A Narrow Endemic or a Species Showing Disjunct Distribution? Studies on Meehania montis-koyae Ohwi (Lamiaceae)
Abstract
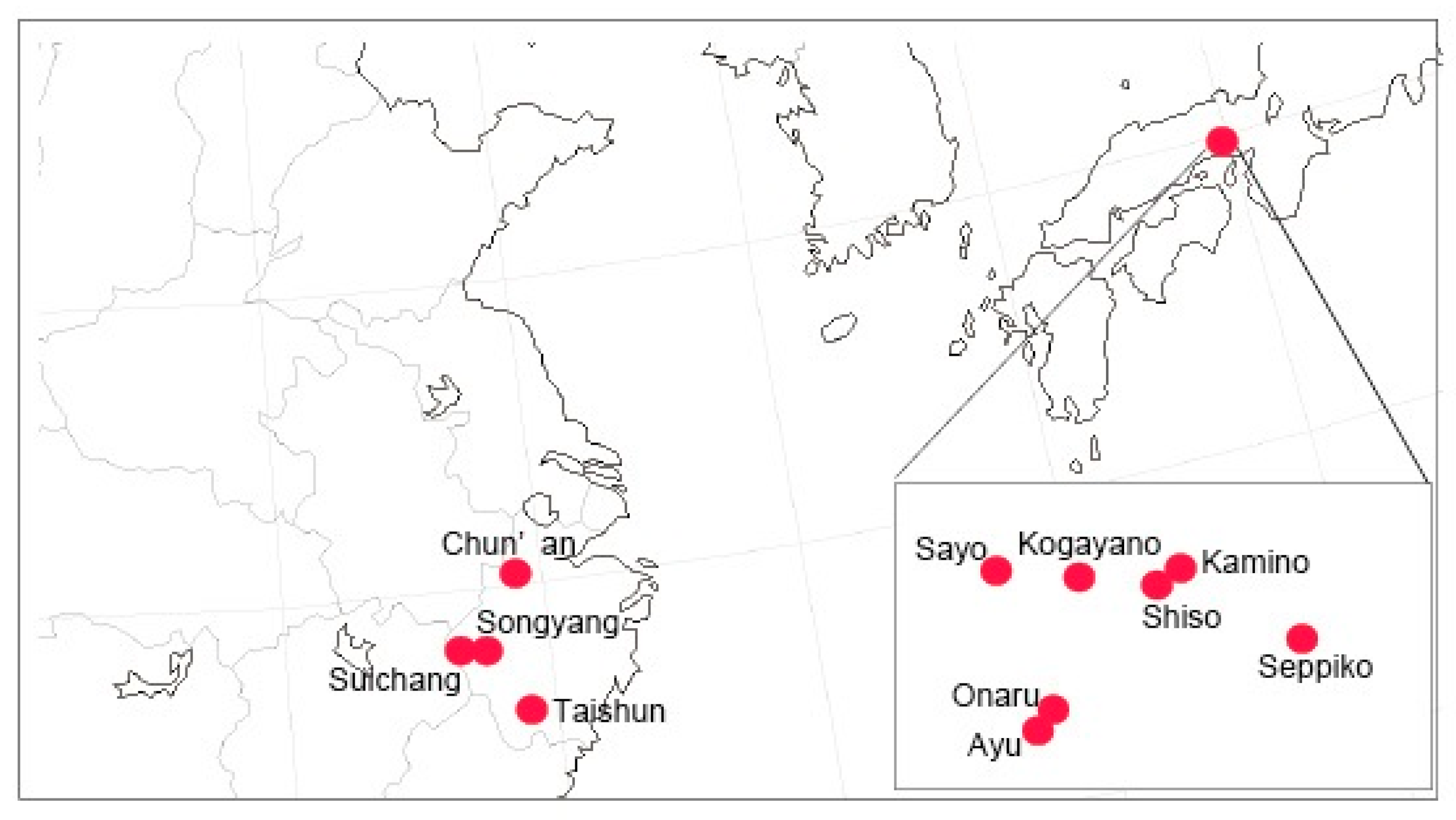
:1. Introduction
2. Results
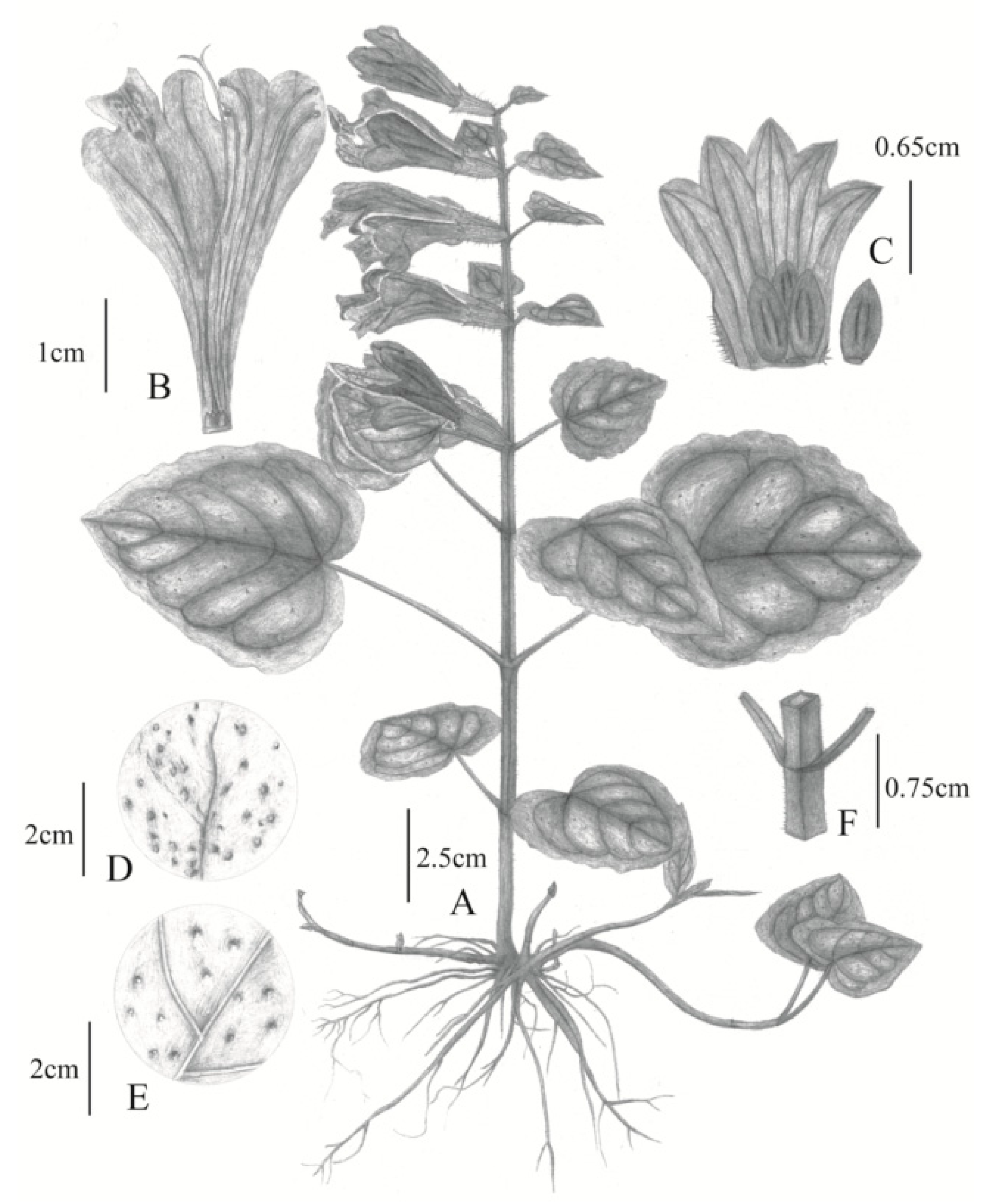
2.1. Morphological Differences between Chinese and Japanese M. montis-koyae
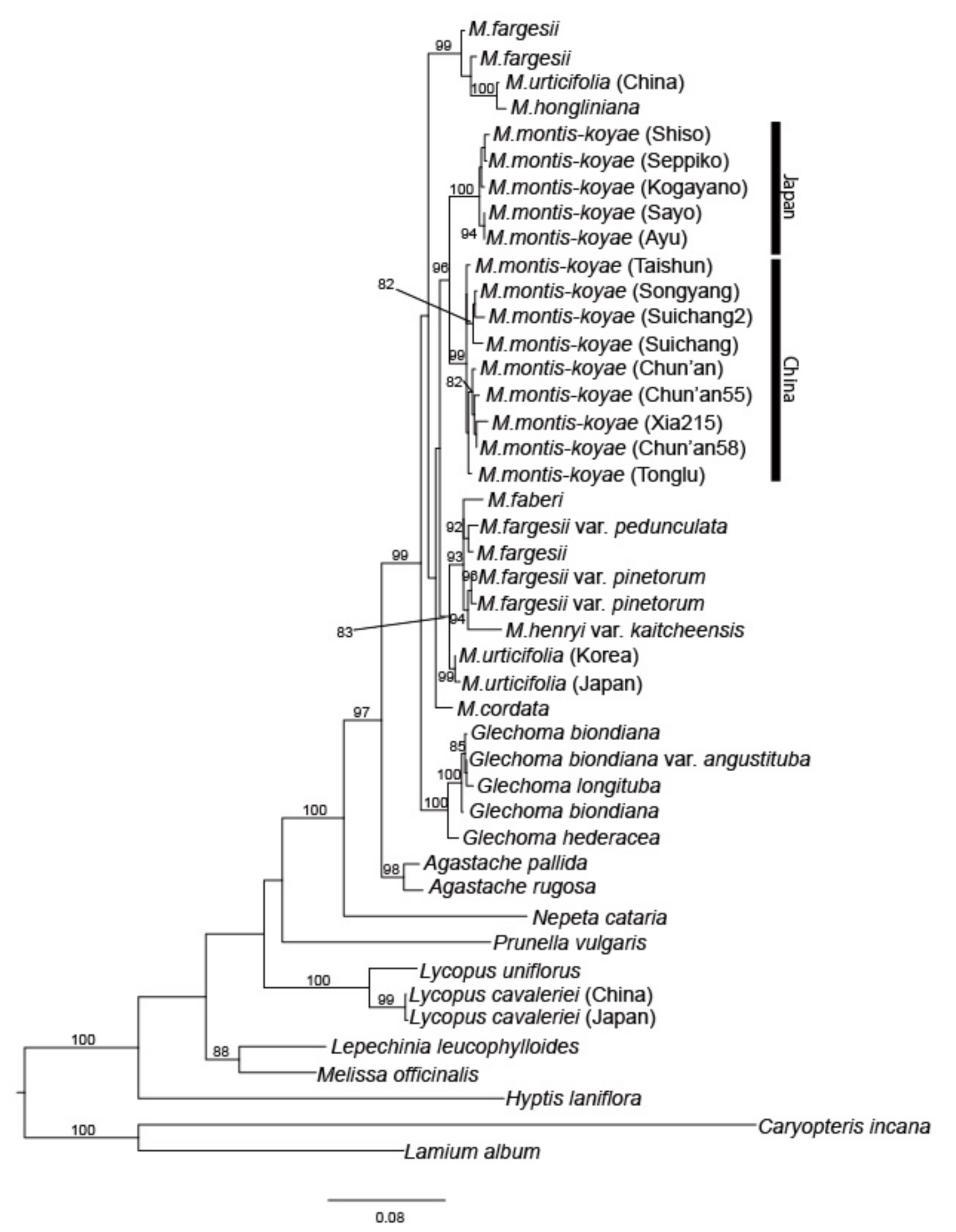
2.2. Maximum Likelihood Analysis by Using nrDNA and MIG-seq Data
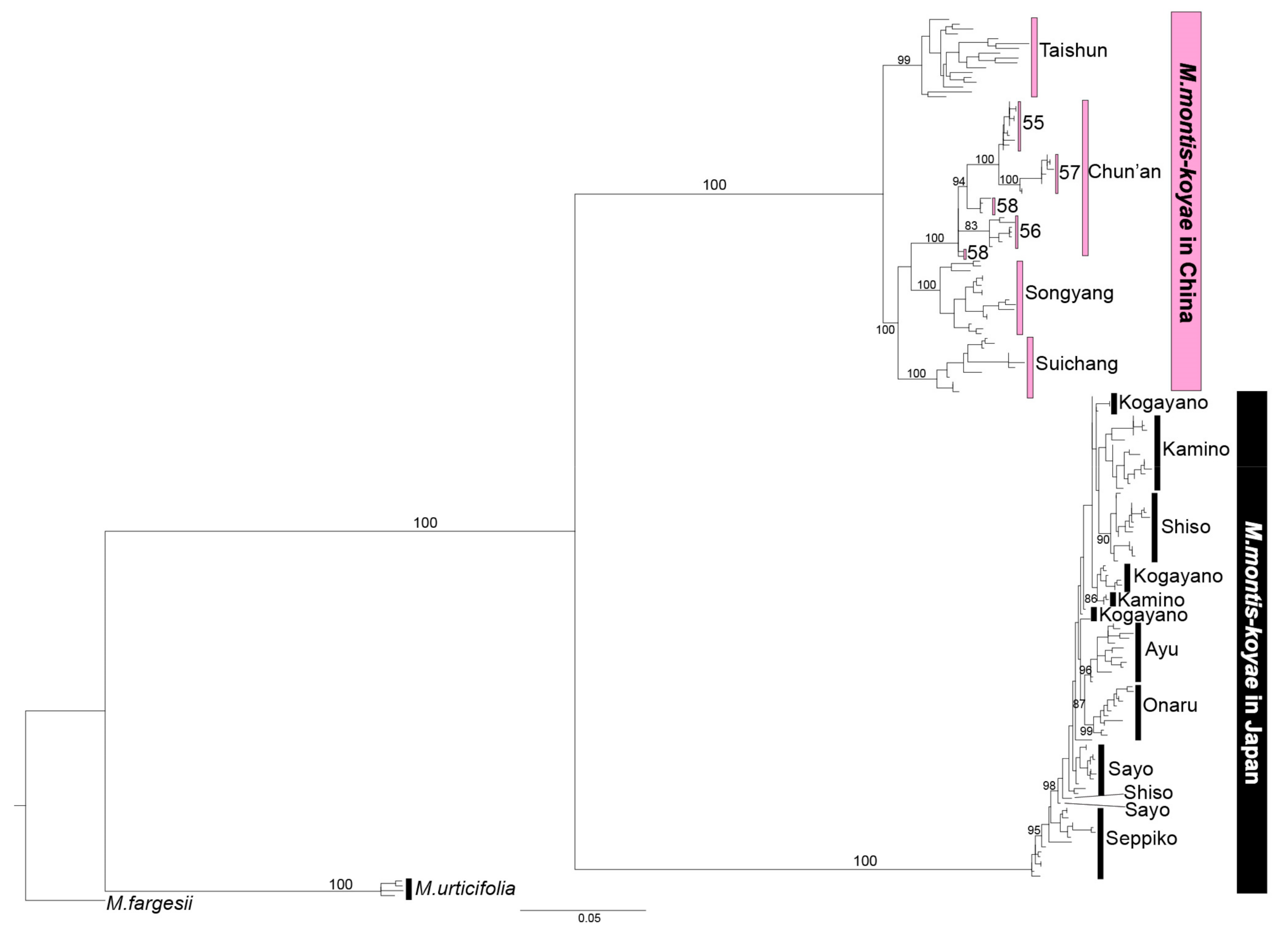
2.3. Population Analysis Using MIG-seq Data
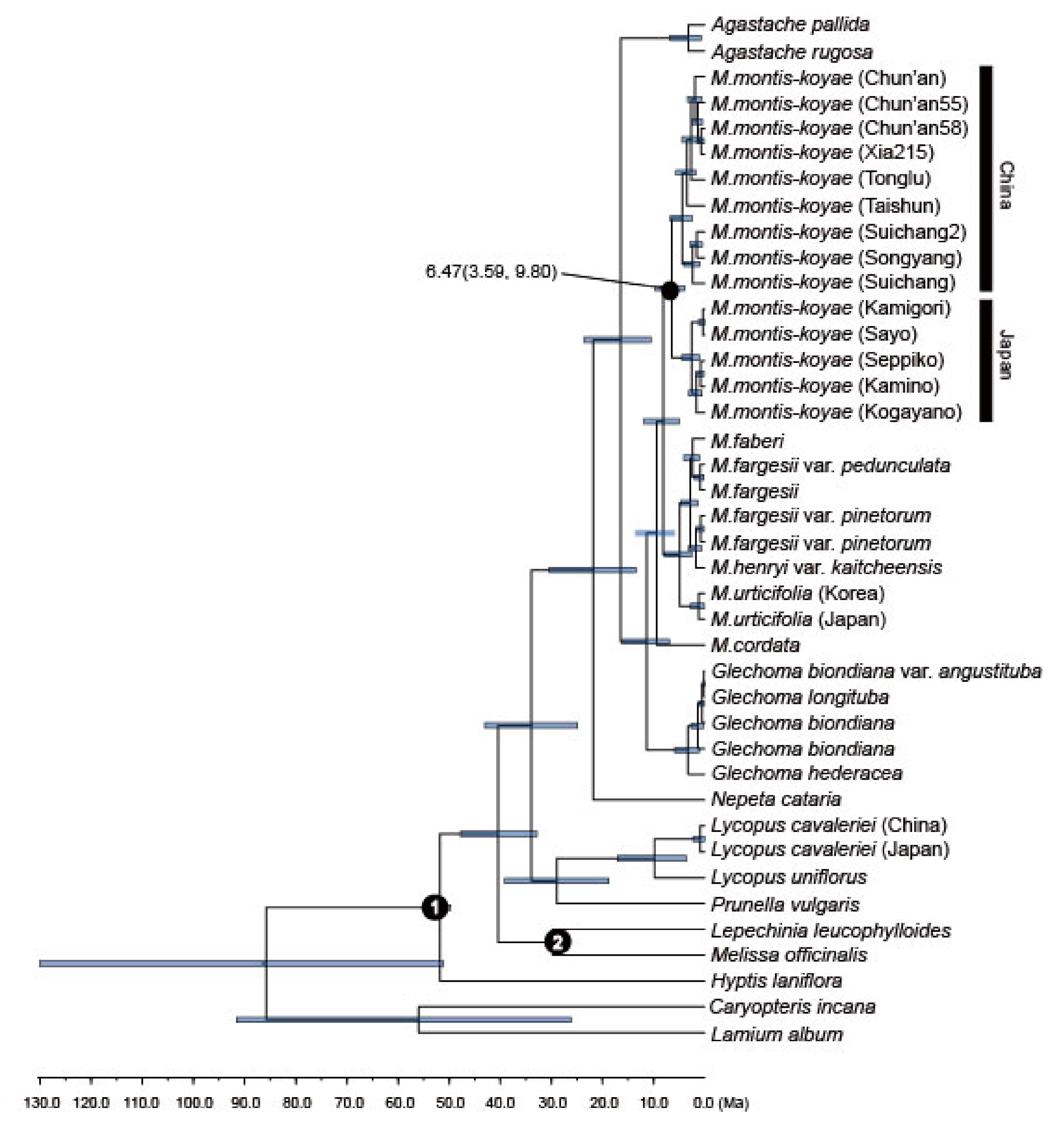
2.4. Divergence Time Analysis of M. montis-koyae
3. Discussion
3.1. Taxonomic Treatment
Meehania zheminensis A. Takano, Pan Li & G.-H. Xia sp. nov. (浙闽龙头草)
3.2. Other Specimens Examined
3.3. Key to the Species of Asian Meehania
- 1a
- Rhizomes stout, short, with slender decumbent leafy stolons after flowering; stems and petioles sparsely pilose; flowers 1 or 2 in leaf axils, forming 2- or 4-flowered verticillasters, generally 3 or 4 verticillasters forming a spike-like raceme, having no smell to the plant body-------------------------------------------------------------------------------------------------------------------------------- (2)
- 1b
- Rhizomes slender, elongate, without stolons; stems and petioles subdensely puberulent; flowers solitary and axillary, forming a pair, with 1 to 4 pairs on a stem, stink-bug-like smell to the plant body ------------------------------------------------------------------------------------------------------- (3)
- 2a
- Leaves ovate, long elliptic to narrowly lanceolate, base slightly cordate or cuneate--------------------------------------------------------------------------------------------------------------------------------------------- (4)
- 2b
- Leaves cordate to ovate, base cordate, rarely truncate to rounded ----------------------------------- (5)
