Bacterial Microbiota Isolated from Cysts of Globodera rostochiensis (Nematoda: Heteroderidae)
Abstract
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. Isolation of Bacteria
3.2. Molecular Study
3.3. Statistical Data Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Schuerger, A.C.; Lee, P. Microbial ecology of a crewed rover traverse in the Arctic: Low microbial dispersal and implications for planetary protection on human Mars missions. Astrobiology 2015, 15, 478–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danovaro, R.; Canals, M.; Tangherlini, M.; Dell’Anno, A.; Gambi, C.; Lastras, G.; Amblas, D.; Sanchez–Vidal, A.; Frigola, J.; Calafat, A.M.; et al. A submarine volcanic eruption leads to a novel microbial habitat. Nat. Ecol. Evol. 2017, 1, 144. [Google Scholar] [CrossRef] [PubMed]
- Gómez, F.; Cavalazzi, B.; Rodríguez, N.; Amils, R.; Ori, G.; Olsson–Francis, K.; Escudero, C.; Martínez, J.; Miruts, H. Ultra–small microorganisms in the polyextreme conditions of the Dallol volcano, Northern Afar, Ethiopia. Sci. Rep. 2019, 9, 7907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Back, M.A.; Haydock, P.P.J.; Jenkinson, P. Disease complexes involving plant parasitic nematodes and soilborne pathogens. Plant Pathol. 2002, 51, 683–697. [Google Scholar] [CrossRef]
- Oro, V. Potato Cyst Nematodes–Morphology, Molecular Characterization and Antagonists. Ph.D. Thesis, Faculty of Biofarming, Backa Topola, Serbia, 8 September 2011. [Google Scholar]
- Ryan, N.A.; Duffy, E.M.; Cassells, A.C.; Jones, P.W. The effect of mycorrhizal fungi on the hatch of potato cyst nematodes. Appl. Soil Ecol. 2000, 15, 233–240. [Google Scholar] [CrossRef]
- Oro, V.; Nikolic, B.; Josic, D. The potato road and biogeographic history of potato cyst nematode populations from different continents. Genetika 2014, 46, 895–904. [Google Scholar] [CrossRef]
- Nour, S.M.; Lawrence, J.R.; Zhu, H.; Swerhone, G.D.W.; Welsh, M.; Welacky, T.W.; Topp, E. Bacteria associated with cysts of the soybean cyst nematode (Heterodera glycines). Appl. Environ. Microbiol. 2003, 69, 607–615. [Google Scholar] [CrossRef] [Green Version]
