Gene Expression Profiles and Flavonoid Accumulation during Salt Stress in Ginkgo biloba Seedlings
Abstract
:1. Introduction
2. Results
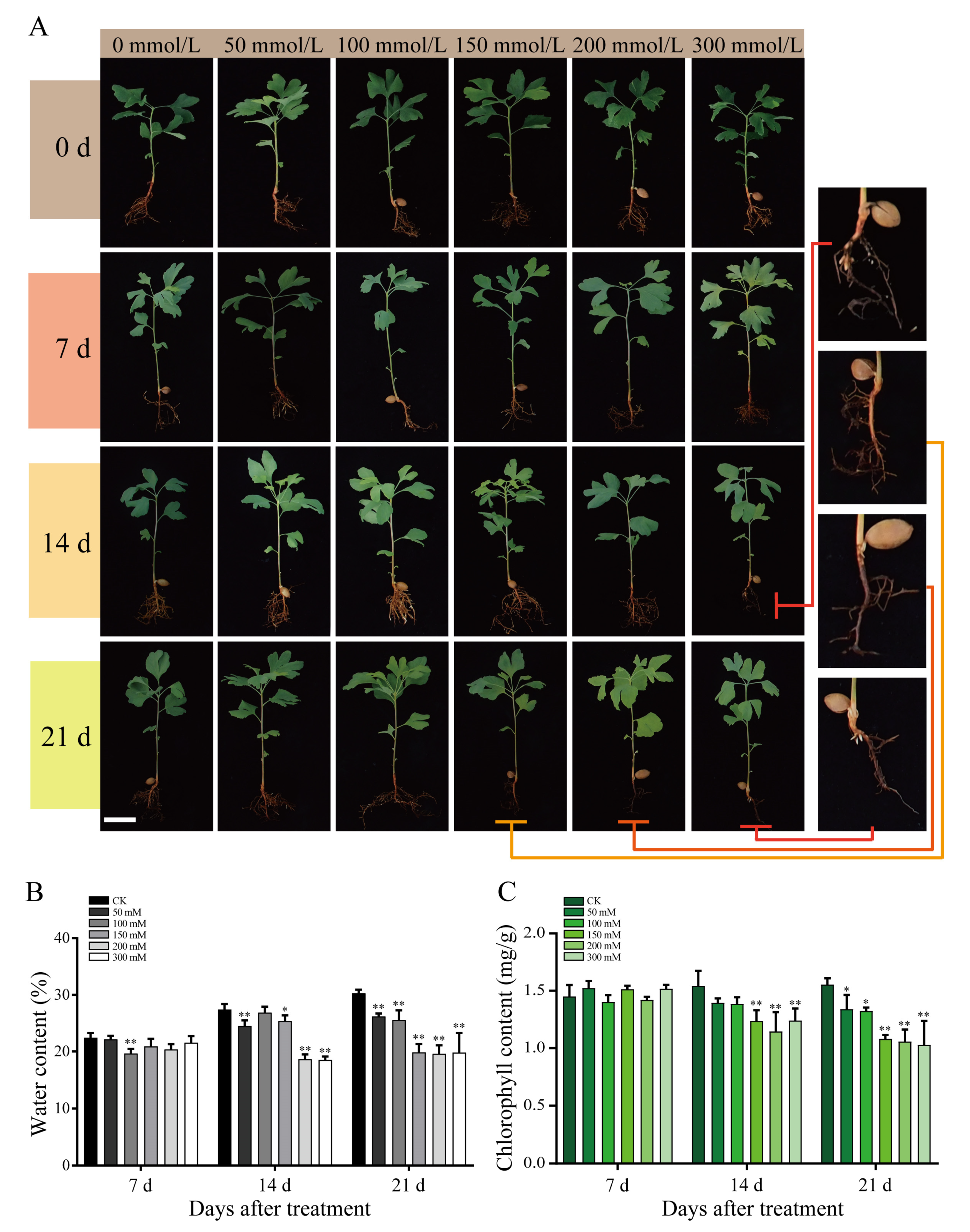
2.1. Salt Treatment-Induced Physiological Changes in G. biloba Leaves
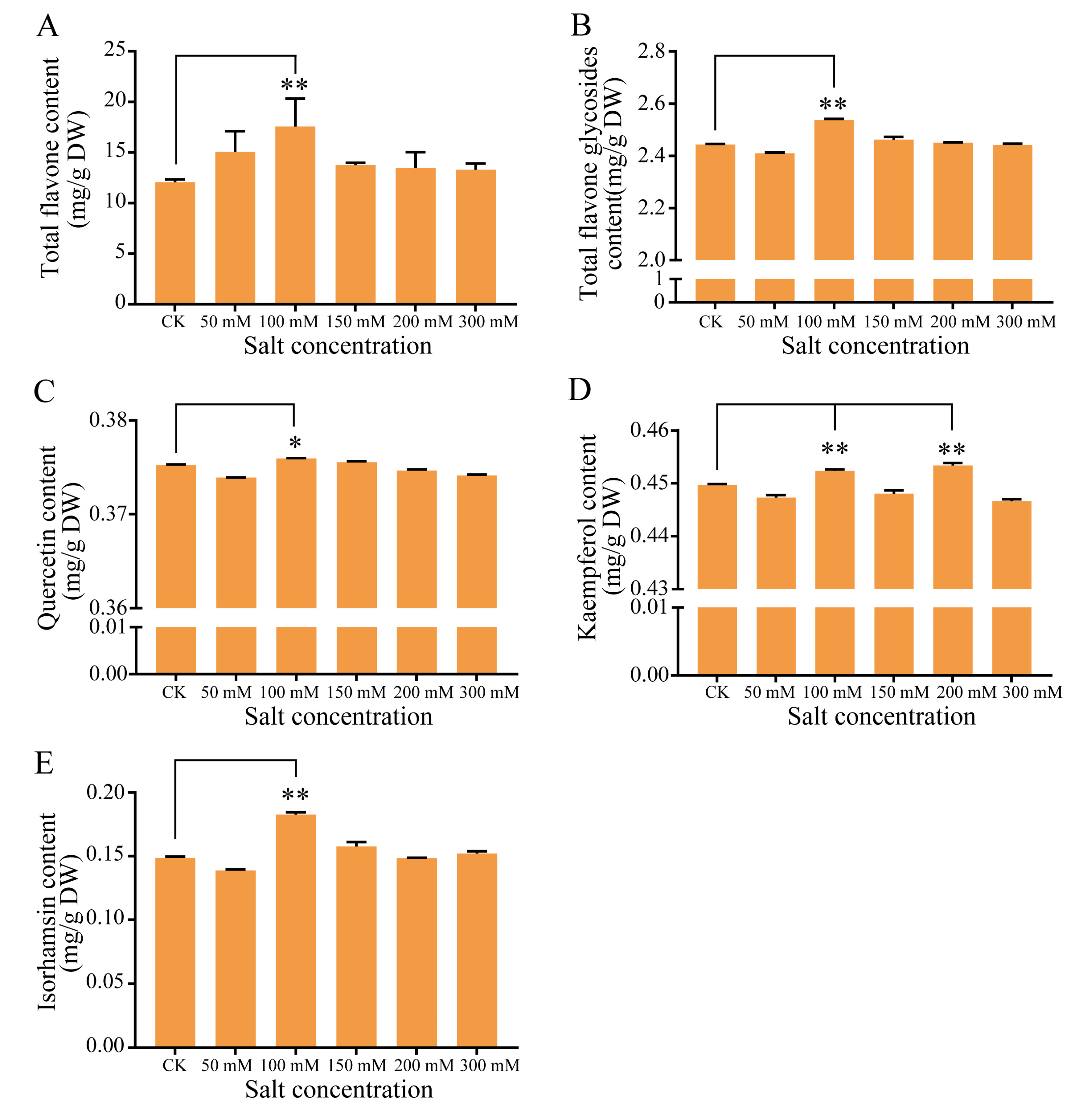
2.2. Flavonoid Accumulation in G. biloba Leaves after NaCl Treatment
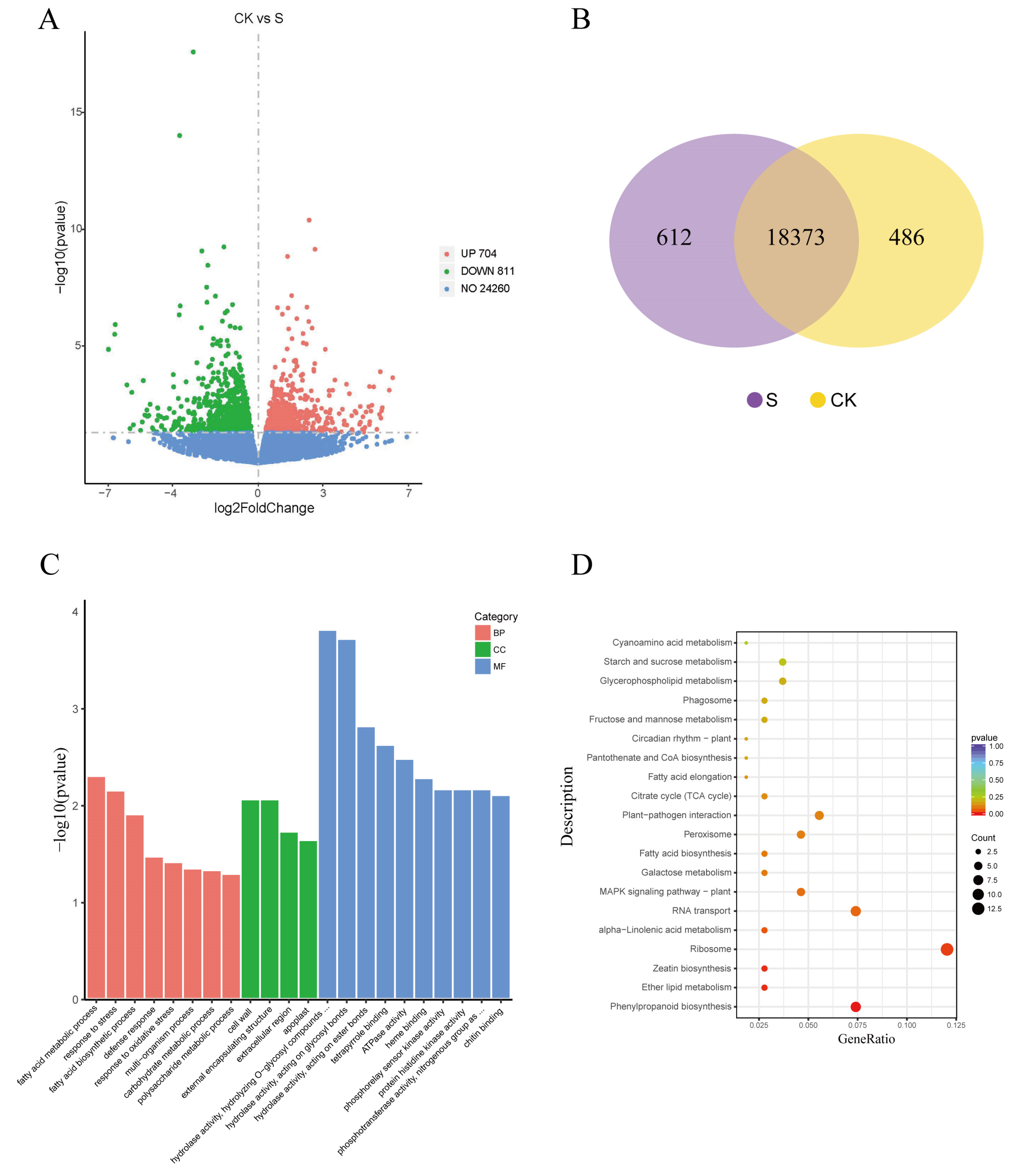
2.3. Differential Gene Expression after NaCl Treatment
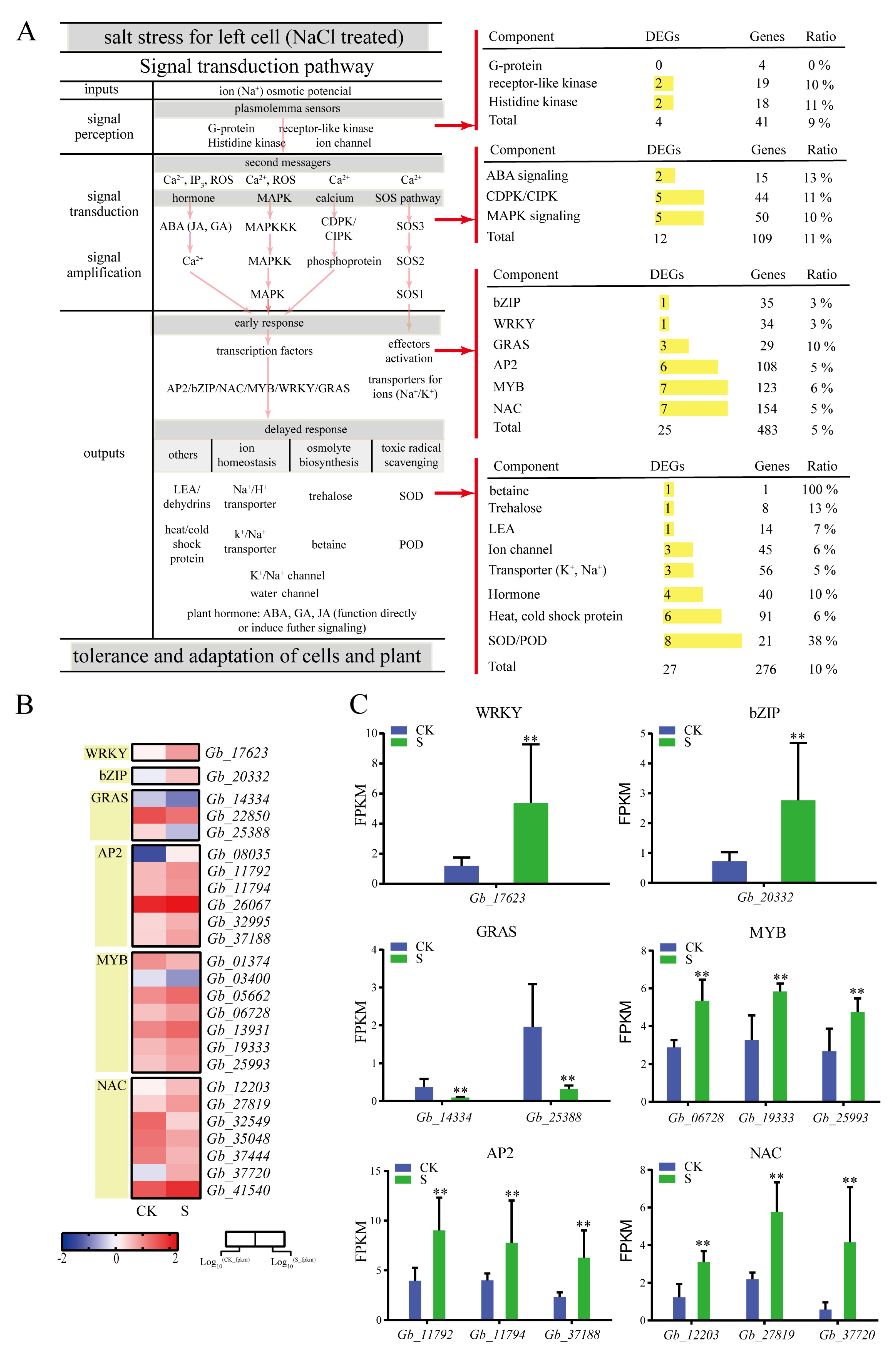
2.4. Identification of Signaling Pathways Associated with Salt Stress
2.5. Identification of Flavonoid Biosynthesis Genes Induced by Salt Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials and NaCl Treatment
4.2. Determination of Flavonoid and Flavonol Content
4.3. Detection of ROS Metabolism
4.4. Transcriptome Sequencing
4.5. Identification and Functional Analysis of Differentially Expressed Genes
4.6. Real-Time Quantitative PCR (RT-qPCR)
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Munns, R. Genes and salt tolerance: Bringing them together. New Phytol. 2005, 167, 645–663. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [Green Version]
- Jithesh, M.N.; Prashanth, S.R.; Sivaprakash, K.R.; Parida, A.K. Antioxidative response mechanisms in halophytes: Their role in stress defence. J. Genet. 2006, 85, 237–254. [Google Scholar] [CrossRef]
