Differential Regulation of Phytoene Synthase PSY1 During Fruit Carotenogenesis in Cultivated and Wild Tomato Species (Solanum section Lycopersicon)
Abstract
:1. Introduction
2. Results
2.1. Identification and Structural Analysis of PSY1 Homologous Genes in Solanum Section Lycopersicon Species
2.2. Structural Analysis of Tomato PSY1 Homologs
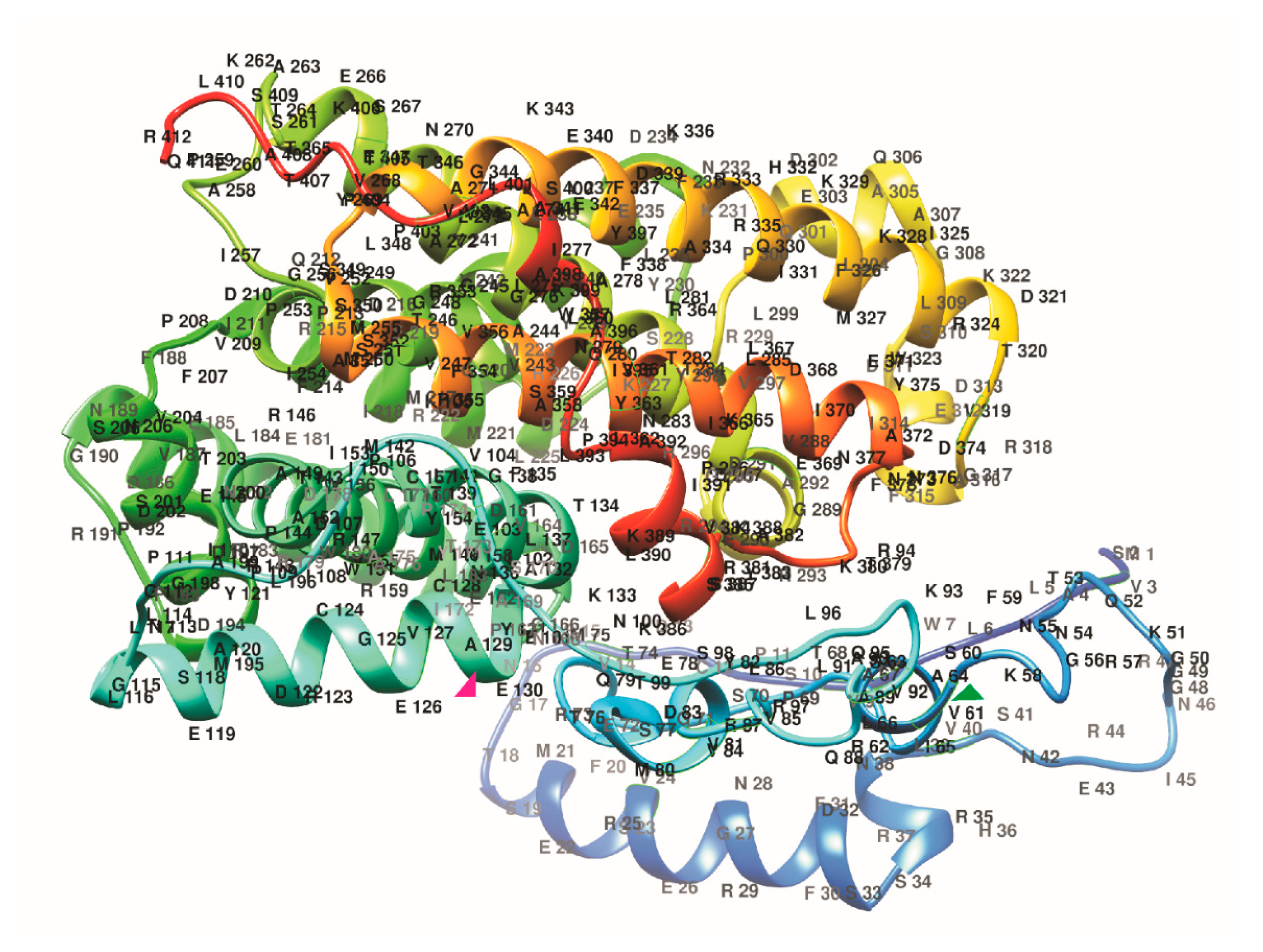
2.3. Spatial Structure of Tomato PSY1 Homologs
2.4. PSY1-Based Phylogeny of Tomato Species
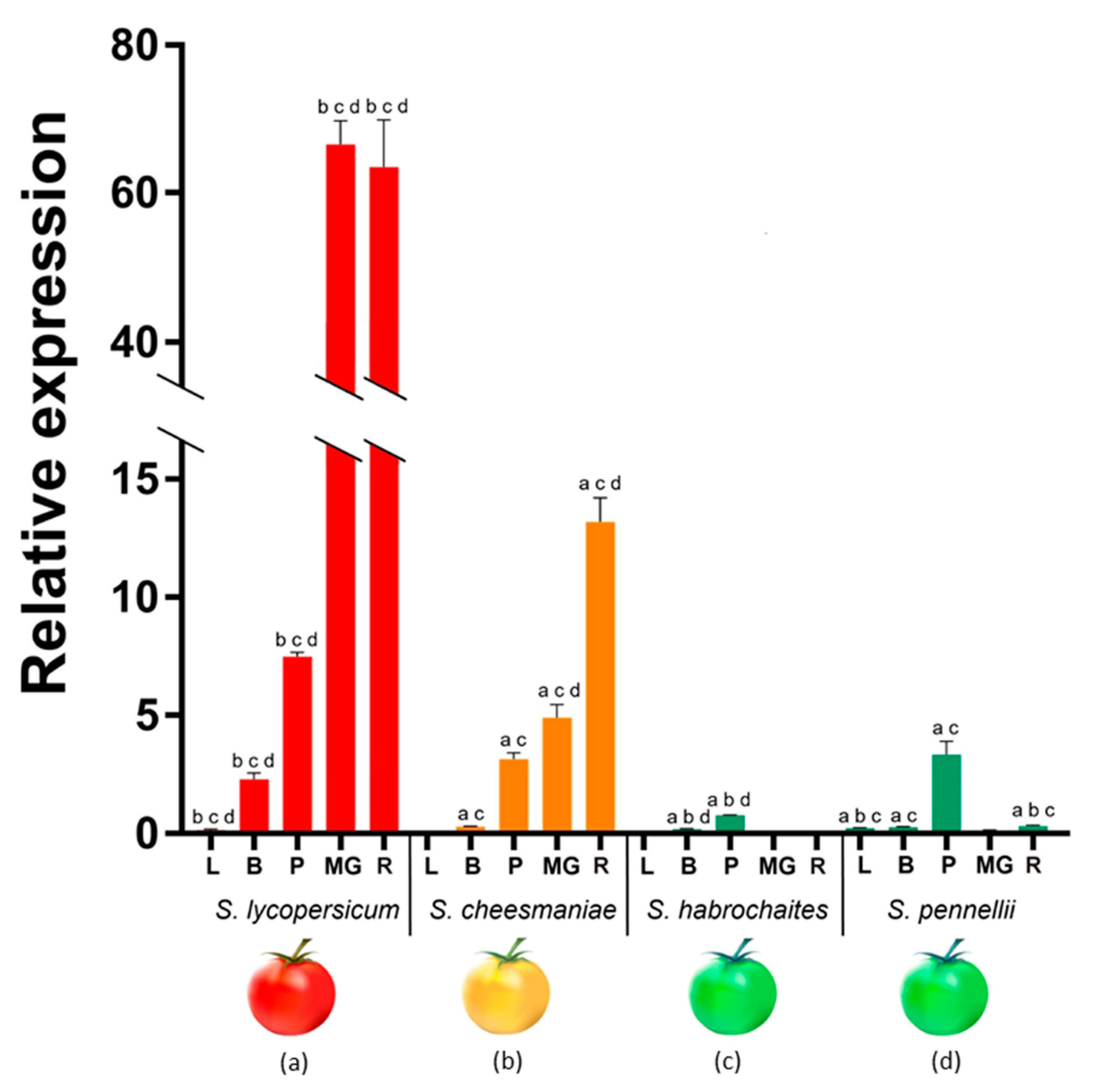
2.5. PSY1 Expression Pattern
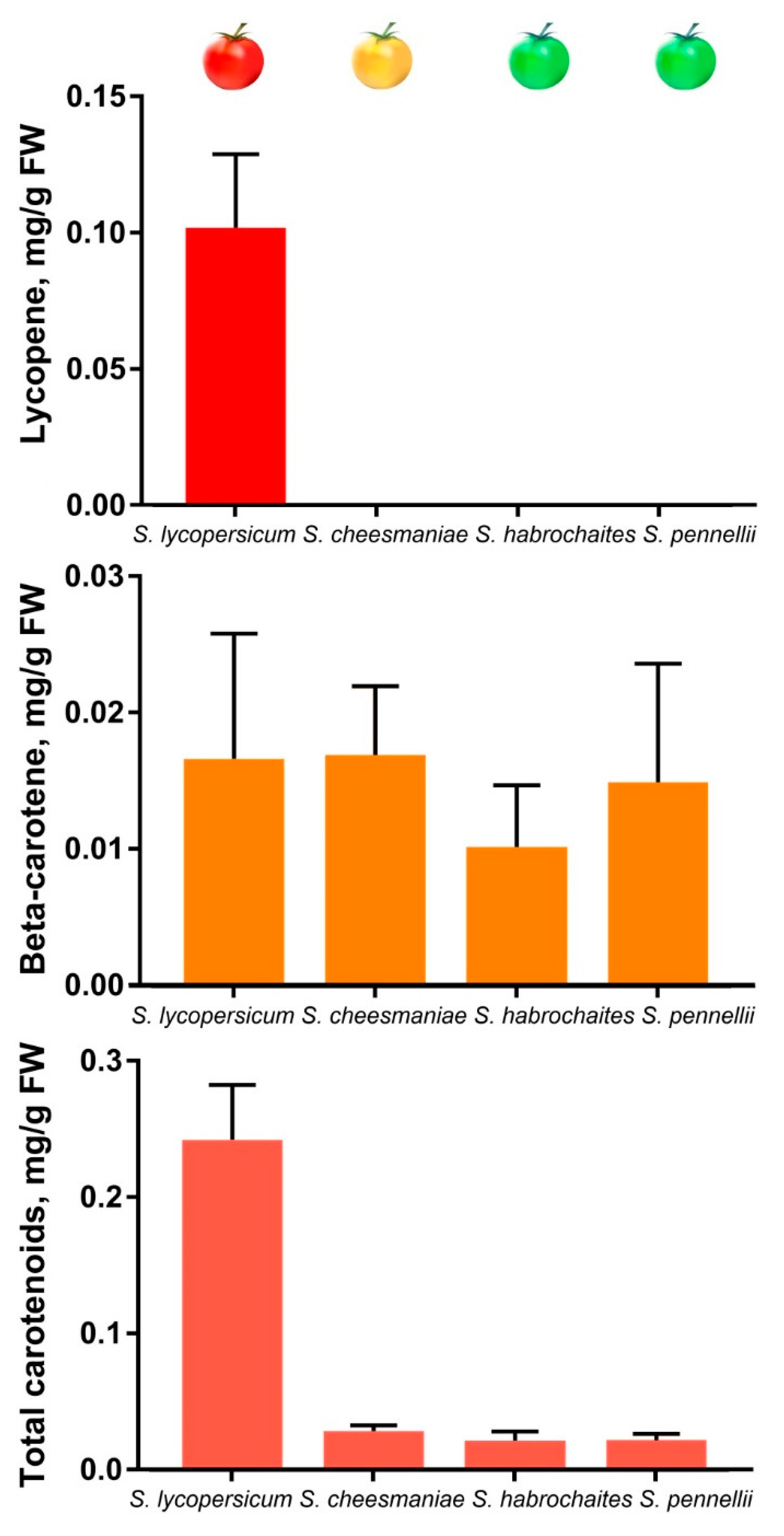
2.6. Carotenoid and Chlorophyll Content
2.7. Promoter and 5′-UTR Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Gene Identification
4.3. Structural and Phylogenetic Analysis
4.4. Gene Expression
4.5. Carotenoid and Chlorophyll Content
Chlorophyll b (µg/mL) = 21.85 (A648 − A750) − 4.53 (A666 − A750)
Total carotenoids (x + c) (µg/mL) = [1000 (A480 − A750) − 1.33 Chl a − 23.93 Chl b]/202
Lycopene (mg/100 mL) = 0.204 A645 − 0.0458 A663 + 0.372 A505 − 0.0806 A453
β-carotene (mg/100 mL) = 0.216 A663 − 1.22 A645 − 0.304 A505 + 0.452 A453
4.6. Promoter and 5′-UTR Analyses
- The search of specific cis-elements in promoters and 5′-UTRs was performed using the PlantCARE database, which provides an evaluation of cis-regulatory elements, enhancers, and repressors [79]; (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/; accessed May 31, 2020).
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yoo, H.J.; Park, W.J.; Lee, G.M.; Oh, C.S.; Yeam, I.; Won, D.C.; Kim, C.K.; Lee, J.M. Inferring the genetic determinants of fruit colors in tomato by carotenoid profiling. Molecules 2017, 22, 764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Shao, Z.; Zhang, M.; Wang, Q. Regulation of carotenoid metabolism in tomato. Mol. Plant 2015, 8, 28–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peralta, I.E.; Knapp, S.; Spooner, D.M. Nomenclature for wild and cultivated tomatoes. Rep. Tomato Genet. Coop. 2006, 56, 6–10. [Google Scholar]
- Peralta, I.E.; Spooner, D.M. History, origin and early cultivation of tomato (solanaceae). Genet. Improv. Solanaceous Crop. 2006, 2, 1–24. [Google Scholar]
- Galpaz, N.; Ronen, G.; Khalfa, Z.; Zamir, D.; Hirschberg, J. A chromoplast-specific carotenoid biosynthesis pathway is revealed by cloning of the tomato white-flower locus. Plant Cell 2006, 18, 1947–1960. [Google Scholar] [CrossRef] [Green Version]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [Green Version]
- Niyogi, K.K.; Truong, T.B. Evolution of flexible non-photochemical quenching mechanisms that regulate light harvesting in oxygenic photosynthesis. Curr. Opin. Plant Biol. 2013, 16, 307–314. [Google Scholar] [CrossRef]
- Al-babili, S.; Bouwmeester, H.J. Strigolactones, a novel plant hormone. Annu. Rev. Plant Biol. 2015, 66, 161–186. [Google Scholar] [CrossRef]
- Hashimoto, H.; Uragami, C.; Cogdell, R.J. Carotenoids and photosynthesis. Sub-Cell. Biochem. 2016, 79, 111–139. [Google Scholar] [CrossRef]
- Cao, H.; Luo, H.; Yuan, H.; Eissa, M.A.; Thannhauser, T.W.; Welsch, R.; Hao, Y.J.; Cheng, L.; Li, L. A neighboring aromatic-aromatic amino acid combination governs activity divergence between tomato phytoene synthases. Plant Physiol. 2019, 180, 1988–2003. [Google Scholar] [CrossRef] [Green Version]
- Giuliano, G. Plant carotenoids: Genomics meets multi-gene engineering. Curr. Opin. Plant Biol. 2014, 19, 111–117. [Google Scholar] [CrossRef] [Green Version]