- Kerry, B.R. Rhizosphere interactions and the exploitation of microbial agents for the biological control of plant–parasitic nematodes. Annu. Rev. Phytopathol. 2000, 38, 423–441. [Google Scholar] [CrossRef] [Green Version]
- Krechel, A.; Faupel, A.; Hallmann, J.; Ulrich, A.; Berg, G. Potato–associated bacteria and their antagonistic potential towards plant–pathogenic fungi and the plant–parasitic nematode Meloidogyne incognita (Kofoid& White) Chitwood. Can. J. Microbiol. 2002, 48, 772–786. [Google Scholar] [CrossRef]
- Ryan, N.A.; Jones, P. The ability of rhizosphere bacteria isolated from nematode host and non–host plants to influence the hatch in vitro of the two potato cyst nematode species Globodera rostochiensis and G. pallida. Nematology. 2004, 6, 375–387. [Google Scholar] [CrossRef]
- Tian, B.Y.; Yang, J.K.; Zhang, K.Q. Bacteria used in biological control of plant–parasitic nematodes: Populations, mechanisms of action, and future prospects. FEMS Microbiol. Ecol. 2007, 61, 197–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Liu, J.; Ding, J.; He, Q.R.; Xiong, R.; Zhang, K. The investigation of nematocidal activity in Stenotrophomonas maltophilia G2 and characterization of a novel virulence serine protease. Can. J. Microbiol. 2009, 55, 934–942. [Google Scholar] [CrossRef] [PubMed]
- Paiva, G.; Proença, D.N.; Francisco, R.; Verissimo, P.; Santos, S.S.; Fonseca, L.; Abrantes, I.M.O.; Morais, P.V. Nematicidal bacteria associated to pinewood nematode produce extracellular proteases. PLoS ONE 2013, 8, e79705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, H.; Qi, G.; Yin, R.; Zhang, H.; Li, C.; Zhao, X. Bacillus cereus strain S2 shows high nematicidal activity against Meloidogyne incognita by producing sphingosine. Sci. Rep. 2016, 6, 28756. [Google Scholar] [CrossRef] [PubMed]