- Luo, M.B.; Liu, F. Salinity-induced oxidative stress and regulation of antioxidant defense system in the marine macroalga. Ulva prolifera. J. Exp. Mar. Biol. Ecol. 2011, 409, 223–228. [Google Scholar] [CrossRef]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How Plant Hormones Mediate Salt Stress Responses. Trends Plant Sci. 2020. [Google Scholar] [CrossRef]
- Chen, M.-X.; Lung, S.-C.; Du, Z.-Y.; Chye, M.-L. Engineering plants to tolerate abiotic stresses. Biocatal. Agric. Biotechnol. 2014, 3, 81–87. [Google Scholar] [CrossRef]
- Zhu, J.-K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003, 6, 441–445. [Google Scholar] [CrossRef]
- Zhu, S.Y.; Yu, X.C.; Wang, X.J.; Zhao, R.; Li, Y.; Fan, R.C.; Shang, Y.; Du, S.Y.; Wang, X.F.; Wu, F.Q.; et al. Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 2007, 19, 3019–3036. [Google Scholar] [CrossRef] [Green Version]
- Peng, J.; Li, Z.; Wen, X.; Li, W.; Shi, H.; Yang, L.; Zhu, H.; Guo, H. Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis. PLoS Genet 2014, 10, e1004664. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Wang, H.; Shao, H.; Tang, X. Recent Advances in Utilizing Transcription Factors to Improve Plant Abiotic Stress Tolerance by Transgenic Technology. Front Plant Sci. 2016, 7, 67. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.Y.; Li, Q.; Bi, K.S. Bioactive flavonoids in medicinal plants- Structure, activity and biological fate. Asian J. Pharm. Sci. 2018. [Google Scholar] [CrossRef]