- Giorio, G.; Stigliani, A.L.; D’Ambrosio, C. Phytoene synthase genes in tomato (Solanumlycopersicum L.)—New data on the structures, the deduced amino acid sequences and the expression patterns. FEBS J. 2008, 275, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Moise, A.R.; Al-Babili, S.; Wurtzel, E.T. Mechanistic aspects of carotenoid biosynthesis. Chem. Rev. 2014, 114, 164–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ducreux, L.J.M.; Morris, W.L.; Hedley, P.E.; Shepherd, T.; Davies, H.V.; Millam, S.; Taylor, M.A. Metabolic engineering of high carotenoid potato tubers containing enhanced levels of β-carotene and lutein. J. Exp. Bot. 2005, 56, 81–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Li, W.; Li, Y.; Feng, X.; Du, K.; Wang, G.; Zhao, L. Identified trans-splicing of YELLOW-FRUITED TOMATO 2 encoding the PHYTOENE SYNTHASE 1 protein alters fruit color by map-based cloning, functional complementation and RACE. Plant Mol. Biol. 2019, 100, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Welsch, R.; Yang, Y.; Álvarez, D.; Riediger, M.; Yuan, H.; Fish, T.; Liu, J.; Thannhauser, T.W.; Li, L. Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. Proc. Natl. Acad. Sci. USA 2015, 112, 3558–3563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallagher, C.E.; Matthews, P.D.; Li, F.; Wurtzel, E.T. Gene duplication in the carotenoid biosynthetic pathway preceded evolution of the grasses. Plant Physiol. 2004, 135, 1776–1783. [Google Scholar] [CrossRef] [Green Version]
- Fu, X.; Feng, C.; Wang, C.; Yin, X.; Lu, P.; Grierson, D.; Xu, C.; Chen, K. Involvement of multiple phytoene synthase genes in tissue- and cultivar-specific accumulation of carotenoids in loquat. J. Exp. Bot. 2014, 65, 4679–4689. [Google Scholar] [CrossRef] [Green Version]
- Bartley, G.E.; Viitanen, P.V.; Bacot, K.O.; Scolnik, P.A. A tomato gene expressed during fruit ripening encodes an enzyme of the carotenoid biosynthesis pathway. J. Biol. Chem. 1992, 267, 5036–5039. [Google Scholar]
- Bartley, G.E.; Scolnik, P.A. cDNA cloning, expression during development and genome mapping of PSY2, a second tomato gene encoding phytoene synthase. J. Biol. Chem. 1993, 268, 25718–25721. [Google Scholar]
- Sato, S.; Tabata, S.; Hirakawa, H.; Asamizu, E.; Shirasawa, K.; Isobe, S.; Kaneko, T.; Nakamura, Y.; Shibata, D.; Aoki, K.; et al. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef] [Green Version]
- Fraser, P.D.; Schuch, W.; Bramley, P.M. Phytoene synthase from tomato (Lycopersicon esculentum) chloroplasts—Partial purification and biochemical properties. Planta 2000, 211, 361–369. [Google Scholar] [CrossRef]
- Stauder, R.; Welsch, R.; Camagna, M.; Kohlen, W.; Balcke, G.U.; Tissier, A.; Walter, M.H. Strigolactone levels in dicot roots are determined by an ancestral symbiosis-regulated clade of the PHYTOENE SYNTHASE gene family. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Vallabhaneni, R.; Wurtzel, E.T. PSY3, a new member of the phytoene synthase gene family conserved in the poaceae and regulator of abiotic stress-induced root carotenogenesis. Plant Physiol. 2008, 146, 1333–1345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dibari, B.; Murat, F.; Chosson, A.; Gautier, V.; Poncet, C.; Lecomte, P.; Mercier, I.; Bergès, H.; Pont, C.; Blanco, A.; et al. Deciphering the genomic structure, function and evolution of carotenogenesis related phytoene synthases in grasses. BMC Genom. 