl. Food Res. 2022, 2, 100065. [Google Scholar] [CrossRef]
- Martinez-Villaluenga, C.; Rupasinghe, S.G.; Schuler, M.A.; de Mejia, E.G. Peptides from purified soybean beta-conglycinin inhibit fatty acid synthase by interaction with the thioesterase catalytic domain. FEBS J. 2010, 277, 1481–1493. [Google Scholar] [CrossRef] [PubMed]
- Lammi, C.; Zanoni, C.; Arnoldi, A.; Vistoli, G. Two Peptides from Soy β-Conglycinin Induce a Hypocholesterolemic Effect in HepG2 Cells by a Statin-Like Mechanism: Comparative in Vitro and in Silico Modeling Studies. J. Agric. Food Chem. 2015, 63, 7945–7951. [Google Scholar] [CrossRef] [PubMed]
- Marques, M.R.; Fontanari, G.G.; Pimenta, D.C.; Soares-Freitas, R.M.; Arêas, J.A.G. Proteolytic hydrolysis of cowpea proteins is able to release peptides with hypocholesterolemic activity. Food Res. Int. 2015, 77, 43–48. [Google Scholar] [CrossRef]
- Guha, S.; Majumder, K. Structural-features of food-derived bioactive peptides with anti-inflammatory activity: A brief review. J. Food Biochem. 2019, 43, e12531. [Google Scholar] [CrossRef]
- Moreno, C.; Mojica, L.; González de Mejía, E.; Camacho Ruiz, R.M.; Luna-Vital, D.A. Combinations of Legume Protein Hydrolysates Synergistically Inhibit Biological Markers Associated with Adipogenesis. Food 2020, 9, 1678. [Google Scholar] [CrossRef]
- Farkas, A.; Maróti, G.; Kereszt, A.; Kondorosi, É. Comparative Analysis of the Bacterial Membrane Disruption Effect of Two Natural Plant Antimicrobial Peptides. Front. Microbiol. 2017, 8, 51. [Google Scholar] [CrossRef]
- Ageitos, J.M.; Sánchez-Pérez, A.; Calo-Mata, P.; Villa, T.G. Antimicrobial peptides (AMPs): Ancient compounds that represent novel weapons in the fight against bacteria. Biochem. Pharmacol. 2017, 133, 117–138. [Google Scholar] [CrossRef]
- Luong, H.X.; Thanh, T.T.; Tran, T.H. Antimicrobial peptides—Advances in development of therapeutic applications. Life Sci. 2020, 260, 118407. [Google Scholar] [CrossRef] [PubMed]
- Benko-Iseppon, A.M.; Galdino, S.L.; Calsa, T., Jr.; Kido, E.A.; Tossi, A.; Belarmino, L.C.; Crovella, S. Overview on plant antimicrobial peptides. Curr. Protein Pept. Sci. 2010, 11, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Mechkarska, M.; Lukic, M.L.; Flatt, P.R. Potential therapeutic applications of multifunctional host-defense peptides from frog skin as anti-cancer, anti-viral, immunomodulatory, and anti-diabetic agents. Peptides 2014, 57, 67–77. [Google Scholar] [CrossRef]
- Aiello, G.; Lammi, C.; Boschin, G.; Zanoni, C.; Arnoldi, A. Exploration of Potentially Bioactive Peptides Generated from the Enzymatic Hydrolysis of Hempseed Proteins. J. Agric. Food Chem. 2017, 65, 10174–10184. [Google Scholar] [CrossRef]
- Osman, A.; Enan, G.; Al-Mohammadi, A.-R.; Abdel-Shafi, S.; Abdel-Hameid, S.; Sitohy, M.Z.; El-Gazzar, N. Antibacterial Peptides Produced by Alcalase from Cowpea Seed Proteins. Antibiotics 2021, 10, 870. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Guan, N.; Li, P.; Li, J.; Luo, J. Monitoring and dietary exposure assessment of pesticide residues incowpea (Vigna unguiculata L. Walp) in Hainan, China. Food Control 2016, 59, 250–255. [Google Scholar] [CrossRef]