- Ku, Y.S.; Ng, M.S.; Cheng, S.S.; Lo, A.W.; Xiao, Z.; Shin, T.S.; Chung, G.; Lam, H.M. Understanding the composition, biosynthesis, accumulation and transport of flavonoids in crops for the promotion of crops as healthy sources of flavonoids for human Consumption. Nutrients 2020, 12, 1717. [Google Scholar] [CrossRef]
- Ni, J.; Hao, J.; Jiang, Z.; Zhan, X.; Dong, L.; Yang, X.; Sun, Z.; Xu, W.; Wang, Z.; Xu, M. NaCl Induces flavonoid biosynthesis through a putative novel pathway in post-harvest Ginkgo leaves. Front. Plant Sci. 2017, 8, 920. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Lin, F.; Yang, H.; Yue, L.; Hu, F.; Wang, J.; Luo, Y.; Cao, F. Effect of varying NaCl doses on flavonoid production in suspension cells of Ginkgo biloba: Relationship to chlorophyll fluorescence, ion homeostasis, antioxidant system and ultrastructure. Acta Physiol. Plant. 2014, 36, 3173–3187. [Google Scholar] [CrossRef]
- Levy, D.; Coleman, W.K.; Veilleux, R.E. Adaptation of potato to water shortage: Irrigation management and enhancement of tolerance to drought and salinity. Am. J. Potato Res. 2013, 90, 186–206. [Google Scholar] [CrossRef]
- Gao, F.; Zhou, J.; Deng, R.Y.; Zhao, H.X.; Li, C.L.; Chen, H.; Suzuki, T.; Park, S.U.; Wu, Q. Overexpression of a tartary buckwheat R2R3-MYB transcription factor gene, FtMYB9, enhances tolerance to drought and salt stresses in transgenic Arabidopsis. J. Plant Physiol. 2017, 214, 81–90. [Google Scholar] [CrossRef]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Xu, M.; Gu, Y.; Xu, L.-A. Differentially expressed gene analysis of Tamarix chinensis provides insights into NaCl-stress response. Trees 2016, 31, 645–658. [Google Scholar] [CrossRef]