- 3a
- 10–40 cm in height, bearing reddish spots on the lower labellum, distributed in China-------------------------------------------------------------------------------------------------- Meehania zheminensis sp. nov.
- 3b
- 10–20 cm in height, bearing no spots on the lower labellum, distributed in Japan-------------------------------------------------------------------------------------------------------------------------------M. montis-koyae
- 4a
- Leaves oblong-elliptic or ovate, 5.0–12.0 cm long, 2.0–5.0 cm wide ---------------------------------- (6)
- 4b
- Leaves narrowly lanceolate, 6.0–12.0 cm long, 1.2–2.5 cm wide ------------------------- M. pinfaensis
- 5a
- Verticillasters in terminal and lateral racemes; calyx tube narrowly tubular-------------- M. henryi
- 5b
- Verticillasters in terminal racemes or 2-flowered, inserted in leaf axils of upper 2 or 3 leaf pairs on stem; calyx campanulate or ± tubular --------------------------------------------------------------------- (7)
- 6a
- having dimorphic (=sterile and fertile) stems ---------------------------------------------- M. hongliniana
- 6b
- having monomorphic stems ---------------------------------------------------------------------------- M. faberi
- 7a
- Calyx campanulate, inconspicuously veined, floccose-villous on veins, teeth triangular, subequal -------------------------------------------------------------------------------------------------- M. urticifolia
- 7b
- Calyx narrow, ± tubular, conspicuously veined, sparsely pubescent on veins, teeth narrowly triangular ---------------------------------------------------------------------------------------------------- M. fargesii
4. Materials and Methods
4.1. Herbarium Studies on Morphology, Phenology, and Habitats of M. montis-koyae
4.2. Sampling for Molecular Analyses
