Dynamics and Functions of Stress Granules and Processing Bodies in Plants
Abstract
1. Introduction
2. Stress Granules
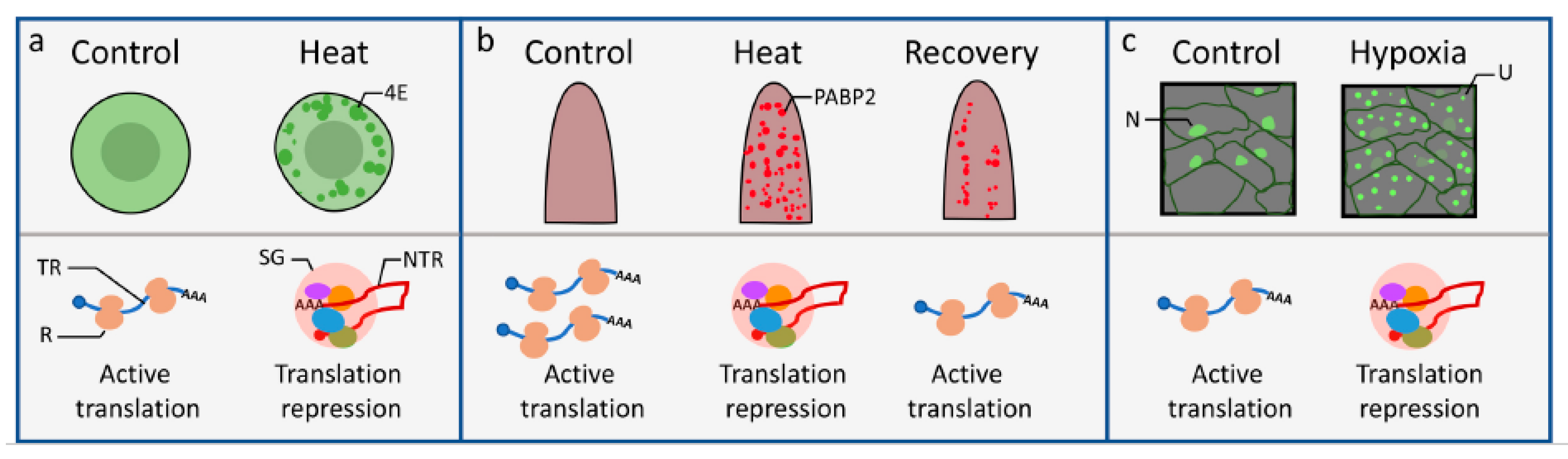
2.1. SG and Heat Stress
2.2. SGs and Hypoxia Stress
2.3. Regulation of SG Dynamics and Movement
3. P-Bodies
3.1. P-Body-Mediated Translation Repression and Developmental Regulation
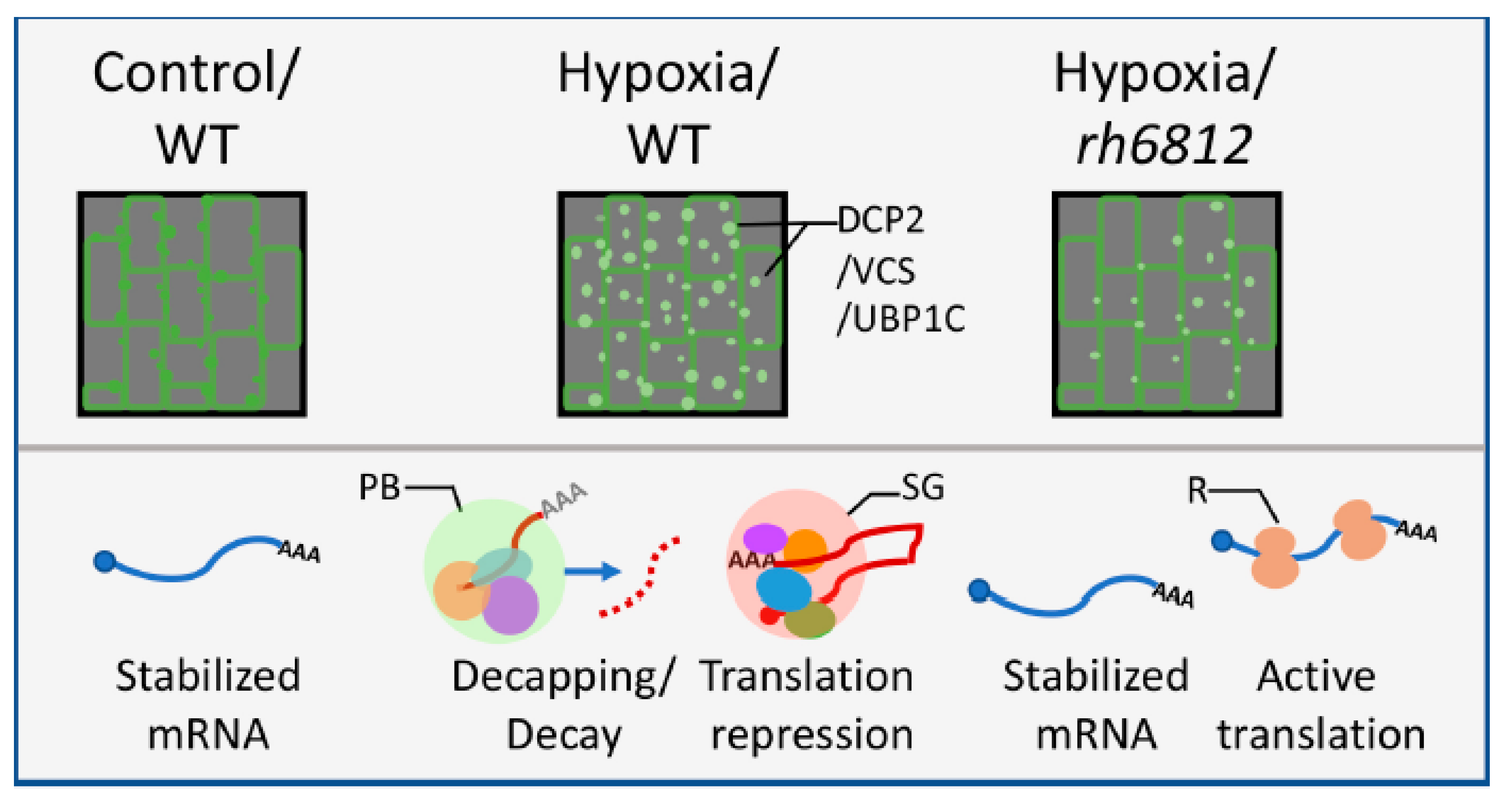
3.2. P-Body-Mediated Decapping/Decay for Abiotic Responses
3.3. P-Body Assembly, mRNA Decay, and Translation Repression in Plant Immunity
4. Future Perspectives
Funding
Conflicts of Interest
References
- Hu, W.; Sweet, T.J.; Chamnongpol, S.; Baker, K.E.; Coller, J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature 2009, 461, 225–229. [Google Scholar] [CrossRef]
- Chen, C.-Y.A.; Shyu, A.-B. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip. Rev. RNA 2011, 2, 167–183. [Google Scholar] [CrossRef] [PubMed]
- Boeynaems, S.; Alberti, S.; Fawzi, N.L.; Mittag, T.; Polymenidou, M.; Rousseau, F.; Schymkowitz, J.; Shorter, J.; Wolozin, B.; Bosch, L.V.D.; et al. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol. 2018, 28, 420–435. [Google Scholar] [CrossRef]
- Luo, Y.; Na, Z.; Slavoff, S.A. P-Bodies: Composition, Properties, and Functions. Biochemistry 2018, 57, 2424–2431. [Google Scholar] [CrossRef] [PubMed]
- Decker, C.J.; Parker, R. P-Bodies and Stress Granules: Possible Roles in the Control of Translation and mRNA Degradation. Cold Spring Harb. Perspect. Biol. 2012, 4, a012286. [Google Scholar] [CrossRef] [PubMed]
- Chantarachot, T.; Bailey-Serres, J. Polysomes, Stress Granules, and Processing Bodies: A Dynamic Triumvirate Controlling Cytoplasmic mRNA Fate and Function. Plant Physiol. 2018, 176, 254–269. [Google Scholar] [CrossRef] [PubMed]
- Maldonado-Bonilla, L.D. Composition and function of P bodies in Arabidopsis thaliana. Front. Plant Sci. 2014, 5, 201. [Google Scholar] [CrossRef] [PubMed]
- Kosmacz, M.; Gorka, M.; Schmidt, S.; Luzarowski, M.; Moreno, J.C.; Szlachetko, J.; Leniak, E.; Sokolowska, E.M.; Sofroni, K.; Schnittger, A.; et al. Protein and metabolite composition of Arabidopsis stress granules. New Phytol. 2019, 222, 1420–1433. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.; Anderson, P. Stress granules: Sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 2002, 30, 963–969. [Google Scholar] [CrossRef]
- Protter, D.S.W.; Parker, R. Principles and Properties of Stress Granules. Trends Cell Biol. 2016, 26, 668–679. [Google Scholar] [CrossRef]
- Nover, L.; Scharf, K.D.; Neumann, D. Formation of cytoplasmic heat shock granules in tomato cell cultures and leaves. Mol. Cell. Biol. 1983, 3, 1648–1655. [Google Scholar] [CrossRef] [PubMed]