- Agarwal, P.K.; Jha, B. Transcription factors in plants and ABA dependent and independent abiotic stress signalling. Biol. Plant. 2010, 54, 201–212. [Google Scholar] [CrossRef]
- Shi, W.Y.; Du, Y.T.; Ma, J.; Min, D.H.; Jin, L.G.; Chen, J.; Chen, M.; Zhou, Y.B.; Ma, Y.Z.; Xu, Z.S.; et al. The WRKY Transcription Factor GmWRKY12 Confers Drought and Salt Tolerance in Soybean. Int. J. Mol. Sci. 2018, 19, 4087. [Google Scholar] [CrossRef] [Green Version]
- Wei, W.; Zhang, Y.; Han, L.; Guan, Z.; Chai, T. A novel WRKY transcriptional factor from Thlaspi caerulescens negatively regulates the osmotic stress tolerance of transgenic tobacco. Plant Cell Rep. 2008, 27, 795–803. [Google Scholar] [CrossRef]
- Tran, L.S.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef] [Green Version]
- Fujita, M.; Fujita, Y.; Maruyama, K.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Tran, L.S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004, 39, 863–876. [Google Scholar] [CrossRef]
- Debeaujon, I.; Nesi, N.; Perez, P.; Devic, M.; Grandjean, O.; Caboche, M.; Lepiniec, L. Proanthocyanidin-accumulating cells in Arabidopsis testa: Regulation of differentiation and role in seed development. Plant Cell 2003, 15, 2514–2531. [Google Scholar] [CrossRef] [Green Version]