- Rocha, A.J.; Sousa, B.L.; Girão, M.S.; Barroso-Neto, I.L.; Monteiro-Júnior, J.E.; Oliveira, J.T.A.; Nagano, C.S.; Carneiro, R.F.; Monteiro-Moreira, A.C.O.; Rocha, B.A.M.; et al. Cloning of cDNA sequences encoding cowpea (Vigna unguiculata) vicilins: Computational simulations suggest a binding mode of cowpea vicilins to chitin oligomers. Int. J. Biol. Macromol. 2018, 117, 565–573. [Google Scholar] [CrossRef]
- Pina-Pérez, M.C.; Ferrús Pérez, M.A. Antimicrobial potential of legume extracts against foodborne pathogens: A review. Trends Food Sci. Technol. 2018, 72, 114–124. [Google Scholar] [CrossRef]
- Ferreira, E.S.; Amaral, A.L.S.; Demonte, A.; Zanelli, C.F.; Capraro, J.; Duranti, M.; Neves, V.A. Hypocholesterolaemic effect of rat-administered oral doses of the isolated 7S globulins from cowpeas and adzuki beans. J. Nutr. Sci. 2015, 4, e7. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Akeson, W.R.; Stahmann, M.A. A pepsin pancreatin digest index of protein quality evaluation. J. Nutr. 1964, 83, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2022, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM Database of Bioactive Peptides: Current Opportunities. Int. J. Mol. Sci 2019, 20, 5978. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Methods for Antimicrobial Broth Dilution and Disk Diffusion Susceptibility Testing of Bacteria Isolated From Aquatic Animals, 2nd ed.; CLSI Guideline VET03; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th ed.; CLSI Standard M07; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- ISO 10993-1:2009; Biological Evaluation of Medical Devices—Part 5: Tests for In Vitro Cytotoxicity. British Standards: London, UK, 2009.
- Severino, P.; Chaud, M.V.; Shimojo, A.; Antonini, D.; Lancelloti, M.; Santana, M.H.A.; Souto, E.B. Sodium alginate-cross-linked polymyxin B sulphate-loaded solid lipid nanoparticles: Antibiotic resistance tests and HaCat and NIH/3T3 cell viability studies. Colloids Surf. B Biointerfaces 2015, 129, 191–197. [Google Scholar] [CrossRef]
- Aluko, R.E.; Girgih, A.T.; He, R.; Malomo, S.; Li, H.; Offengenden, M.; Wu, J. Structural and functional characterization of yellow field pea seed (Pisum sativum L.) protein-derived antihypertensive peptides. Food Res. Int. 2015, 77, 10–16. [Google Scholar] [CrossRef]
- Freitas, R.L.; Teixeira, A.R.; Ferreira, R.B. Characterization of the proteins from Vigna unguiculata seeds. J. Agric. Food Chem. 2004, 52, 1682–1687. [Google Scholar] [CrossRef]
- Gonçalves, A.; Goufo, P.; Barros, A.; Domínguez-Perles, R.; Trindade, H.; Rosa, E.A.; Ferreira, L.; Rodrigues, M. Cowpea (Vigna unguiculata L. Walp), a renewed multipurpose crop for a more sustainable agri-food system: Nutritional advantages and constraints. J. Sci. Food Agric. 2016, 96, 2941–2951. [Google Scholar] [CrossRef]