- Collier, N.C.; Heuser, J.; Levy, M.A.; Schlesinger, M.J. Ultrastructural and biochemical analysis of the stress granule in chicken embryo fibroblasts. J. Cell Biol. 1988, 106, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Hoyle, N.P.; Castelli, L.M.; Campbell, S.G.; Holmes, L.E.; Ashe, M.P. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J. Cell Biol. 2007, 179, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Beltran, E.; Moschou, P.N.; Smertenko, A.; Bozhkov, P.V. Tudor Staphylococcal Nuclease Links Formation of Stress Granules and Processing Bodies with mRNA Catabolism in Arabidopsis. Plant Cell 2015, 27, 926–943. [Google Scholar] [CrossRef] [PubMed]
- Pomeranz, M.C.; Hah, C.; Lin, P.-C.; Kang, S.G.; Finer, J.J.; Blackshear, P.J.; Jang, J.-C. The Arabidopsis Tandem Zinc Finger Protein AtTZF1 Traffics between the Nucleus and Cytoplasmic Foci and Binds Both DNA and RNA. Plant Physiol. 2010, 152, 151–165. [Google Scholar] [CrossRef] [PubMed]
- Sorenson, R.; Bailey-Serres, J. Selective mRNA sequestration by OLIGOURIDYLATE-BINDING PROTEIN 1 contributes to translational control during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 2373–2378. [Google Scholar] [CrossRef]
- Weber, C.; Nover, L.; Fauth, M. Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J. 2008, 56, 517–530. [Google Scholar] [CrossRef]
- Yan, C.; Yan, Z.; Wang, Y.; Yan, X.; Han, Y. Tudor-SN, a component of stress granules, regulates growth under salt stress by modulating GA20ox3 mRNA levels in Arabidopsis. J. Exp. Bot. 2014, 65, 5933–5944. [Google Scholar] [CrossRef]
- Haslbeck, M.; Vierling, E. A First Line of Stress Defense: Small Heat Shock Proteins and Their Function in Protein Homeostasis. J. Mol. Biol. 2015, 427, 1537–1548. [Google Scholar] [CrossRef]
- Merret, R.; Carpentier, M.-C.; Favory, J.-J.; Picart, C.; Descombin, J.; Bousquet-Antonelli, C.; Tillard, P.; Lejay, L.; Deragon, J.-M.; Charng, Y.-Y. Heat Shock Protein HSP101 Affects the Release of Ribosomal Protein mRNAs for Recovery after Heat Shock. Plant Physiol. 2017, 174, 1216–1225. [Google Scholar] [CrossRef]
- Cherkasov, V.; Hofmann, S.; Druffel-Augustin, S.; Mogk, A.; Tyedmers, J.; Stoecklin, G.; Bukau, B. Coordination of Translational Control and Protein Homeostasis during Severe Heat Stress. Curr. Biol. 2013, 23, 2452–2462. [Google Scholar] [CrossRef] [PubMed]