- Albert, N.W.; Lewis, D.H.; Zhang, H.; Irving, L.J.; Jameson, P.E.; Davies, K.M. Light-induced vegetative anthocyanin pigmentation in Petunia. J. Exp. Bot. 2009, 60, 2191–2202. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Stanton, J.D.; Tolson, A.H.; Luo, Y.; Wang, H. Bioactive terpenoids and flavonoids from Ginkgo biloba extract induce the expression of hepatic drug-metabolizing enzymes through pregnane X receptor, constitutive androstane receptor, and aryl hydrocarbon receptor-mediated pathways. Pharm. Res. 2009, 26, 872–882. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Yang, S.; Lu, Z.; He, Z.; Ye, Y.; Zhao, B.; Wang, L.; Jin, B. Cytological, physiological, and transcriptomic analyses of golden leaf coloration in Ginkgo biloba L. Hortic. Res. 2018, 5, 12. [Google Scholar] [CrossRef] [Green Version]
- Walia, H.; Wilson, C.; Condamine, P.; Liu, X.; Ismail, A.M.; Zeng, L.; Wanamaker, S.I.; Mandal, J.; Xu, J.; Cui, X.; et al. Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol. 2005, 139, 822–835. [Google Scholar] [CrossRef] [Green Version]
- Wahid, A.; Ghazanfar, A. Possible involvement of some secondary metabolites in salt tolerance of sugarcane. J. Plant Physiol. 2006, 163, 723–730. [Google Scholar] [CrossRef]
- Liu, A. Effects of Salt Stress on the Growth and the Photosynthesis in Alternanthera philoxeroides. Acta Bot. Yunnanica 2007, 29, 85–90. [Google Scholar]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Zhao, B.; Wang, L.; Pang, S.; Jia, Z.; Wang, L.; Li, W.; Jin, B. UV-B promotes flavonoid synthesis in Ginkgo biloba leaves. Ind. Crops Prod. 2020, 151, 112483. [Google Scholar] [CrossRef]