2012, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahrazem, O.; Diretto, G.; Argandoña Picazo, J.; Fiore, A.; Rubio-Moraga, Á.; Rial, C.; Varela, R.M.; Macías, F.A.; Castillo, R.; Romano, E.; et al. The specialized roles in carotenogenesis and apocarotenogenesis of the phytoene synthase gene family in saffron. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arango, J.; Wüst, F.; Beyer, P.; Welsch, R. Characterization of phytoene synthases from cassava and their involvement in abiotic stress-mediated responses. Planta 2010, 232, 1251–1262. [Google Scholar] [CrossRef]
- Costa, M.G.C.; Moreira, C.D.; Melton, J.R.; Otoni, W.C.; Moore, G.A. Characterization and developmental expression of genes encoding the early carotenoid biosynthetic enzymes in Citrus paradisi Macf. Mol. Biol. Rep. 2012, 39, 895–902. [Google Scholar] [CrossRef]
- Soltis, D.E.; Albert, V.A.; Leebens-Mack, J.; Palmer, J.D.; Wing, R.A.; DePamphilis, C.W.; Ma, H.; Carlson, J.E.; Altman, N.; Kim, S.; et al. The Amborella genome: An evolutionary reference for plant biology. Genome Biol. 2008, 9. [Google Scholar] [CrossRef] [Green Version]
- Giovannoni, J.; Nguyen, C.; Ampofo, B.; Zhong, S.; Fei, Z. The epigenome and transcriptional dynamics of fruit ripening. Annu. Rev. Plant Biol. 2017, 68, 61–84. [Google Scholar] [CrossRef]
- Meléndez-Martínez, A.J.; Fraser, P.D.; Bramley, P.M. Accumulation of health promoting phytochemicals in wild relatives of tomato and their contribution to in vitro antioxidant activity. Phytochemistry 2010, 71, 1104–1114. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.; Scossa, F.; Bolger, M.E.; Lanz, C.; Maumus, F.; Tohge, T.; Quesneville, H.; Alseekh, S.; Sørensen, I.; Lichtenstein, G.; et al. The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat. Genet. 2014, 46, 1034–1038. [Google Scholar] [CrossRef] [PubMed]
- Kilambi, H.V.; Manda, K.; Rai, A.; Charakana, C.; Bagri, J.; Sharma, R.; Sreelakshmi, Y. Green-fruited Solanum habrochaites lacks fruit-specific carotenogenesis due to metabolic and structural blocks. J. Exp. Bot. 2017, 68, 4803–4819. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Zhang, J.; Li, J.; Yang, C.; Wang, T.; Ouyang, B.; Li, H.; Giovannoni, J.; Ye, Z. A STAY-GREEN protein SlSGR1 regulates lycopene and $β$-carotene accumulation by interacting directly with SlPSY1 during ripening processes in tomato. New Phytol. 2013, 198, 442–452. [Google Scholar] [CrossRef]
- Osorio, C.E. The role of orange gene in carotenoid accumulation: Manipulating chromoplasts toward a colored future. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Tanksley, S.D.; McCouch, S.R. Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 1997, 277, 1063–1066. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Zhao, W.; Qu, H.; Wang, Q.; Zhao, L. The yellow-fruited tomato 1 (yft1) mutant has altered fruit carotenoid accumulation and reduced ethylene production as a result of a genetic lesion in ETHYLENE INSENSITIVE2. Theor. Appl. Genet. 2016, 129, 717–728. [Google Scholar] [CrossRef]
- Blanca, J.; Cañizares, J.; Cordero, L.; Pascual, L.; Diez, M.J.; Nuez, F. Variation revealed by SNP genotyping and morphology provides insight into the origin of the Tomato. PLoS ONE 2012, 7, e48198. [Google Scholar] [CrossRef] [Green Version]