- Merret, R.; Descombin, J.; Juan, Y.-T.; Favory, J.-J.; Carpentier, M.-C.; Chaparro, C.; Charng, Y.-Y.; Deragon, J.-M.; Bousquet-Antonelli, C. XRN4 and LARP1 Are Required for a Heat-Triggered mRNA Decay Pathway Involved in Plant Acclimation and Survival during Thermal Stress. Cell Rep. 2013, 5, 1279–1293. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, M.; Winkelhaus, S.; Thierfelder, J.M.; Nover, L. Transient expression and heat-stress-induced co-aggregation of endogenous and heterologous small heat-stress proteins in tobacco protoplasts. Plant J. 2000, 24, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Nover, L.; Scharf, K.D.; Neumann, D. Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol. Cell. Biol. 1989, 9, 1298–1308. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, M.E.; Chang, I.-F.; Gong, F.; Galbraith, D.W.; Bailey-Serres, J. Immunopurification of Polyribosomal Complexes of Arabidopsis for Global Analysis of Gene Expression. Plant Physiol. 2005, 138, 624–635. [Google Scholar] [CrossRef] [PubMed]
- Juntawong, P.; Girke, T.; Bazin, J.; Bailey-Serres, J. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, E203–E212. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.L.; Gupta, M.; Li, W.; Miller, I.; Anderson, P.J. RNA-Binding Proteins Tia-1 and Tiar Link the Phosphorylation of Eif-2α to the Assembly of Mammalian Stress Granules. J. Cell Biol. 1999, 147, 1431–1442. [Google Scholar] [CrossRef] [PubMed]
- Gilks, N.; Kedersha, N.; Ayodele, M.; Shen, L.; Stoecklin, G.; Dember, L.M.; Anderson, P. Stress Granule Assembly Is Mediated by Prion-like Aggregation of TIA-1. Mol. Biol. Cell 2004, 15, 5383–5398. [Google Scholar] [CrossRef]
- Hamada, T.; Yako, M.; Minegishi, M.; Sato, M.; Kamei, Y.; Yanagawa, Y.; Toyooka, K.; Watanabe, Y.; Hara-Nishimura, I. Stress granule formation is induced by a threshold temperature rather than a temperature difference inArabidopsis. J. Cell Sci. 2018, 131, jcs216051. [Google Scholar] [CrossRef]
- Jackson, E.K.; Ren, J.; Mi, Z. Extracellular 2′,3′-cAMP Is a Source of Adenosine. J. Biol. Chem. 2009, 284, 33097–33106. [Google Scholar] [CrossRef]
- Verrier, J.D.; Jackson, T.C.; Bansal, R.; Kochanek, P.; Puccio, A.M.; Okonkwo, D.O.; Jackson, E.K. The brain in vivo expresses the 2′,3′-cAMP-adenosine pathway. J. Neurochem. 2012, 122, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, T.; Blancquaert, D.; Couturon, P.; Van Der Straeten, D.; Sandra, P.; Lynen, F. Wounding stress causes rapid increase in concentration of the naturally occurring 2′,3′-isomers of cyclic guanosine- and cyclic adenosine monophosphate (cGMP and cAMP) in plant tissues. Phytochemistry 2014, 103, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Kosmacz, M.; Luzarowski, M.; Kerber, O.; Leniak, E.; Gutierrez-Beltran, E.; Moreno, J.C.; Gorka, M.; Szlachetko, J.; Veyel, D.; Graf, A.; et al. Interaction of 2′,3′-cAMP with Rbp47b Plays a Role in Stress Granule Formation. Plant Physiol. 2018, 177, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Chua, N.-H. Processing bodies and plant development. Curr. Opin. Plant Biol. 2011, 14, 88–93. [Google Scholar] [CrossRef]
