Keep Calm and Survive: Adaptation Strategies to Energy Crisis in Fruit Trees under Root Hypoxia
Abstract
:1. Introduction
2. Edaphic Conditions that Promote O2 Deficiency
3. Fruit Tree Responses to O2 Deficiency
3.1. Physiological and Biochemical Response of Fruit Trees under O2 Deficiency
3.2. Morpho-Anatomical Changes in Fruit Trees under O2 Deficiency
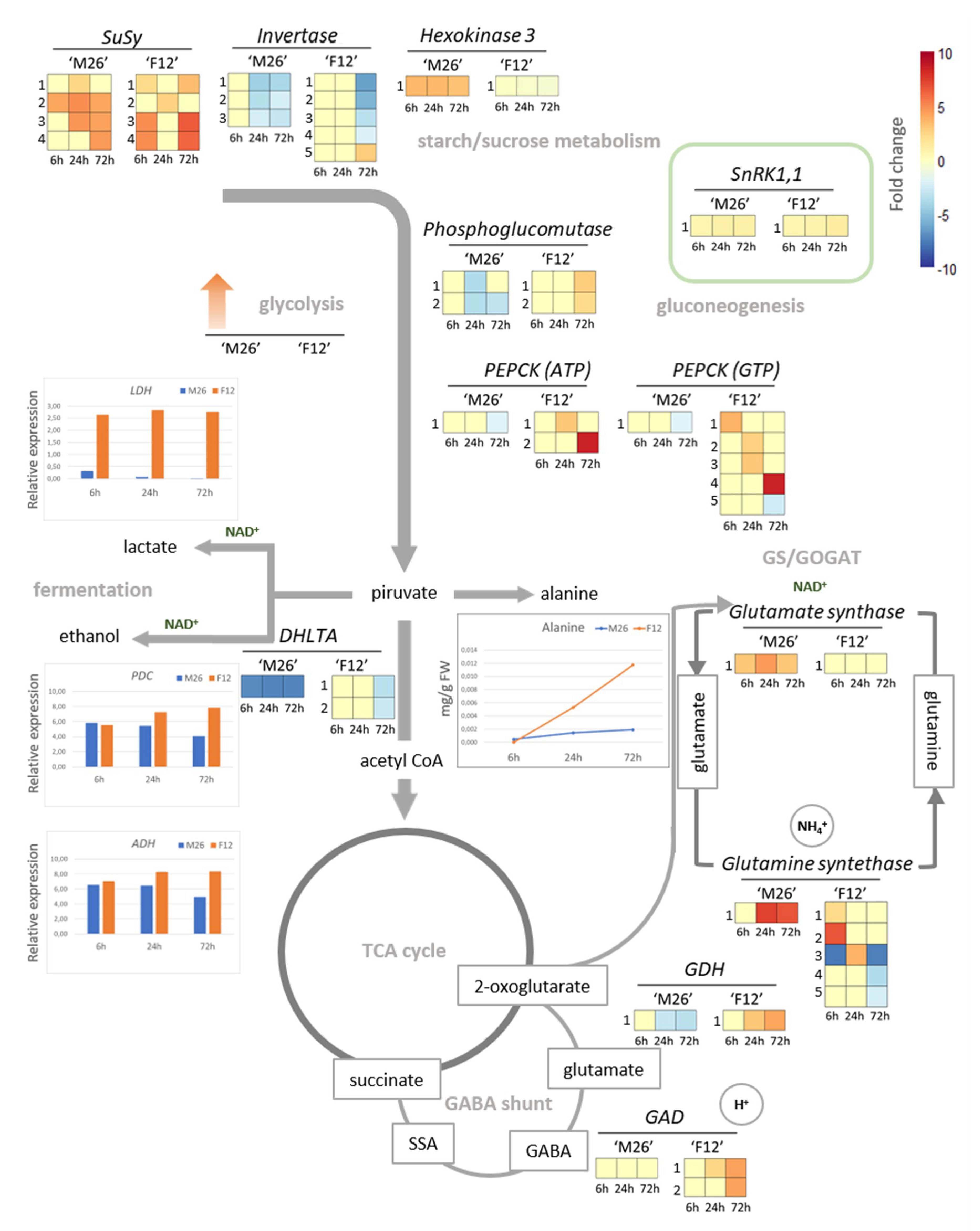
4. Transcriptomic Reprogramming of Principal Pathways Involved in Energy Metabolism under O2 Deficiency
5. Root Respiration under O2 Deficiency
6. Conclusions and Perspectives in Fruit Trees Research
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Loreti, E.; Perata, P. The many facets of hypoxia in plants. Plants 2020, 9, 745. [Google Scholar] [CrossRef]
- FAOSTAT. Crops Production in 2018. 2018. Available online: http://faostat.fao.org/ (accessed on 5 June 2019).