4.3. DNA Extraction, Amplification, and Sequencing
4.4. MIG-seq Preparation
4.5. Phylogenetic Analysis Using Combined nrDNA Sequence and MIG-seq Data
4.6. Population Genetic Analysis Using MIG-seq Data
4.7. Divergence Time Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Takhtajan, A. Flowering Plants, 2nd ed.; Springer: Amsterdam, The Netherlands, 2009. [Google Scholar]
- Ying, T.S.; Zhang, Z.S. Endemism in the flora of China—Studies on the endemic genera. Acta Phytotax. Sin. 1984, 22, 259–268. [Google Scholar]
- Xie, G.-W. Phytogeographical affinities in the forest floras of eastern China and Japan. J. For. Res. 1997, 8, 87–90. [Google Scholar]
- Murata, G.; Yamazaki, T. Meehania Britt. In Flora of Japan IIIa; Iwatsuki, K., Yamazaki, T., Boufford, D.E., Ohba, H., Eds.; Kodansha: Tokyo, Japan, 1993; pp. 289–290. [Google Scholar]
- Li, X.W.; Hedge, I.C. Meehania Britton. In Flora of China; Wu, Z.Y., Raven, P.H., Eds.; Science Press: Beijing, China; Missouri Botanical Garden: St. Louis, MO, USA, 1994; Volume 17, pp. 122–124. [Google Scholar]
- Wen, J. Evolution of eastern Asian and eastern North American disjunct distributions in flowering plants. Ann. Rev. Ecol. Syst. 1999, 30, 421–455. [Google Scholar] [CrossRef]
- Xiang, Q.Y.; Soltis, D.E.; Soltis, P.S.; Manchester, S.R.; Crawford, D.J. Timing the eastern Asian-Eastern North American floristic disjunction: Molecular clock corroborates paleontological estimates. Mol. Phylogenet. Evol. 2000, 15, 462–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, L.; Wagner, W.L.; Ree, R.H.; Berry, P.E.; Wen, J. Molecular phylogeny, divergence time estimates, and historical biogeography of Circaea (Onagraceae) in the Northern Hemisphere. Mol. Phylogenet. Evol. 2009, 53, 995–1009. [Google Scholar] [CrossRef] [PubMed]