- Johnson, P.R.; Ecker, J.R. The Ethylene Gas Signal Transduction Pathway: A Molecular Perspective. Annu. Rev. Genet. 1998, 32, 227–254. [Google Scholar] [CrossRef]
- Li, W.; Ma, M.; Feng, Y.; Li, H.; Wang, Y.; Ma, Y.; Li, M.; An, F.; Guo, H. EIN2-Directed Translational Regulation of Ethylene Signaling in Arabidopsis. Cell 2015, 163, 670–683. [Google Scholar] [CrossRef]
- Jang, G.-J.; Yang, J.-Y.; Hsieh, H.-L.; Wu, S.-H. Processing bodies control the selective translation for optimal development of Arabidopsis young seedlings. Proc. Natl. Acad. Sci. USA 2019, 116, 6451–6456. [Google Scholar] [CrossRef]
- Xu, J.; Yang, J.-Y.; Niu, Q.-W.; Chua, N.-H. Arabidopsis DCP2, DCP1, and VARICOSE Form a Decapping Complex Required for Postembryonic Development. Plant Cell 2006, 18, 3386–3398. [Google Scholar] [CrossRef]
- Xu, J.; Chua, N.-H. Arabidopsis Decapping 5 Is Required for mRNA Decapping, P-Body Formation, and Translational Repression during Postembryonic Development. Plant Cell 2009, 21, 3270–3279. [Google Scholar] [CrossRef]
- Perea-Resa, C.; Carrasco-López, C.; Catala, R.; Turečková, V.; Novak, O.; Zhang, W.; Sieburth, L.E.; Jiménez-Gómez, J.M.; Salinas, J. The LSM1-7 Complex Differentially Regulates Arabidopsis Tolerance to Abiotic Stress Conditions by Promoting Selective mRNA Decapping. Plant Cell 2016, 28, 505–520. [Google Scholar] [CrossRef]
- Walley, J.W.; Kelley, D.R.; Nestorova, G.; Hirschberg, D.L.; Dehesh, K. Arabidopsis Deadenylases AtCAF1a and AtCAF1b Play Overlapping and Distinct Roles in Mediating Environmental Stress Responses. Plant Physiol. 2010, 152, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, B.; Jang, G.-J.; Jiang, S.; Jiang, D.; Jang, J.-C.; Wu, S.-H.; Shan, L.; He, P. Orchestration of Processing Body Dynamics and mRNA Decay in Arabidopsis Immunity. Cell Rep. 2019, 28, 2194–2205. [Google Scholar] [CrossRef]
- Tabassum, N.; Eschen-Lippold, L.; Athmer, B.; Baruah, M.; Brode, M.; Maldonado-Bonilla, L.D.; Hoehenwarter, W.; Hause, G.; Scheel, D.; Lee, J.; et al. Phosphorylation-dependent control of an RNA granule-localized protein that fine-tunes defence gene expression at a post-transcriptional level. Plant J. 2020, 101, 1023–1039. [Google Scholar] [CrossRef] [PubMed]
- Roux, M.; Rasmussen, M.W.; Palma, K.; Lolle, S.; Regué, A.M.; Bethke, G.; Glazebrook, J.; Zhang, W.; Sieburth, L.; Larsen, M.R.; et al. The mRNA decay factor PAT 1 functions in a pathway including MAP kinase 4 and immune receptor SUMM 2. EMBO J. 2015, 34, 593–608. [Google Scholar] [CrossRef]