- Kreuzwieser, J.; Rennenberg, H. Molecular and physiological responses of trees to waterlogging stress. Plant Cell Environ. 2014, 37, 2245–2259. [Google Scholar] [CrossRef] [PubMed]
- Herrera, A.; Tezara, W.; Marín, O.; Rengifo, E. Stomatal and non-stomatal limitations of photosynthesis in trees of a tropical seasonally flooded forest. Physiol. Plant. 2008, 134, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Calvo-Polanco, M.; Senorans, J.; Zwiazek, J. Role of adventitious roots in water relations of tamarack (Larix laricina) seedlings exposed to flooding. BMC Plant Biol. 2012, 12, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrera, A. Responses to flooding of plant water relations and leaf gas exchange in tropical tolerant trees of a black-water wetland. Front. Plant Sci. 2013, 4, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Pimentel, P.; Almada, R.; Salvatierra, A.; Toro, G.; Arismendi, M.J.; Pino, M.T.; Sagredo, B.; Pinto, M. Physiological and morphological responses of Prunus species with different degree of tolerance to long-term root hypoxia. Sci. Hortic. 2014, 180, 14–23. [Google Scholar] [CrossRef]
- Warschefsky, E.J.; Klein, L.L.; Frank, M.H.; Chitwood, D.H.; Londo, J.P.; Von Wettberg, E.J.B.; Miller, A.J. Rootstocks: Diversity, domestication, and impacts on shoot phenotypes. Trends Plant Sci. 2016, 21, 418–437. [Google Scholar] [CrossRef]
- Martínez-Ballesta, M.C.; Alcaraz-López, C.; Muries, B.; Mota-Cadenas, C.; Carvajal, M. Physiological aspects of rootstock–scion interactions. Sci. Hort. 2010, 127, 112–118. [Google Scholar] [CrossRef]
- Mizutani, F.; Yamada, M.; Tomana, T. Differential water tolerance and ethanol accumulation in Prunus species under flooded conditions. J. Jpn. Soc. Hortic. Sci. 1982, 51, 29–34. [Google Scholar] [CrossRef] [Green Version]
- Ranney, T.G. Differential tolerance of eleven Prunus taxa to root zone flooding. J. Environ. Hortic. 1994, 12, 138–141. [Google Scholar] [CrossRef]
- Pinochet, J. ‘Replantpac’ (Rootpac® R), a plum–almond hybrid rootstock for replant situations. HortScience 2010, 45, 299–301. [Google Scholar] [CrossRef] [Green Version]
- Rubio Cabetas, M.J.; Pons, C.; Amador Delgado, M.L.; Marti, C.; Granell, A. Transcriptomic analysis of two Prunus genotypes differing in waterlogging response reveals the importance of ANP and hypoxia-associated oxidative response. In Proceedings of the Thirteenth Eucarpia Symposium on Fruit Breeding and Genetics, Warsaw, Poland, 11 September 2011. [Google Scholar]
- Iacona, C.; Cirilli, M.; Zega, A.; Frioni, E.; Silvestri, C.; Muleo, R. A somaclonal myrobalan rootstock increases waterlogging tolerance to peach cultivar in controlled conditions. Sci. Hort. 2013, 156, 1–8. [Google Scholar] [CrossRef]
- Aguilera, E.; Díaz-Gaona, C.; García-Laureano, R.; Reyes-Palomo, C.; Guzmán, G.I.; Ortolani, L.; Sánchez-Rodríguez, M.; Rodríguez-Estévez, V. Agroecology for adaptation to climate change and resource depletion in the Mediterranean region. A review. Agric. Syst. 2020, 181, 102809. [Google Scholar] [CrossRef]
- Venkatramanan, V.; Shah, S.; Prasad, R. Global Climate Change and Environmental Policy; Springer: Singapore, 2020. [Google Scholar]
- Hillel, D. Introduction to Environmental Soil Physics. Eur. J. Soil Sci. 2004, 56, 684. [Google Scholar] [CrossRef]
- Bhattarai, S.; Su, N.; Midmore, D. Oxygation unlocks yield potentials of crops in oxygen-limited soil environments. Adv. Agron. 2005, 88, 313–377. [Google Scholar] [CrossRef]