- Mata-Nicolás, E.; Montero-Pau, J.; Gimeno-Paez, E.; Garcia-Carpintero, V.; Ziarsolo, P.; Menda, N.; Mueller, L.A.; Blanca, J.; Cañizares, J.; van der Knaap, E.; et al. Exploiting the diversity of tomato: The development of a phenotypically and genetically detailed germplasm collection. Hortic. Res. 2020, 7, 66. [Google Scholar] [CrossRef]
- Bai, Y.; Lindhout, P. Domestication and breeding of tomatoes: What have we gained and what can we gain in the future? Ann. Bot. 2007, 100, 1085–1094. [Google Scholar] [CrossRef]
- Sim, S.C.; Robbins, M.D.; Van Deynze, A.; Michel, A.P.; Francis, D.M. Population structure and genetic differentiation associated with breeding history and selection in tomato (Solanum lycopersicum L.). Heredity 2011, 106, 927–935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The 100 Tomato Genome Sequencing Consortium. Exploring genetic variation in the tomato (Solanum section Lycopersicon) clade by whole-genome sequencing. Plant J. 2014, 80, 136–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, M.H.; Zhu, J.; Jiang, J.G. Carotenoids biosynthesis and cleavage related genes from bacteria to plants. Crit. Rev. Food Sci. Nutr. 2018, 58, 2314–2333. [Google Scholar] [CrossRef]
- Shumskaya, M.; Bradbury, L.M.T.; Monaco, R.R.; Wurtzela, E.T. Plastid localization of the key carotenoid enzyme phytoene synthase is altered by isozyme, allelic variation and activity. Plant Cell 2011, 24, 3725–3741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Silva Mendes, A.F.; Fontes Soares, V.n.L.c.; Cardoso Costa, M.G. Carotenoid biosynthesis genomics. Pigment. Fruits Veg. 2015, 9–29. [Google Scholar]
- Särkinen, T.; Bohs, L.; Olmstead, R.G.; Knapp, S. A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): A dated 1000-tip tree. BMC Evol. Biol. 2013, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peralta, I.E.; Spooner, D.M.; Knapp, S. Taxonomy of wild tomatoes and their relatives (Solanum sect. Lycopersicoides, sect. Juglandifolia, sect. Lycopersicon; Solanaceae). Syst. Bot. Monogr. 2008, 84, 1–186. [Google Scholar]
- Ronen, G.; Carmel-Goren, L.; Zamir, D.; Hirschberg, J. An alternative pathway to $β$-carotene formation in plant chromoplasts discovered by map-based cloning of Beta and old-gold color mutations in tomato. Proc. Natl. Acad. Sci. USA 2000, 97, 11102–11107. [Google Scholar] [CrossRef] [Green Version]
- Paran, I.; Van Der Knaap, E. Genetic and molecular regulation of fruit and plant domestication traits in tomato and pepper. J. Exp. Bot. 2007, 58, 3841–3852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palaisa, K.A.; Morgante, M.; Williams, M.; Rafalski, A. Contrasting effects of selection on sequence diversity and linkage disequilibrium at two phytoene synthase loci. Plant Cell 2003, 15, 1795–1806. [Google Scholar] [CrossRef] [Green Version]
- Ye, X.; Beyer, P. Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 2000, 287, 303–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lätari, K.; Wüst, F.; Hübner, M.; Schaub, P.; Beisel, K.G.; Matsubara, S.; Beyer, P.; Welsch, R. Tissue-specific apocarotenoid glycosylation contributes to carotenoid homeostasis in Arabidopsis leaves. Plant Physiol. 2015, 168, 1550–1562. [Google Scholar] [CrossRef] [PubMed]