- Deng, T.; Nie, Z.-L.; Drew, B.T.; Volis, S.; Kim, C.; Xiang, C.-L.; Zhang, J.-W.; Wang, Y.-H.; Sun, H. Does the Arcto-Tertiary Biogeographic Hypothesis Explain the Disjunct Distribution of Northern Hemisphere Herbaceous Plants? The Case of Meehania (Lamiaceae). PLoS ONE 2015, 10, e0117171. [Google Scholar] [CrossRef] [Green Version]
- Kadota, Y. Meehania. In Endemic plants of Japan, a Book Series from the National Museum of Nature and Science, No. 11; Kato, M., Ebihara, A., Eds.; Tokai University Press: Tokyo, Japan, 2011; p. 216. (In Japanese) [Google Scholar]
- Murata, G. Meehania montis-koyae. In Red Data Plants; Yahara, T., Fujii, S., Ito, M., Ebihara, A., Eds.; Yama-Kei Publishers: Tokyo, Japan, 2015; p. 133. (In Japanese) [Google Scholar]
- Xia, G.-H.; Li, G.-Y. Meehania montis-koyae, a new record of Lamiaceae from China. Guihaia 2011, 31, 581–583. [Google Scholar]
- Takano, A.; Sakoda, M.; Kurosaki, N. Phenology of Meehania montis-koyae Ohwi (Lamiaceae) in May 2009-February 2010. Plants Hyogo 2010, 20, 37–40. (In Japanese) [Google Scholar]
- Holbourn, A.E.; Wolfgang, K.; Clemens, S.C.; Kochhann, K.G.D.; Jöhnck, J.; Lübbers, J.; Andersen, N. Late Miocene climate cooling and intensification of southeast Asian winter monsoon. Nat. Commun. 2018, 9, 1584. [Google Scholar] [CrossRef]
- Kimura, M. Paleogeography of the Ryukyu Islands. Tropics 2000, 10, 5–24. [Google Scholar] [CrossRef] [Green Version]
- Harrison, S.P.; Yu, G.; Takahara, H.; Prentice, I.C. Palaeovegetaion: Diversity of temperate plants in East Asia. Nature 2001, 413, 129–130. [Google Scholar] [CrossRef] [PubMed]
- Millien-Parra, A.; Jaeger, J.-J. Island biogeography of the Japanese terrestrial mammal assemblages: An example of a relict fauna. J. Biogeogr. 1999, 26, 959–972. [Google Scholar] [CrossRef]