- Batey, T. Soil compaction and soil management—A Review. Soil Use Manag. 2009, 25, 335–345. [Google Scholar] [CrossRef]
- Morales-Olmedo, M.; Ortiz, M.; Sellés, G. Effects of transient soil waterlogging and its importance for rootstock selection. Chil. J. Agric. Res. 2015, 75, 45–56. [Google Scholar] [CrossRef] [Green Version]
- Ellies, A.; Horn, R.; Smith, R. Effect of management of a volcanic ash soil on structural properties. Int. Agrophys 2000, 14, 377–384. [Google Scholar]
- Seguel, O.; Farías, E.; Luzio, W.; Casanova, M.; Pino, I.; Parada, M.; Videla, X.; Nario, A. Changes in soil physical properties on hillsides vineyard (Vitis vinifera). In Proceedings of the ISTRO 18th Triennial conference, Izmir, Turkey, 15–19 June 2009; pp. 15–19. [Google Scholar]
- Becerra, A.T.; Botta, G.F.; Bravo, X.L.; Tourn, M.; Melcon, F.B.; Vazquez, J.; Rivero, D.; Linares, P.; Nardon, G. Soil compaction distribution under tractor traffic in almond (Prunus amigdalus L.) orchard in Almería España. Soil Tillage Res. 2010, 107, 49–56. [Google Scholar] [CrossRef]
- Cook, F.J.; Knight, J.H. Oxygen transport to plant roots. Soil Sci. Soc. Am. J. 2003, 67, 20–31. [Google Scholar] [CrossRef]
- Dexter, A.R. Advances in characterization of soil structure. Soil Tillage Res. 1988, 11, 199–238. [Google Scholar] [CrossRef]
- Kawase, M. Anatomical and morphological adaptation of plants to waterlogging. Hort. Sci. 1981, 16, 8–12. [Google Scholar]
- Shabala, S. Physiological and cellular aspects of phytotoxicity tolerance in plants: The role of membrane transporters and implications for crop breeding for waterlogging tolerance. New Phytol. 2011, 190, 289–298. [Google Scholar] [CrossRef] [Green Version]
- Unger, I.M.; Kennedy, A.C.; Muzika, R.-M. Flooding effects on soil microbial communities. Appl. Soil Ecol. 2009, 42, 1–8. [Google Scholar] [CrossRef]
- Vepraskas, M.J.; Faulkner, S.; Richardson, J. Redox chemistry of hydric soils. In Wetland Soils: Genesis, Hydrology, Landscapes, and Classification; CRC Press: Boca Raton, FL, USA, 2001; pp. 85–106. [Google Scholar]
- Ponnamperuma, F. Flooding and plant growth. Effects of flooding on soils. In Flooding and Plant Growth; NRC Research Press: Ottawa, ON, Canada, 1984; pp. 9–45. [Google Scholar]
- McKee, W.H., Jr.; McKevlin, M.R. Geochemical processes and nutrient uptake by plants in hydric soils. Environ. Toxicol. Chem. 1993, 12, 2197–2207. [Google Scholar] [CrossRef]
- Drew, M. Oxygen deficiency and root metabolism: Injury and acclimation under hypoxia and anoxia. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 223–250. [Google Scholar] [CrossRef]
- Sanclemente, M.A.; Schaffer, B.; Gil, P.M.; Vargas, A.I.; Davies, F.S. Pruning after flooding hastens recovery of flood-stressed avocado (Persea americana Mill.) trees. Sci. Hort. 2014, 169, 27–35. [Google Scholar] [CrossRef]
- Savé, R.; Serrano, L. Some physiological and growth responses of kiwi fruit (Actinidia chinensis) to flooding. Physiol. Plant. 1986, 66, 75–78. [Google Scholar] [CrossRef]
- Vu, J.; Yelenosky, G. Photosynthetic responses of citrus trees to soil flooding. Physiol. Plant. 1991, 81, 7–14. [Google Scholar] [CrossRef]
- Hossain, Z.; López-Climent, M.F.; Arbona, V.; Pérez-Clemente, R.M.; Gómez-Cadenas, A. Modulation of the antioxidant system in citrus under waterlogging and subsequent drainage. J. Plant Physiol. 2009, 166, 1391–1404. [Google Scholar] [CrossRef]
- Arbona, V.; López-Climent, M.F.; Pérez-Clemente, R.M.; Gómez-Cadenas, A. Maintenance of a high photosynthetic performance is linked to flooding tolerance in citrus. Environ. Exp. Bot. 2009, 66, 135–142. [Google Scholar] [CrossRef]