- Fraser, P.D.; Enfissi, E.M.A.; Halket, J.M.; Truesdale, M.R.; Yu, D.; Gerrish, C.; Bramley, P.M. Manipulation of phytoene levels in tomato fruit: Effects on isoprenoids, plastids and intermediary metabolism. Plant Cell 2007, 19, 4131–4132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Andrea, L.; Rodriguez-Concepcion, M. Manipulation of plastidial protein quality control components as a new strategy to improve carotenoid contents in Tomato fruit. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Romer, S.; Hugueney, P.; Bouvier, F.; Camara, B.; Kuntz, M. Expression of the genes encoding the early carotenoid biosynthetic enzymes in Capsicum annuum. Biochem. Biophys. Res. Commun. 1993, 196, 1414–1421. [Google Scholar] [CrossRef]
- Zhang, W.; Dubcovsky, J. Association between allelic variation at the Phytoene synthase 1 gene and yellow pigment content in the wheat grain. Theor. Appl. Genet. 2008, 116, 635–645. [Google Scholar] [CrossRef] [Green Version]
- Welsch, R.; Arango, J.; Bär, C.; Salazar, B.; Al-Babili, S.; Beltrán, J.; Chavarriaga, P.; Ceballos, H.; Tohme, J.; Beyer, J. Provitamin A accumulation in cassava (Manihot esculenta) roots driven by a single nucleotide polymorphism in a phytoene synthase gene. Plant Cell 2010, 22, 3348–3356. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Reimer, S.; Pozniak, C.J.; Clarke, F.R.; Clarke, J.M.; Knox, R.E.; Singh, A.K. Allelic variation at Psy1-A1 and association with yellow pigment in durum wheat grain. Theor. Appl. Genet. 2009, 118, 1539–1548. [Google Scholar] [CrossRef]
- Qin, X.; Coku, A.; Inoue, K.; Tian, L. Expression, subcellular localization and cis-regulatory structure of duplicated phytoene synthase genes in melon (Cucumis melo L.). Planta 2011, 234, 737–748. [Google Scholar] [CrossRef]
- Dong, T.; Park, Y.; Hwang, I. Abscisic acid: Biosynthesis, inactivation, homoeostasis and signalling. Essays Biochem. 2015, 58, 29–48. [Google Scholar] [CrossRef]
- Bürger, M.; Chory, J. Stressed Out about hormones: How plants orchestrate immunity. Cell Host Microbe 2019, 26, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Fraser, P.D.; Truesdale, M.R.; Bird, C.R.; Schuch, W.; Bramley, P.M. Carotenoid biosynthesis during tomato fruit development. Evidence for tissue-specific gene expression. Plant Physiol. 1994, 105, 405–413. [Google Scholar] [CrossRef] [Green Version]
- Grumet, R.; Fobes, J.F.; Herner, R.C. Ripening behavior of wild tomato species. Plant Physiol. 1981, 68, 1428–1432. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, C.; Nafati, M.; Mathieu-Rivet, E.; Bourdon, M.; Frangne, N.; Cheniclet, C.; Renaudin, J.P.; Gvaudant, F.; Hernould, M. Elucidating the functional role of endoreduplication in tomato fruit development. Ann. Bot. 2011, 107, 1159–1169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puchooa, D. A simple, rapid and efficient method for the extraction of genomic DNA from lychee (Litchi chinensis Sonn.). Afr. J. Biotechnol. 2004, 3, 253–255. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0. molecular biology and evolution. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Kozlowski, L.P. IPC—Isoelectric Point Calculator. Biol. Direct 2016, 11, 55. [Google Scholar] [CrossRef] [Green Version]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.; Sims, G.E.; Murphy, S.; Miller, J.R.; Chan, A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 2012, 7, e46688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar] [PubMed]
- Expósito-Rodríguez, M.; Borges, A.A.; Borges-Pérez, A.; Pérez, J.A. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 2008, 8, 131. [Google Scholar] [CrossRef] [Green Version]