- Aizawa, M.; Yoshimasru, H.; Saito, H.; Katsuki, T.; Kawahara, T.; Kitamura, K.; Shi, F.; Kaji, M. Phytogeography of a northeast Asian spruce, Picea jezoensis, inferred from genetic variation observed in organelle DNA markers. Mol. Ecol. 2007, 16, 3393–3405. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, R.; Jae-Hong, P.; Takahashi, H.; Maki, M. Disjunct distribution of chloroplast DNA haplotypes in the understory perennial Veratrum album ssp. oxysepalum (Melanthiaceae) in Japan as a result of ancient introgression. New Phytol. 2010, 188, 879–891. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.-Y.; Sun, Y.; Zhang, X.-P.; Lee, J.; Fu, C.-X.; Comes, H.P. Molecular phytogeography of East Asian Kirengeshoma (Hydrangeaceae) in relation to Quaternary climate change and land bridge configurations. New Phytol. 2009, 183, 480–495. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.-K.; Landrein, S.; Barrett, R.L.; Sakaguchi, S.; Maki, M.; Mu, W.-X.; Yang, T.; Zhu, Z.-X.; Liu, H.; Wang, H.-F. Phylogeographic analysis and genetic structure of an endemic Sino-Japanese disjunctive genus Diabelia (Caprifoliaceae). Front. Plant Sci. 2019, 10, 913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takano, A.; Sakoda, M.; Kurosaki, N. Mating system of Meehania montis-koyae Ohwi (Lamiaceae): Evidence from crossing-tests. Bunrui 2014, 14, 161–168. (In Japanese) [Google Scholar]
- Doyle, J.J.; Doyle, D. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Elsevier: Amsterdam, The Netherlands, 1990; Volume 18, pp. 315–322. [Google Scholar]
- Drew, B.T.; Sytsma, K.J. Testing the monophyly and placement of Lepechinia in the tribe Mentheae (Lamiaceae). Syst. Bot. 2011, 36, 1038–1049. [Google Scholar] [CrossRef]
- Baldwin, B.; Markos, S. Phylogenetic utility of the external transcribed spacers (ETS) of 18S-26S rDNA: Congruence of ETS and ITS trees of Calycadenia (Compositae). Mol. Phylogenet. Evol. 1998, 10, 449–463. [Google Scholar] [CrossRef]