| Gene ID | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| Gb_14024 | CTTGCTCTCGGCGTCGAATCTC | ACTTGGAGTCCTGGCACATCATTG |
| Gb_14029 | TCCTGAGCAACGGCAAGTTCAAG | GAATACCGGCCACGACATCCTTAC |
| Gb_00285 | TGGTGGATGAAGGCATTGACAGC | ACAACAGCAGATCGGCGTTCAC |
| Gb_06667 | GCCCTCAGAATGGGAACGGA | AGCATCTGAAGCGAGGCCTT |
| Gb_13074 | GTCGTCACAGATAGCCGCTTGG | AGTGTCTCCTTGGCAACGCAATC |
| Gb_21115 | AGGAAGGGCTGGCTTTGGTAAATG | TCGCAGAACATAGCAGACGCAATC |
| Gb_10949 | AATGTGCTTCCTGTGCTGTCTGAG | ATCCTGCTTGGGTTTGGGTTTCTG |
| Gb_26459 | TGAGTCGCAGGACCCAGAGAAC | CGCTTGACAGACTTTGCCTTTGC |
| Gb_19792 | GTGTGGTGCCGAGCTTGAGATAC | ATGAGGAGGAGGAAGAGGTTGCC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, N.; Liu, S.; Lu, Z.; Pang, S.; Wang, L.; Wang, L.; Li, W. Gene Expression Profiles and Flavonoid Accumulation during Salt Stress in Ginkgo biloba Seedlings. Plants 2020, 9, 1162. https://doi.org/10.3390/plants9091162
Xu N, Liu S, Lu Z, Pang S, Wang L, Wang L, Li W. Gene Expression Profiles and Flavonoid Accumulation during Salt Stress in Ginkgo biloba Seedlings. Plants. 2020; 9(9):1162. https://doi.org/10.3390/plants9091162
Chicago/Turabian StyleXu, Ningtao, Sian Liu, Zhaogeng Lu, Siyu Pang, Lu Wang, Li Wang, and Weixing Li. 2020. "Gene Expression Profiles and Flavonoid Accumulation during Salt Stress in Ginkgo biloba Seedlings" Plants 9, no. 9: 1162. https://doi.org/10.3390/plants9091162