- Costa, L.S.A.S.; Campos, V.P.; Terra, W.C.; Pfenning, L.H. Microbiota from Meloidogyne exigua egg masses and evidence for the effect of volatiles on infective juvenile survival. J. Nematol. 2015, 17, 715–724. [Google Scholar] [CrossRef]
- Ali, B. Functional and genetic diversity of bacteria associated with the surfaces of agronomic plants. Plants 2019, 8, 91. [Google Scholar] [CrossRef] [Green Version]
- Fernández–González, A.J.; Martínez–Hidalgo, P.; Cobo–Díaz, J.F.; Villadas, P.J.; Martínez–Molina, E.; Toro, N.; Tringe, S.G.; Fernández–López, M. The rhizosphere microbiome of burned holm–oak: Potential role of the genus Arthrobacter in the recovery of burned soils. Sci. Rep. 2017, 7, 6008. [Google Scholar] [CrossRef]
- Elhady, A.; Gine, A.; Topalovic, O.; Jacquiod, S.; Sørensen, S.J.; Sorribas, F.J.; Heuer, H. Microbiomes associated with infective stages of root–knot and lesion nematodes in soil. PLoS ONE 2017, 12, e0177145. [Google Scholar] [CrossRef] [Green Version]
- Beesley, C.A.; Vanner, C.L.; Helsel, L.O.; Gee, J.E.; Hoffmaster, A.R. Identifcation and characterization of clinical Bacillus spp. isolates phenotypically similar to Bacillus anthracis. FEMS Microbiol. Lett. 2010, 313, 47–53. [Google Scholar] [CrossRef] [Green Version]
- Tong, X.; Yuan, L.; Luo, L.; Yin, X. Characterization of a selenium–tolerant rhizosphere strain from a novel Se–hyperaccumulating plant Cardamine hupingshanesis. Sci. World J. 2014, 108562. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Li, X.; Yin, L.; Liu, C.; Zou, H.; Wu, Z.; Zhang, Z. Analysis of the complete genome sequence of Brevibacterium frigoritolerans ZB201705 isolated from drought– and salt–stressed rhizosphere soil of maize. Ann. Microbiol. 2019, 69, 1489–1496. [Google Scholar] [CrossRef]
- Singh, P.; Kim, Y.J.; Singh, H.; Wang, C.; Hwang, K.H.; Farh, M.E.-A.; Yang, D.C. Biosynthesis, characterization, and antimicrobial applications of silver nanoparticles. Int. J. Nanomed. 2015, 10, 2567–2577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.-H.; Liu, B.; Wang, J.-P.; Che, J.