- Suyama, Y.; Matsuki, Y. MIG-seq: An effective PCR-based method for genome-wide single-nucleotide polymorphism genotyping using the next-generation sequencing platform. Sci. Rep. 2015, 5, 16963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lassmann, T.; Hayashizaki, Y.; Daub, C.O. TagDust—A program to eliminate artifacts from next generation sequencing data. Bioinformatics 2009, 25, 2839–2840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eaton, D.A. PyRAD: Assembly of de novo RADseq loci for phylogenetic analyses. Bioinformatics 2014, 30, 1844–1849. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Edger, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Hall, T. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 1999, 2, 60–61. [Google Scholar]
- Guindon, S.; Gascuel, O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Silvestro, D.; Michalak, I. RAXMLGUI: A graphical front-end for RAxML. Organ. Divers. Evol. 2012, 12, 335–337. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree 1.4.0. 2012. Available online: http://tree.bio.ed.ac.uk (accessed on 25 November 2019).
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Evanno, G.; Regnaut, S.; Gouder, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kar, R.K. On the Indian origin of Ocimum (Lamiaceae): A palynological approach. Palaeobotanist 1996, 43, 45–50. [Google Scholar]
- Martínez-Millán, M. Fossil record and age of the Asteridae. Bot. Rev. 2010, 76, 83–135. [Google Scholar] [CrossRef]