- Bemer, M.; Karlova, R.; Ballester, A.R.; Tikunov, Y.M.; Bovy, A.G.; Wolters-Arts, M.; de Barros Rossetto, P.; Angenent, G.C.; de Maagd, R.A. The tomato fruitfull homologs tdr4/ful1 and mbp7/ful2 regulate ethylene-independent aspects of fruit ripening. Plant Cell 2012, 24, 4437–4451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folch, J.; Lees, M.; Sloane, G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1953, 226, 497–509. [Google Scholar]
- Solovchenko, A.E.; Chivkunova, O.B.; Merzlyak, M.N.; Reshetnikova, I.V. A spectrophotometric analysis of pigments in apples. Russ. J. Plant Physiol. 2001, 48, 693–700. [Google Scholar] [CrossRef]
- Nagata, M.; Yamashita, I. A simple method for simultaneous determination of chlorophyll and carotenoids in tomato fruit. J. Jpn. Soc. Food Sci. Technol. 1992, 39, 925–928. [Google Scholar] [CrossRef] [Green Version]
- Meléndez-Martínez, A.J.; Mapelli-Brahm, P.; Benítez-González, A.; Stinco, C.M. A comprehensive review on the colorless carotenoids phytoene and phytofluene. Arch. Biochem. Biophys. 2015, 572, 188–200. [Google Scholar] [CrossRef] [Green Version]
- Rombauts, S.; Déhais, P.; Van Montagu, M.; Rouzé, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef] [Green Version]




| Accession | TGRC Collection Number | Origin | Ripe Fruit Color * | NCBI Gene ID/Solyc No. | Gene, bp | cDNA, bp | Protein, aa | pI | MW, kDa |
|---|---|---|---|---|---|---|---|---|---|
| PSY1 genes identified in this study | |||||||||
| S. lycopersicum L. cv. Heinz 1706-BG (Lycopersicon group) | LA4345 | Red | MT664042 | 4871 | 1239 | 412 | 8.1 | 46.6 | |
| S. cheesmaniae (L. Riley) Fosberg (Esculenthum group/Lycopersicon group) | LA0421 | San Cristobal: cliff East of Wreck Bay, Galapagos Islands, Ecuador | Yellow | MN782521 MN782522 | 4883/4876 | 1239 | 412 | 7.74/7.9 | 46.6/45.5 |
| S. chilense (Dunal) Reiche (Peruvianum group/Eriopersicon group) | LA1963 | Rio Caplina, Tacna, Peru | Green to whitish green with purple stripes | MN782523 MN812838 | 4858/4840 | 1239 | 412 | 7.74/7.29 | 46.6/46.3 |
| LA2884 | Ayaviri, Antofagasta, Chile | MN782524 MN782525 | 4878/4876 | 1239 | 412 | 7.93/7.90 | 46.5/46.5 | ||
| S. habrochaites S. Knapp & D. M. Spooner (Hirsutum group/Eriopersicon group) | LA1771 | Rio Casma, Ancash, Peru | Green with darker green stripes | MN782526 MN782527 | 4914/4916 | 1239 | 412 | 8.24 | 46.6 |
| LA2144 | Chanchan, Chimborazo, Ecuador | MN782528 MN782529 | 4906/4903 | 1239 | 412 | 8.1 | 46.6 | ||
| S. pennellii Correll (Hirsutum group/Neolycopersicon group) | LA1926 | Agua Pertida, Ica, Peru | Green | MN782530 MN782531 | 4898/4901 | 1239 | 412 | 8.5/8.1 | 46.7/46.6 |
| LA0716 | Atico, Arequipa, Peru | MN782532 | 4886 | 1239 | 412 | 8.1 | 46.5 | ||
| PSY1 genes available in NCBI GenBank | |||||||||
| S. lycopersicum cv. Heinz 1706 | Red | 543988, NC_015440.3 (4350836..4355976); Solyc03g031860.2.1 | 4872 | 1239 | 412 | 8.1 | 46.6 | ||
| S. pimpinellifolium | LA1589 | La Libertad, Peru | Red | AGFK01024844.1 (1289..7095) | 4872 | 1239 | 412 | 8.1 | 46.6 |
| S. arcanum | LA2157 | Tunel Chotano, Cajamarica, Peru | Green | CBYQ010012533.1 (26020..31886) | 4880 | 1239 | 412 | 8.1 | 46.6 |
| S. habrochaites | LYC4 | Peru/Ecuador | Green with dark-green stripes | CBYS010011028.1 (46212..52055) | 4910 | 1239 | 412 | 8.1 | 46.6 |