-M.; Li, P.F. Reclassification of Brevibacterium frigoritolerans DSM 8801T as Bacillus frigoritolerans comb. nov. based on genome analysis. Curr. Microbiol. 2020, 77, 1916–1923. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.R.; Choi, G.J.; Choi, Y.H.; Jang, K.S.; Sung, N.-D.; Kang, M.S.; Moon, Y.; Lee, S.K.; Kim, J.-C. Suppression of pine wilt disease by an antibacterial agent, oxolinic acid. Pest. Manag. Sci. 2010, 66, 634–639. [Google Scholar] [CrossRef]
- Selvakumar, G.; Sushil, S.; Stanley, J.; Mohan, M.; Deol, A.; Rai, D.; Ramkewal; Bhatt, J.C.; Gupta, S.H. Brevibacterium frigoritolerans a novel entomopathogen of Anomala dimidiata and Holotrichia longipennis (Scarabaeidae: Coleoptera). Biocontrol Sci. Technol. 2011, 21, 821–827. [Google Scholar] [CrossRef]
- Mehan, S.; Guo, T.; Gitau, M.W.; Flanagan, D.C. Comparative study of different stochastic weather generators for long–term climate data simulation. Climate 2017, 5, 26. [Google Scholar] [CrossRef]
- Li, L.; Xu, M.; Eyakub Ali, M.; Zhang, W.; Duan, Y.; Li, D. Factors affecting soil microbial biomass and functional diversity with the application of organic amendments in three contrasting cropland soils during a field experiment. PLoS ONE 2018, 13, e0203812. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Banin, A.; Borochovitch, A. Effect of potassium on soil structure in relation to hydraulic conductivity. Geoderma 1983, 30, 135–147. [Google Scholar] [CrossRef]
- Marshall, K.C. Clay mineralogy in relation to survival of soil bacteria. Annu. Rev. Phytopathol. 1975, 13, 357–373. [Google Scholar] [CrossRef]
- Lee, Y.S.; Kim, K.Y. Antagonistic potential of Bacillus pumilus L1 against root–knot nematode, Meloidogyne arenaria. J. Phytopathol. 2016, 164, 29–33. [Google Scholar] [CrossRef]
- Padgham, J.L.; Sikora, R.A. Biological control potential and modes of action of Bacillus megaterium against Meloidogyne graminicola on rice. J. Crop Prot. 2007, 26, 971–977. [Google Scholar] [CrossRef]