- Drew, B.T.; Sytsma, K.J. Phylogenetics, biogeography, and staminal evolution in the tribe Mentheae (Lamiaceae). Am. J. Bot. 2012, 99, 933–953. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Qi, Z.-C.; Liu, L.-X.; Ohi-Toma, T.; Lee, J.; Hsieh, T.-H.; Fu, C.-X.; Cameron, K.M.; Qiu, Y.-Y. Molecular phylogenetics and biogeography of the mint tribe Elsholtzieae (Nepetoideae, Lamiaceae), with emphasis on its diversification in East Asia. Sci. Rep. 2017, 7, 2057. [Google Scholar] [CrossRef] [Green Version]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [Green Version]
- Drummond, A.; Rambaut, A.; Bouckkaert, R. Divergence dating tutorial with BEAST 2.0. PLoS Comput. Biol. 2014. [Google Scholar] [CrossRef]


| Koga-yano | Ayu | Seppi-ko | Ona-ru | Sayo | Kami-no | Shiso | Sui-chang | Song-yang | Ta-ishun | Chun’-an | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Kogayano | 0.000 | ||||||||||
| Ayu | 0.315 | 0.000 | |||||||||
| Seppiko | 0.308 | 0.309 | 0.000 | ||||||||
| Onaru | 0.304 | 0.306 | 0.297 | 0.000 | |||||||
| Sayo | 0.304 | 0.304 | 0.296 | 0.293 | 0.000 | ||||||
| Kamino | 0.309 | 0.310 | 0.302 | 0.300 | 0.301 | 0.000 | |||||
| Shiso | 0.304 | 0.308 | 0.295 | 0.292 | 0.292 | 0.301 | 0.000 | ||||
| Suichang | 0.573 | 0.579 | 0.579 | 0.570 | 0.565 | 0.581 | 0.575 | 0.000 | |||
| Songyang | 0.592 | 0.597 | 0.597 | 0.588 | 0.585 | 0.600 | 0.593 | 0.426 | 0.000 | ||
| Taishun | 0.563 | 0.559 | 0.563 | 0.558 | 0.556 | 0.569 | 0.563 | 0.427 | 0.425 | 0.000 | |
| Chun’an | 0.563 | 0.563 | 0.564 | 0.560 | 0.557 | 0.571 | 0.565 | 0.415 | 0.432 | 0.406 | 0.000 |
| Pop Code | N | I | Ho | He | |
|---|---|---|---|---|---|
| Kogayano | Mean | 8.840 | 0.008 | 0.007 | 0.005 |
| (n = 13) | SE | 0.306 | 0.004 | 0.004 | 0.003 |
| Ayu | Mean | 7.805 | 0.012 | 0.007 | 0.007 |
| (n = 13) | SE | 0.288 | 0.004 | 0.003 | 0.003 |
| Seppiko | Mean | 9.858 | 0.007 | 0.004 | 0.004 |
| (n = 15) | SE | 0.333 | 0.003 | 0.002 | 0.002 |
| Onaru | Mean | 7.266 | 0.011 | 0.007 | 0.007 |
| (n = 11) | SE | 0.247 | 0.004 | 0.003 | 0.003 |
| Sayo | Mean | 8.308 | 0.011 | 0.008 | 0.007 |