| S. pennellii | LA0716 | Atico, Arequipa, Peru | Green | CCXL01009615.1 (3669..9559) | 4949 | 1239 | 412 | 8.1 | 46.6 |
| Accession | Ripe Fruit Pigment Content, µg/g FW | Leaf Pigment Content, µg/g FW | |||||
|---|---|---|---|---|---|---|---|
| Chlorophyll (a + b) | Lycopene | Total Carotenoids (x + c) | β-carotene | Other x + c (-β-carotene) | Total Carotenoids (x + c) | Chlorophyll (a + b) | |
| Solanum lycopersicum cv. Heinz 1706 | N/D | 0.09 ± 0.02 | 0.24 ± 0.030 | 0.01 ± 0.008 | ~0.23 | 0.94 ± 0.098 | 3.19 ± 0.390 |
| Solanum cheesmaniae LA 0421 | N/D | N/D | 0.03 ± 0.004 | 0.02 ± 0.003 | ~0.01 | 0.96 ± 0.210 | 3.33 ± 0.820 |
| Solanum chilense LA 1963 | 0.03 ± 0.004 | N/D | 0.01 ± 0.004 | 0.01 ± 0.001 | ~0.00 | 0.89 ± 0.070 | 2.78 ± 0.260 |
| Solanum habrochaites LA 2144 | 0.06 ± 0.010 | N/D | 0.02 ± 0.006 | 0.01 ± 0.003 | ~0.00 | 0.93 ± 0.004 | 3.15 ± 0.060 |
| Solanum pennellii LA 0716 | 0.09 ± 0.050 | N/D | 0.02 ± 0.005 | 0.02 ± 0.004 | ~0.00 | 0.96 ± 0.110 | 3.51 ± 0.400 |
| No | Type | Sequence | S. lycopersicum cv. Heinz 1706 | S. pennellii LA0716 | Comments | ||
|---|---|---|---|---|---|---|---|
| Strand | Position | Strand | Position | ||||
| 1 | chs-Unit 1 m1 | n/d | - | −1110 | Part of a light responsive element | ||
| 2 | Box II | ACACGTAGA | - | −1713 | - | −1735 | |
| 3 | GATA-motif | AAGGATAAGG | + | −2207 | n/d | ||
| 4 | GTGGC-motif | GATTCTGTGGC | + | −564 | + | −566 | |
| 5 | TCT-motif | n/d | + | −2419 | |||
| 6 | AE-box | AGAAACAA | - | −528 | + | −530 | Part of a conserved DNA module involved in light response |
| 7 | Box 4 | ATTAAT | + | −2373 | + | −2402 | |
| + | −1685 | + | −1707 | ||||
| 8 | GA-motif | ATAGATAA | n/d | - | −2143 | ||
| 9 | I-box | TAGATAACC | n/d | + | −28 | ||
| 10 | 3-AF3 binding site | CACTATCTAAC | + | −2321 | n/d | Light response | |
| 11 | GT1-motif | GGTTAA | + | −1101 | + | −1102 | |
| n/d | + | −1079 | |||||
| 12 | ABRE | TACGTGTC | + | −1711 | + | −1733 | The binding sites for AREB/ABF factors involved in the abscisic acid response |
| ACGTG | + | −675 | + | −677 | |||
| + | −1710 | + | −1732 | ||||
| 13 | ABRE3a | TACGTG | + | −1711 | + | −1733 | |
| + | −676 | + | −678 | ||||
| 14 | ABRE4 | CACGTA | - | −1711 | - | −1733 | |
| - | −676 | - | −678 | ||||
| 15 | AT~ABRE | TACGTGTC | + | −1711 | + | −1733 | |
| 16 | ERE | ATTTTAAA | + | −1272 | n/d | Ethylene-response | |
| 17 | TGA-element | AACGAC | + | −2241 | + | −2270 | Auxin-response |
| + | −2219 | + | −2248 | ||||
| 18 | CGTCA-motif | CGTCA | - | −1846 | - | −1876 | MeJA response |
| 19 | STRE | AGGGG | + | −1384 | n/d | Defense and stress response | |
| - | −1459 | ||||||
| + | −712 | + | −714 | ||||
| 20 | TC-rich repeats | GTTTTCTTAC | + | −16 | + | −16 | |
| 21 | ARE | AAACCA | - | −1988 | n/d | Essential for the anaerobic induction | |
| + | −1964 | ||||||
| - | −265 | - | −265 | ||||
| + | −1527 | + | −1560 | ||||
| - | −204 | - | −204 | ||||
| - | −682 | - | −684 | ||||
| 22 | WUN-motif | AAATTACT | - | −102 | - | −102 | Wounding response |
| 23 | CAT-box | GCCACT | - | −1496 | - | −1495 | Related to meristem-specific expression |
| - | −1221 | n/d | |||||
| n/d | - | −1528 | |||||
| 24 | AT~TATA-box | TATATA | + | −1391 | n/d | Enriched near transcription start. TATA-box-like, putative TBP-binding | |
| - | −973 | ||||||
| - | −1007 | - | −1007 | ||||
| - | −1159 | - | −1164 | ||||