- Abd–El–Khair, H.; Wafaa, M.A.; El–Nagdi, W.M.A.; Mahmoud, M.A.; Youssef, M.M.A.; Abd–Elgawad, M.M.M.; Dawood, M.G. Protective effect of Bacillus subtilis, B. pumilus, and Pseudomonas fluorescens isolates against root knot nematode Meloidogyne incognita on cowpea. Bull. Natl. Res. Cent. 2019, 43, 64. [Google Scholar] [CrossRef] [Green Version]
- Enright, M.R.; Griffin, C.T. Effects of Paenibacillus nematophilus on the entomopathogenic nematode Heterorhabditis megidis. J. Invertebr. Pathol. 2005, 88, 40–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pham, V.H.T.; Jeong, S.-W.; Kim, J. Psychrobacillus soli sp. nov., capable of degrading oil, isolated from oil–contaminated soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 3046–3052. [Google Scholar] [CrossRef]
- Das, G.; Patra, J.K.; Choi, J.; Baek, K.-H. Anticandidal effect of endophytic bacteria isolated from Equisetum arvense L. against Candida albicans and Candida glabrata. Braz. Arch. Biol. Technol. 2017, 60, e17160433. [Google Scholar] [CrossRef]
- Oliveira, V.F.; Abreu, Y.J.L.; Fleming, L.R.; Nascimento, J.S. Anti–staphylococcal and antifungal substances produced by endospore–forming bacilli. J. Appl. Pharm. Sci. 2012, 2, 154–157. [Google Scholar] [CrossRef] [Green Version]
- Talwar, C.; Nagar, S.; Kumar, R.; Scaria, J.; Lal, R.; Negi, R.K. Defining the environmental adaptations of genus Devosia: Insights into its expansive short peptide transport system and positively selected genes. Sci. Rep. 2020, 10, 1151. [Google Scholar] [CrossRef]
- Wackett, L.P. Arthrobacter and related genera: An annotated selection of World Wide Web sites relevant to the topics in environmental microbiology. Microb. Biotechnol. 2016, 9, 136–138. [Google Scholar] [CrossRef]
- Jones, D.; Keddie, R.M. The Genus Arthrobacter. Prokaryotes 2006, 3, 945–960. [Google Scholar] [CrossRef]
- Xu, Y.; Lu, H.; Wang, X.; Zhang, K.; Li, G. Effect of volatile organic compounds from bacteria on nematodes. Chem. Biodivers. 2015, 12, 1415–1421. [Google Scholar] [CrossRef]