- Awika, J.M.; Duodu, K.G. Bioactive polyphenols and peptides in cowpea (Vigna unguiculata) and their health promoting properties: A review. J. Funct. Foods 2017, 38, 686–697. [Google Scholar] [CrossRef]
- Holetz, F.B.; Pessini, G.L.; Sanches, N.R.; Cortez, D.A.G.; Nakamura, C.V.; Dias Filho, B.P. Screening of some plants used in the Brazilian folk medicine for the treatment of infectious diseases. Memórias Instituo Oswaldo Cruz 2002, 97, 1027–1031. [Google Scholar] [CrossRef]
- Carvalho, A.F.; Silva, D.M.I.; Silva, T.R.C.; Scarcelli, E.; Manhani, M.R. Evaluation of the antibacterial activity of ethanolic and cyclohexane extracts of chamomile flowers (Matricaria chamomilla L.). Rev. Bras. Plantas Med. 2014, 16, 521–526. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D.; Kubicek-Sutherland, J.Z. Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist. Updates 2016, 26, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Bahar, A.A.; Ren, D. Antimicrobial Peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef] [PubMed]
- Salas, C.E.; Badillo-Corona, J.A.; Ramírez-Sotelo, G.; Oliver-Salvador, C. Biologically active and antimicrobial peptides from plants. BioMed Res. Int. 2015, 2015, 102129. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.; Souza, C.; Philadelpho, B.O.; Cunha, M.; Batista, F.P.R.; Silva, J.R.D.; Druzian, J.I.; Castilho, M.S.; Cilli, E.M.; Ferreira, E.S. In vitro and in silico studies of 3-hydroxy-3-methyl-glutaryl coenzyme A reductase inhibitory activity of the cowpea Gln-Asp-Phe peptide. Food Chem. 2018, 259, 270–277. [Google Scholar] [CrossRef]
- Fassini, P.G.; Noda, R.W.; Ferreira, E.S.; Silva, M.A.; Neves, V.A.; Demonte, A. Soybean glycinin improves HDL-C and suppresses the effects of rosuvastatin on hypercholesterolemic rats. Lipids Health Dis. 2011, 10, 165. [Google Scholar] [CrossRef]
- Xiang, N.; Lyu, Y.; Zhu, X.; Bhunia, A.K.; Narsimhan, G. Methodology for identification of pore forming antimicrobial peptides from soy protein subunits β-conglycinin and glycinin. Peptides 2016, 85, 27–40. [Google Scholar] [CrossRef]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef]
- Mangoni, M.L.; Shai, Y. Short native antimicrobial peptides and engineered ultrashort lipopeptides: Similarities and differences in cell specificities and modes of action. Cell. Mol. Life Sci. 2011, 68, 2267–2280. [Google Scholar] [CrossRef]


| Peptides Fractions | Chymotrypsin (mg/mL) | Pepsin (mg/mL) |
|---|---|---|
| >30 kDa | 33.7 | 45.0 |
| 30–10 kDa | 2.48 | 3.9.0 |
| 10–3 kDa | 2.48 | 5.39 |
| Peptide | Localization | Molecular Mass | Charge | Hydrophobicity | Ip | Sequence |
|---|---|---|---|---|---|---|
| 1 | 1–10 | 1083.72 | 0.0 | −1.5 | 5.70 | VPLLLLGVLF |
| 2 | 11–18 | 823.46 | 0.0 | −0.9 | 5.70 | LASLSVSF |
| 3 | 19–41 | 2632.22 | −1.8 | 0.6 | 5.52 | GIVHRGHQESQEESEPRGQNNPF |