| (n = 13) | SE | 0.288 | 0.004 | 0.003 | 0.003 |
| Kamino | Mean | 10.630 | 0.012 | 0.007 | 0.008 |
| (n = 19) | SE | 0.397 | 0.004 | 0.003 | 0.003 |
| Shiso | Mean | 11.024 | 0.004 | 0.004 | 0.002 |
| (n = 16) | SE | 0.366 | 0.002 | 0.003 | 0.002 |
| Suichang | Mean | 6.672 | 0.017 | 0.007 | 0.012 |
| (n = 12) | SE | 0.272 | 0.006 | 0.004 | 0.004 |
| Songyang | Mean | 7.855 | 0.014 | 0.004 | 0.009 |
| (n = 16) | SE | 0.355 | 0.005 | 0.002 | 0.003 |
| Taishun | Mean | 8.704 | 0.020 | 0.012 | 0.013 |
| (n = 17) | SE | 0.370 | 0.006 | 0.005 | 0.004 |
| Chun’an | Mean | 18.769 | 0.014 | 0.006 | 0.010 |
| (nn = 33) | SE | 0.743 | 0.005 | 0.003 | 0.003 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takano, A.; Sakaguchi, S.; Li, P.; Matsuo, A.; Suyama, Y.; Xia, G.-H.; Liu, X.; Isagi, Y. A Narrow Endemic or a Species Showing Disjunct Distribution? Studies on Meehania montis-koyae Ohwi (Lamiaceae). Plants 2020, 9, 1159. https://doi.org/10.3390/plants9091159
Takano A, Sakaguchi S, Li P, Matsuo A, Suyama Y, Xia G-H, Liu X, Isagi Y. A Narrow Endemic or a Species Showing Disjunct Distribution? Studies on Meehania montis-koyae Ohwi (Lamiaceae). Plants. 2020; 9(9):1159. https://doi.org/10.3390/plants9091159
Chicago/Turabian StyleTakano, Atsuko, Shota Sakaguchi, Pan Li, Ayumi Matsuo, Yoshihisa Suyama, Guo-Hua Xia, Xi Liu, and Yuji Isagi. 2020. "A Narrow Endemic or a Species Showing Disjunct Distribution? Studies on Meehania montis-koyae Ohwi (Lamiaceae)" Plants 9, no. 9: 1159. https://doi.org/10.3390/plants9091159
APA StyleTakano, A., Sakaguchi, S., Li, P., Matsuo, A., Suyama, Y., Xia, G.-H., Liu, X., & Isagi, Y. (2020). A Narrow Endemic or a Species Showing Disjunct Distribution? Studies on Meehania montis-koyae Ohwi (Lamiaceae). Plants, 9(9), 1159. https://doi.org/10.3390/plants9091159