- Coyne, D.L.; Nicol, J.M.; Claudius-Cole, B. Practical Plant Nematology: A Field and Laboratory Guide, 2nd ed.; SP-IPM Secretariat International Institute of Tropical Agriculture (IITA): Cotonou, Benin, 2014; pp. 25–29. [Google Scholar]
- Spears, J.F. The Golden Nematode Handbook-Survey, Laboratory, Control and Quarantine Procedures, Agriculture Handbook 353; USDA, Agricultural Research Service: Washington, DC, USA, 1968; pp. 1–82. [Google Scholar]
- Heungens, K.; Mugniery, D.; Van Montagu, M.; Gheysen, G.; Niebel, A. A method to obtain disinfected Globodera infective juveniles directly from cysts. Fundam. Appl. Nematol. 1996, 19, 91–93. [Google Scholar]
- Brown, A.; Smith, H. Benson’s Microbiological Applications: Laboratory Manual in General Microbiology, 13th ed.; McGraw-Hill Education: New York, NY, USA, 2015; pp. 73–80. [Google Scholar]
- Goldenberger, D.; Perschil, I.; Ritzler, M.; Altwegg, M. Simple “universal” DNA extraction procedure using SDS and proteinase K is compatible with direct PCR amplification. Genome Res. 1995, 4, 368–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picard, C.; Di Cello, F.; Ventura, M.; Fani, R.; Guckert, A. Frequency and biodiversity of 2,4–Diacetylphloroglucinol producing bacteria isolated from the maize rhizosphere at different stages of plant growth. Appl. Environ. Microbiol. 2000, 66, 948–955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gunidon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huelsenbeck, J.P.; Ronquist, F. Bayesian analysis of molecular evolution using MrBayes. In Statistical Methods in Molecular Evolution: Statistics for Biology and Health; Springer: New York, NY, USA, 2005; pp. 183–226. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- SRPS ISO 10390:2007. Soil Quality-Determination of pH; Institute for Standardisation of Republic of Serbia: Belgrade, Serbia, 2007. [Google Scholar]
- SRPS ISO 10694:2005. Soil Quality-Determination of Organic and Total Carbon after Dry Combustion (“Elemental Analysis”); Institute for Standardization: Belgrade, Serbia, 2005. [Google Scholar]