- Soma, F.; Mogami, J.; Yoshida, T.; Abekura, M.; Takahashi, F.; Kidokoro, S.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-unresponsive SnRK2 protein kinases regulate mRNA decay under osmotic stress in plants. Nat. Plants 2017, 3, 16204. [Google Scholar] [CrossRef] [PubMed]
- Steffens, A.; Bräutigam, A.; Jakoby, M.; Hülskamp, M. The BEACH Domain Protein SPIRRIG Is Essential for Arabidopsis Salt Stress Tolerance and Functions as a Regulator of Transcript Stabilization and Localization. PLoS Biol. 2015, 13, e1002188. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Li, C.; Liu, F.; Jiang, H.; Li, S.; Sun, J.; Wu, X.; Li, C. The Arabidopsis homologs of CCR4-associated factor 1 show mRNA deadenylation activity and play a role in plant defence responses. Cell Res. 2008, 19, 307–316. [Google Scholar] [CrossRef]
- Chantarachot, T.; Sorenson, R.S.; Hummel, M.; Ke, H.; Kettenburg, A.T.; Chen, D.; Aiyetiwa, K.; Dehesh, K.; Eulgem, T.; Sieburth, L.E.; et al. DHH1/DDX6-like RNA helicases maintain ephemeral half-lives of stress-response mRNAs. Nat. Plants 2020, 6, 675–685. [Google Scholar] [CrossRef]
- Mäkinen, K.; Lõhmus, A.; Pollari, M. Plant RNA Regulatory Network and RNA Granules in Virus Infection. Front. Plant Sci. 2017, 8, 2093. [Google Scholar] [CrossRef]
- Hondele, M.; Sachdev, R.; Heinrich, S.; Wang, J.; Vallotton, P.; Fontoura, B.M.A.; Weis, K. DEAD-box ATPases are global regulators of phase-separated organelles. Nature 2019, 573, 144–148. [Google Scholar] [CrossRef]
- Aizer, A.; Kalo, A.; Kafri, P.; Shraga, A.; Ben-Yishay, R.; Jacob, A.I.; Kinor, N.; Shav-Tal, Y. Quantifying mRNA targeting to P-bodies in living human cells reveals their dual role in mRNA decay and storage. J. Cell Sci. 2014, 127, 4443–4456. [Google Scholar] [CrossRef] [PubMed]
- Horvathova, I.; Voigt, F.; Kotrys, A.V.; Zhan, Y.; Artus-Revel, C.G.; Eglinger, J.; Stadler, M.B.; Giorgetti, L.; Chao, J.A. The Dynamics of mRNA Turnover Revealed by Single-Molecule Imaging in Single Cells. Mol. Cell 2017, 68, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Hubstenberger, A.; Courel, M.; Benard, M.; Souquère, S.; Ernoult-Lange, M.; Chouaib, R.; Yi, Z.; Morlot, J.-B.; Munier, A.; Fradet, M.; et al. P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Mol. Cell 2017, 68, 144–157. [Google Scholar] [CrossRef]
- Wheeler, J.R.; Jain, S.; Khong, A.; Parker, R. Isolation of yeast and mammalian stress granule cores. Methods 2017, 126, 12–17. [Google Scholar] [CrossRef]
- Markmiller, S.; Soltanieh, S.; Server, K.L.; Mak, R.; Jin, W.; Fang, M.Y.; Luo, E.-C.; Krach, F.; Yang, D.; Sen, A.; et al. Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 2018, 172, 590–604. [Google Scholar] [CrossRef] [PubMed]