| 4 | 44–48 | 678.28 | −1.0 | 1.2 | 3.71 | DSDRW |
| 7 | 58–66 | 1125.66 | 2.1 | −0.2 | 12.50 | GHLRVLQRF |
| 8 | 67–79 | 1635.81 | 0.0 | 0.6 | 6.57 | DQRSKQIQNLENY |
| 9 | 80–84 | 649.37 | 0.0 | 0.1 | 6.36 | RVVEF |
| 10 | 85–101 | 1903.97 | −0.8 | 0.0 | 6.18 | QSKPNTLLLPHHADADF |
| 11 | 102–123 | 2398.35 | 0.0 | −0.3 | 6.05 | LLVVLNGRAILTLVNPDGRDSY |
| 12 | 124–139 | 1698.88 | 0.0 | −0.3 | 7.20 | ILEQGHAQKTPAGTTF |
| 13 | 141–165 | 2923.56 | 0.2 | 0.1 | 7.37 | LVNHDDNENLRIVKLAVPVNNPHRF |
| 14 | 170–179 | 1113.51 | −1.0 | −0.1 | 3.85 | LSSTEAQQSY |
| 15 | 180–183 | 464.25 | 0.0 | −1.0 | 5.70 | LQGF |
| 16 | 184–192 | 1008.54 | 0.0 | 0.0 | 5.91 | SKNILEASF |
| 17 | 193–196 | 483.17 | −2.0 | 1.0 | −0.01 | DSDF |
| 18 | 197–204 | 1018.60 | 1.0 | 0.2 | 9.00 | KEINRVLF |
| 19 | 205–252 | 5612.82 | −1.9 | 0.9 | 5.35 | GEEEQKQQDEESQQEGVIVQLKREQIRELMKHAKSTSKKSLSTQNEPF |
| 20 | 253–261 | 1118.63 | 2.0 | 0.1 | 10.30 | NLRSQKPIY |
| 22 | 266–285 | 2377.26 | −0.9 | 0.5 | 5.55 | GRLHEITPEKNPQLRDLDVF |
| 23 | 286–301 | 1749.91 | −1.0 | −0.2 | 4.13 | LTSVDIKEGGLLMPNY |
| 24 | 302–335 | 3893.02 | −2.0 | 0.2 | 4.69 | NSKAIVILVVNKGEANIELVGQREQQQQQQEESW |
| 25 | 336–340 | 694.35 | 0.0 | 0.5 | 6.36 | EVQRY |
| 26 | 341–350 | 1152.52 | −3.0 | 0.9 | 2.92 | RAEVSDDDVF |
| 27 | 351–368 | 1878.00 | 0.0 | −0.8 | 5.69 | VIPASYPVAITATSNLNF |
| 28 | 372–382 | 1276.60 | 0.0 | 0.2 | 6.36 | GINAENNQRNF |
| 29 | 383–422 | 4403.16 | −5.9 | 0.4 | 4.10 | LAGEEDNVMSEIPTEVLDVTFPASGEKVEKLINKQSDSHF |
| 30 | 423–433 | 1343.67 | 0.1 | 1.5 | 7.21 | TDHSSKREERV |
| Peptide | Localization | Molecular Mass | Charge | Hydrophobicity | Ip | Sequence |
|---|---|---|---|---|---|---|
| 1 | 19–41 | 2632.75 | −1.8 | 0.6 | 5.40 | GIVHRGHQESQEESEPRGQNNPF |
| 2 | 44–49 | 824.85 | −1.0 | 0.6 | 4.21 | DSDRWF |
| 3 | 54–60 | 886.97 | 1.1 | −0.2 | 8.75 | RNQYGHL |
| 4 | 67–76 | 1229.36 | 1.0 | 0.6 | 8.75 | DQRSKQIQNL |
| 5 | 77–84 | 1055.16 | −1.0 | 0.2 | 4.53 | ENYRVVEF - |
| 6 | 85–91 | 786.88 | 1.0 | 0.2 | 8.75 | QSKPNTL |
| 7 | 94–101 | 908.93 | −1.8 | 0.2 | 5.05 | PHHADADF |
| 8 | 107–112 | 642.76 | 1.0 | −0.1 | 9.75 | NGRAIL |
| 9 | 115–125 | 1248.36 | −1.0 | 0.2 | 4.21 | VNPDGRDSYIL |
| 10 | 126–139 | 1472.58 | 0.1 | 0.1 | 6.85 | EQGHAQKTPAGTTF |
| 11 | 142–150 | 1069.05 | −2.9 | 0.6 | 4.02 | VNHDDNENL |
| 12 | 151–155 | 627.83 | 2.0 | 0.2 | 11.00 | RIVKL |
| 13 | 156–165 | 1150.31 | 1.1 | −0.3 | 9.80 | AVPVNNPHRF |
| 14 | 171–180 | 1113.15 | −1.0 | −0.1 | 4.00 | SSTEAQQSYL |
| 16 | 197–203 | 871.05 | 1.0 | 0.6 | 8.75 | KEINRVL |
| 17 | 205–225 | 2430.52 | −6.0 | 0.9 | 3.77 | GEEEQKQQDEESQQEGVIVQL |
| 18 | 226–233 | 1071.24 | 1.0 | 1.4 | 8.75 | KREQIREL |
| 19 | 234–245 | 1345.62 | 4.1 | 0.7 | 10.48 | MKHAKSTSKKSL |
| 20 | 246–252 | 821.84 | −1.0 | 0.1 | 4.00 | STQNEPF |
| 21 | 255–265 | 1367.57 | 3.0 | 0.3 | 10.29 | RSQKPIYSNKF |
| 22 | 269–279 | 1305.45 | −0.9 | 0.4 | 5.40 | HEITPEKNPQL |
| 23 | 287–296 | 1018.13 | −1.0 | 0.4 | 4.37 | TSVDIKEGGL |