- SRPS ISO 13878:2005. Soil Quality-Determination of Total Nitrogen Content by Dry Combustion (“Elemental Analysis”); Institute for Standardization: Belgrade, Serbia, 2005. [Google Scholar]
- Egnér, H.; Riehm, H.; Domingo, W.R. Untersuchungen über die chemische bodenanalyse als grundlage für die beurteilung des nährstoff-zustandes der böden II. Chemische extraktions methoden zur phosphor- und kalium bestimmung. Kungl. Lantbrukshögsk. Ann. 1960, 26, 199–215. [Google Scholar]
- Hadzic, V.; Belic, M.; Nesic, L. Determination of the mechanical composition (texturally gravimetric) of the soil. In Methods of Research and Determination of Physical Properties of the Soil; Bosnjak, D.J., Ed.; JDPZ: Novi Sad, Serbia, 1997; pp. 17–32. [Google Scholar]
- Onofri, A. Routine statistical analyses of field experiments by using an Excel extension. In Proceedings of the 6th National Conference of the Italian Biometric Society: “La Statistica nelle Scienze della Vita e Dell’ambiente”, Pisa, Italy, 20–22 June 2007; pp. 93–96. [Google Scholar]
- Shen, Y.; Fu, Y.; Yu, Y.; Zhao, J.; Li, J.; Li, Y.; Wang, X.; Zhang, J.; Xiang, W. Psychrobacillus lasiicapitis sp. nov., isolated from the head of an ant (Lasius fuliginosus). Int. J. Syst. Evol. Microbiol. 2017, 67, 4462–4467. [Google Scholar] [CrossRef]
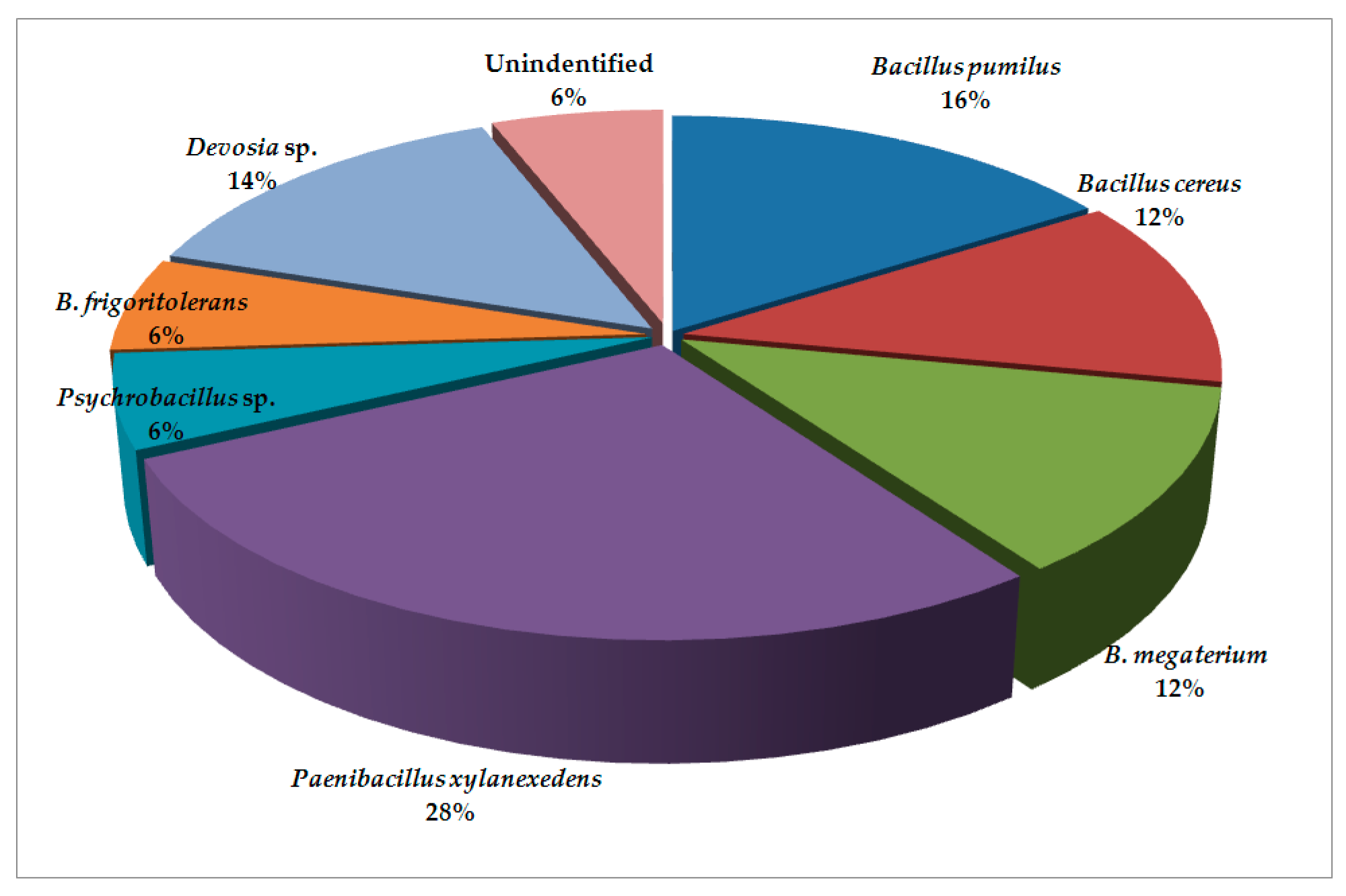
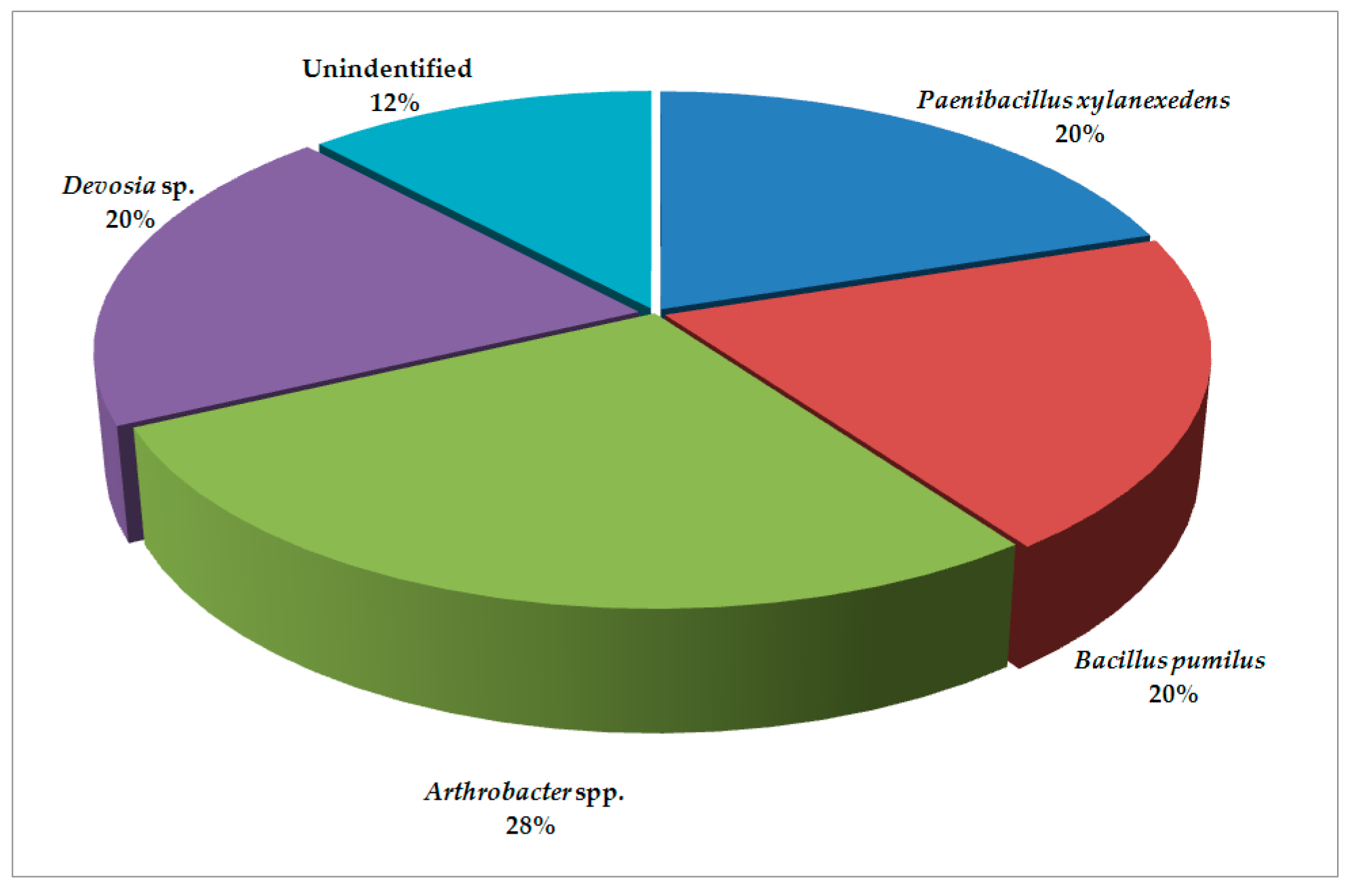

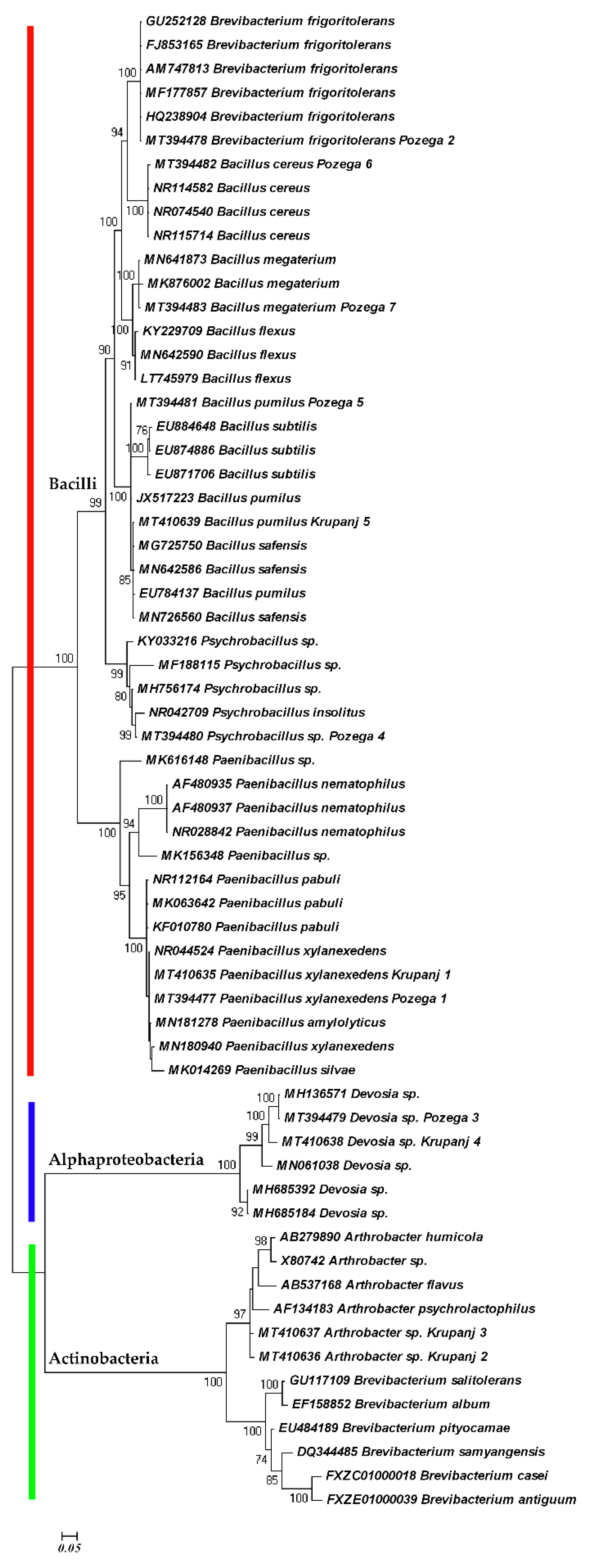
| Climate Factors (Units) | Locations | Means | SD | Range | HSD (p = 0.05) |
|---|---|---|---|---|---|
| Optimum Air Temperature (°C) | Pozega | 10.5 | 0.6 | 9.0–10.9 | a |
| Krupanj | 12.6 | 0.8 | 10.6–13.4 | a | |
| Maximum Air Temperature (°C) | Pozega | 17.4 | 0.7 | 15.0–18.0 | a |
| Krupanj | 18.6 | 1.0 | 15.7–19.7 | a | |
| Minimum Air Temperature (°C) | Pozega | 5.1 | 0.7 | 3.4–6.6 | a |
| Krupanj | 7.7 | 0.7 | 5.0–8.8 | a | |
| Relative Humidity (%) | Pozega | 82.1 | 3.1 | 74–85 | a |
| Krupanj | 77.8 | 4.0 | 67–86 | a | |
| Insolation *(h) | Pozega | 1634.6 | 253.0 | 1110.5–2064.2 | b |