| 25 | CAAT-box | CAAT/CAAAT | 43 repeats | 42 repeats | Common cis-acting element in promoter and enhancer | ||
| 26 | TATA-box | TATAAAAT; TATAAATA; TATAAAT; TATAAA; TATAA; TATA | multiple repeats | multiple repeats | Core promoter element | ||
| 27 | W box | TTGACC | - | −1894 | - | −1924 | WRKY TF binding site |
| 28 | MBS | CAACTG | n/d | - | −1825 | MYB TF binding site; drought response | |
| 29 | MRE | AACCTAA | - | −2275 | - | −2286 | MYB TF binding site; light response |
| n/d | - | −580 | |||||
| + | −2304 | ||||||
| 30 | MYB | TAACTG | + | −551 | + | −553 | MYB TF binding site |
| n/d | - | −1825 | |||||
| + | −289 | n/d | |||||
| CAACAG | - | −517 | - | −519 | |||
| n/d | - | −1538 | |||||
| CAACCA | + | −24 | n/d | ||||
| TAACCA | - | −309 | - | −311 | |||
| n/d | + | −24 | |||||
| 31 | MYC | CATGTG | - | −787 | - | −788 | |
| - | −278 | - | −278 | ||||
| 32 | AS-1 (activation sequence-1) | TGACG | + | −1846 | + | −1876 | Originally found in some viral and bacterial T-DNA promoters. Pathogen-inducible |
| 33 | AT1-motif | AATTATTTTTTATT | - | −2111 | n/d | Binding site of AT-rich DNA binding protein (ATBP-1) | |
| 34 | G-box | TACGTG | + | −1711 | + | −1733 | Multifunctional |
| + | −676 | + | −678 | ||||
| 35 | Unnamed__2 | AACCTAACCT | - | −1107 | n/d | Unknown function | |
| 36 | Unnamed__4 | CTCC | 13 repeats | 13 repeats | |||
| 37 | AAGAA-motif | GAAAGAA | n/d | + | −1515 | ||
| Primer | Sequence (5′→3’) | Application |
|---|---|---|
| PSY1geneF | AGTGGGAATCTACTAGGAGT | Gene amplification and sequencing |
| PSY1geneR | TTATCTTTGAAGAGAGACAGTTT | |
| tPSY_F6 | CTATCTGGGCAATATATGGTG | Gene sequencing |
| tPSY_F7 | TCTCGTCCTAGATACTACAC | |
| tPSY_F8 | CAGTGACGAGCCATGATC | |
| tPSY_F9 | TTGAGCTTGTCGTTCTCAGT | |
| PSY1rtF | CTGAGATCTACCAATGAGTTAG | qRT-PCR, PSY1 |
| PSY1rtR | TCTCGGGAGTCATTAGCATAG | |
| prPSY1-F | GTTGGATTTGCATGTAGACC | Promoter/5′-UTR amplification and sequencing |
| tPSY_F2 | GGCTAAATCGAAAATYGAATC | |
| tPSY_F3 | TAACTTTCTATTGCTTTGCTAGTG | |
| tPSY_F4 | TGGTAGGTAATATTGCTGATTTTG | |
| Actin 2/7-F | CATTGTGCTCAGTGGTGGTTC | qRT-PCR, reference genes |
| Actin 2/7F-R | TCTGCTGGAAGGTGCTAAGTG | |
| Expressed-F | GCTAAGAACGCTGGACCTAATG | |
| Expressed-R | TGGGTGTGCCTTTCTGAATG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Efremov, G.I.; Slugina, M.A.; Shchennikova, A.V.; Kochieva, E.Z. Differential Regulation of Phytoene Synthase PSY1 During Fruit Carotenogenesis in Cultivated and Wild Tomato Species (Solanum section Lycopersicon). Plants 2020, 9, 1169. https://doi.org/10.3390/plants9091169
Efremov GI, Slugina MA, Shchennikova AV, Kochieva EZ. Differential Regulation of Phytoene Synthase PSY1 During Fruit Carotenogenesis in Cultivated and Wild Tomato Species (Solanum section Lycopersicon). Plants. 2020; 9(9):1169. https://doi.org/10.3390/plants9091169
Chicago/Turabian StyleEfremov, Gleb I., Maria A. Slugina, Anna V. Shchennikova, and Elena Z. Kochieva. 2020. "Differential Regulation of Phytoene Synthase PSY1 During Fruit Carotenogenesis in Cultivated and Wild Tomato Species (Solanum section Lycopersicon)" Plants 9, no. 9: 1169. https://doi.org/10.3390/plants9091169
APA StyleEfremov, G. I., Slugina, M. A., Shchennikova, A. V., & Kochieva, E. Z. (2020). Differential Regulation of Phytoene Synthase PSY1 During Fruit Carotenogenesis in Cultivated and Wild Tomato Species (Solanum section Lycopersicon). Plants, 9(9), 1169. https://doi.org/10.3390/plants9091169