- Youn, J.-Y.; Dunham, W.H.; Hong, S.J.; Knight, J.D.; Bashkurov, M.; Chen, G.I.; Bagci, H.; Rathod, B.; MacLeod, G.; Eng, S.W.M.; et al. High-Density Proximity Mapping Reveals the Subcellular Organization of mRNA-Associated Granules and Bodies. Mol. Cell 2018, 69, 517–532. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, M.; Porter, D.F.; Khavari, P.A. Methods to study RNA-protein interactions. Nat. Methods 2019, 16, 225–234. [Google Scholar] [CrossRef]
- Ries, R.J.; Zaccara, S.; Klein, P.; Olarerin-George, A.; Namkoong, S.; Pickering, B.F.; Patil, D.P.; Kwak, H.; Lee, J.H.; Jaffrey, S.R. m6A enhances the phase separation potential of mRNA. Nature 2019, 571, 424–428. [Google Scholar] [CrossRef]
- Scutenaire, J.; Deragon, J.-M.; Jean, V.; Benhamed, M.; Raynaud, C.; Favory, J.-J.; Merret, R.; Bousquet-Antonelli, C. The YTH Domain Protein ECT2 Is an m6A Reader Required for Normal Trichome Branching in Arabidopsis. Plant Cell 2018, 30, 986–1005. [Google Scholar] [CrossRef]
- Aizer, A.; Brody, Y.; Ler, L.W.; Sonenberg, N.; Singer, R.H.; Shav-Tal, Y. The Dynamics of Mammalian P Body Transport, Assembly, and Disassembly In Vivo. Mol. Biol. Cell 2008, 19, 4154–4166. [Google Scholar] [CrossRef]
- Nadezhdina, E.; Lomakin, A.; Shpilman, A.A.; Chudinova, E.M.; Ivanov, P.A. Microtubules govern stress granule mobility and dynamics. Biochim. Biophys. Acta BBA Mol. Cell Res. 2010, 1803, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Hamada, T.; Tominaga, M.; Fukaya, T.; Nakamura, M.; Nakano, A.; Watanabe, Y.; Hashimoto, T.; Baskin, T.I. RNA Processing Bodies, Peroxisomes, Golgi Bodies, Mitochondria, and Endoplasmic Reticulum Tubule Junctions Frequently Pause at Cortical Microtubules. Plant Cell Physiol. 2012, 53, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Steffens, A.; Jaegle, B.; Tresch, A.; Hülskamp, M.; Jakoby, M. Processing-Body Movement in Arabidopsis Depends on an Interaction between Myosins and DECAPPING PROTEIN1. Plant Physiol. 2014, 164, 1879–1892. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, G.-J.; Jang, J.-C.; Wu, S.-H. Dynamics and Functions of Stress Granules and Processing Bodies in Plants. Plants 2020, 9, 1122. https://doi.org/10.3390/plants9091122
Jang G-J, Jang J-C, Wu S-H. Dynamics and Functions of Stress Granules and Processing Bodies in Plants. Plants. 2020; 9(9):1122. https://doi.org/10.3390/plants9091122
Chicago/Turabian StyleJang, Geng-Jen, Jyan-Chyun Jang, and Shu-Hsing Wu. 2020. "Dynamics and Functions of Stress Granules and Processing Bodies in Plants" Plants 9, no. 9: 1122. https://doi.org/10.3390/plants9091122
APA StyleJang, G.-J., Jang, J.-C., & Wu, S.-H. (2020). Dynamics and Functions of Stress Granules and Processing Bodies in Plants. Plants, 9(9), 1122. https://doi.org/10.3390/plants9091122