| Krupanj | 2107.2 | 188.8 | 1701.4–2381.7 | a | |
| Cloudiness | Pozega | 6.5 | 0.5 | 5.3–7.1 | a |
| Krupanj | 5.8 | 0.4 | 4.7–6.4 | a | |
| Precipitation *(mm) | Pozega | 762.3 | 154.4 | 460.6–1121.5 | b |
| Krupanj | 902.5 | 160.7 | 529.2–1242.4 | a |
| Physicochemical Parameters | Locations | Values | HSD (p = 0.05) |
|---|---|---|---|
| pH (H2O) | Pozega | 7.73 | a |
| Krupanj | 7.01 | a | |
| pH (1M KCl) | Pozega | 6.71 | a |
| Krupanj | 6.26 | a | |
| Soil organic matter (%) | Pozega | 5.24 | a |
| Krupanj | 3.33 | a | |
| N (%) | Pozega | 0.22 | a |
| Krupanj | 0.23 | a | |
| P2O5 (mg/100 g) | Pozega | 28.21 | a |
| Krupanj | 29.80 | a | |
| K2O * (mg/100 g) | Pozega | 24.50 | b |
| Krupanj | 61.88 | a | |
| Sand particles (>0.2 mm)% | Pozega | 2.4 | a |
| Krupanj | 1.5 | a | |
| Sand particles (0.02–0.2 mm)% | Pozega | 19.9 | a |
| Krupanj | 17.6 | a | |
| Silt (0.002–0.02 mm)% | Pozega | 40.6 | a |
| Krupanj | 35.6 | a | |
| Clay (<0.002 mm)% | Pozega | 37.1 | a |
| Krupanj | 45.3 | a | |
| Silt+Clay (<0.02 mm)% | Pozega | 77.7 | a |
| Krupanj | 80.9 | a |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oro, V.; Knezevic, M.; Dinic, Z.; Delic, D. Bacterial Microbiota Isolated from Cysts of Globodera rostochiensis (Nematoda: Heteroderidae). Plants 2020, 9, 1146. https://doi.org/10.3390/plants9091146
Oro V, Knezevic M, Dinic Z, Delic D. Bacterial Microbiota Isolated from Cysts of Globodera rostochiensis (Nematoda: Heteroderidae). Plants. 2020; 9(9):1146. https://doi.org/10.3390/plants9091146
Chicago/Turabian StyleOro, Violeta, Magdalena Knezevic, Zoran Dinic, and Dusica Delic. 2020. "Bacterial Microbiota Isolated from Cysts of Globodera rostochiensis (Nematoda: Heteroderidae)" Plants 9, no. 9: 1146. https://doi.org/10.3390/plants9091146
APA StyleOro, V., Knezevic, M., Dinic, Z., & Delic, D. (2020). Bacterial Microbiota Isolated from Cysts of Globodera rostochiensis (Nematoda: Heteroderidae). Plants, 9(9), 1146. https://doi.org/10.3390/plants9091146