| 24 | 298–309 | 1362.65 | 1.0 | −0.6 | 8.34 | MPNYNSKAIVIL |
| 25 | 310–320 | 1185.34 | −1.0 | 0.2 | 4.53 | VVNKGEANIEL |
| 26 | 321–350 | 3697.85 | −5.0 | 0.7 | 4.12 | VGQREQQQQQQEESWEVQRYRAEVSDDDVF |
| 27 | 351–366 | 1616.87 | 0.0 | −0.8 | 5.49 | VIPASYPVAITATSNL |
| 28 | 372–382 | 1276.33 | 0.0 | 0.2 | 6.00 | GINAENNQRNF |
| 29 | 384–399 | 1732.88 | −5.0 | 0.4 | 3.45 | AGEEDNVMSEIPTEVL |
| 30 | 404–413 | 1057.21 | 0.0 | 0.9 | 6.56 | PASGEKVEKL |
| 31 | 414–422 | 1075.15 | 0.1 | 0.2 | 6.74 | INKQSDSHF |
| 32 | 423–437 | 1.343.42 | 0.1 | 1.5 | 6.43 | TDHSSKREERV |
| Strains | Samples | Chymotrypsin | Pepsin |
|---|---|---|---|
| Staphylococcus aureus | >30 kDa | 512 | 512.00 |
| 30–10 kDa | 512 | 512 | |
| 10–3 kDa | 512 | 512 | |
| Ciprofloxacin | 0.125 | 0.125 | |
| Pseudomonas aeruginosa | >30 kDa | 512 | 512 |
| 30–10 kDa | 512 | 512 | |
| 10–3 kDa | 512 | 512 | |
| Ciprofloxacin | 0.125 | 0.125 |
| Bacteria | Chymotrypsin | Pepsin |
|---|---|---|
| 250 µg/mL | ||
| Staphylococcus aureus | 9.00 | 11.11 |
| Pseudomonas aeruginosa | 10.80 | 9.99 |
| 500 µg/mL | ||
| Staphylococcus aureus | 11.00 | 12.10 |
| Pseudomonas aeruginosa | 10.90 | 13.66 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almeida, T.S.; da Cruz Souza, C.A.; de Cerqueira e Silva, M.B.; Batista, F.P.R.; Ferreira, E.S.; Santos, A.L.S.; Silva, L.N.; Melo, C.R.; Bani, C.; Bianconi, M.L.; et al. Extraction and Characterization of β-Viginin Protein Hydrolysates from Cowpea Flour as a New Manufacturing Active Ingredient. Technologies 2022, 10, 89. https://doi.org/10.3390/technologies10040089
Almeida TS, da Cruz Souza CA, de Cerqueira e Silva MB, Batista FPR, Ferreira ES, Santos ALS, Silva LN, Melo CR, Bani C, Bianconi ML, et al. Extraction and Characterization of β-Viginin Protein Hydrolysates from Cowpea Flour as a New Manufacturing Active Ingredient. Technologies. 2022; 10(4):89. https://doi.org/10.3390/technologies10040089
Chicago/Turabian StyleAlmeida, Taline S., Caio A. da Cruz Souza, Mariana B. de Cerqueira e Silva, Fabiana P. R. Batista, Ederlan S. Ferreira, André L. S. Santos, Laura N. Silva, Carlisson R. Melo, Cristiane Bani, M. Lucia Bianconi, and et al. 2022. "Extraction and Characterization of β-Viginin Protein Hydrolysates from Cowpea Flour as a New Manufacturing Active Ingredient" Technologies 10, no. 4: 89. https://doi.org/10.3390/technologies10040089
APA StyleAlmeida, T. S., da Cruz Souza, C. A., de Cerqueira e Silva, M. B., Batista, F. P. R., Ferreira, E. S., Santos, A. L. S., Silva, L. N., Melo, C. R., Bani, C., Bianconi, M. L., Cardoso, J. C., de Albuquerque-Júnior, R. L. C., de Melo Barbosa, R., Pereira, M. M., Souto, E. B., Soares, C. M. F., & Severino, P. (2022). Extraction and Characterization of β-Viginin Protein Hydrolysates from Cowpea Flour as a New Manufacturing Active Ingredient. Technologies, 10(4), 89. https://doi.org/10.3390/technologies10040089