- García-Sánchez, F.; Syvertsen, J.P.; Gimeno, V.; Botía, P.; Perez-Perez, J.G. Responses to flooding and drought stress by two citrus rootstock seedlings with different water-use efficiency. Physiol. Plant. 2007, 130, 532–542. [Google Scholar] [CrossRef]
- Kallestad, J.C.; Sammis, T.W.; Mexala, J.G.; Gutschick, V. The impact of prolonged flood-irrigation on leaf gas exchange in mature pecans in an orchard setting. Int. J. Plant Prod. 2012, 1, 163–178. [Google Scholar] [CrossRef]
- Belloni, V.; Mapelli, S. Effects of drought or flooding stresses on photosynthesis xylem flux and stem radial growth. Acta Hortic. 2001, 544, 327–333. [Google Scholar] [CrossRef]
- Ruperti, B.; Botton, A.; Populin, F.; Eccher, G.; Brilli, M.; Quaggiotti, S.; Trevisan, S.; Cainelli, N.; Guarracino, P.; Schievano, E.; et al. Flooding responses on grapevine: A physiological, transcriptional, and metabolic perspective. Front. Plant Sci. 2019, 10, 339. [Google Scholar] [CrossRef] [Green Version]
- Striegler, R.K.; Howell, G.S.; Flore, J.A. Influence of rootstock on the response of Seyval grapevines to flooding stress. Am. Soc. Enol. Vitic. 1993, 44, 313–319. [Google Scholar]
- Olmo-Vega, A.; García-Sánchez, F.; Simón-Grao, S.; Simón, I.; Lidón, V.; Nieves, M.; Martínez-Nicolás, J.J. Physiological responses of three pomegranate cultivars under flooded conditions. Sci. Hort. 2017, 224, 171–179. [Google Scholar] [CrossRef]
- Bhusal, N.; Kim, H.S.; Han, S.-G.; Yoon, T.-M. Photosynthetic traits and plant–water relations of two apple cultivars grown as bi-leader trees under long-term waterlogging conditions. Environ. Exp. Bot. 2020, 176, 104111. [Google Scholar] [CrossRef]
- Domingo, R.; Pérez-Pastor, A.; Ruiz-Sánchez, M.C. Physiological responses of apricot plants grafted on two different rootstocks to flooding conditions. J. Plant Physiol. 2002, 159, 725–732. [Google Scholar] [CrossRef] [Green Version]
- Xiloyannis, C.; Celano, G.; Vicinanza, L.; Esmenjaud, D.; Gómez-Aparisi, J.; Salesses, G.; Dichio, B. Performance of new selections of Prunus rootstocks, resistant to root knot nematodes, in waterlogging conditions. In Proceedings of the I International Symposium on Rootstocks for Deciduous Fruit Tree Species 658, Zaragoza, Spain, 11–14 June 2002; pp. 403–405. [Google Scholar]
- Salvatierra, A.; Pimentel, P.; Almada, R.; Hinrichsen, P. Exogenous GABA application transiently improves the tolerance to root hypoxia on a sensitive genotype of Prunus rootstock. Environ. Exp. Bot. 2016, 125, 52–66. [Google Scholar] [CrossRef]
- Iacona, C.; Pistelli, L.; Cirilli, M.; Gatti, L.; Mancinelli, R.; Ripa, M.N.; Muleo, R. Day-length is involved in flooding tolerance response in wild type and variant genotypes of rootstock Prunus cerasifera L. Front. Plant Sci. 2019, 10, 546. [Google Scholar] [CrossRef] [PubMed]
- Klumb, E.; Rickes, L.; Braga, E.; Bianchi, V. Evaluation of gas exchanges in different Prunus spp. rootstocks under drought and flooding stress. Rev. Bras. Frutic. 2017, 39, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Schaffer, B.; Andersen, P.C.; Ploetz, R.C. Responses of fruit crops to flooding. Hortic. Rev. 1992, 13, 257–313. [Google Scholar]
- Parent, C.; Capelli, N.; Berger, A.; Crèvecoeur, M.; Dat, J.F. An overview of plant responses to soil waterlogging. Plant Stress 2008, 2, 20–27. [Google Scholar]
- Pistelli, L.; Iacona, C.; Miano, D.; Cirilli, M.; Colao, M.C.; Mensuali-Sodi, A.; Muleo, R. Novel Prunus rootstock somaclonal variants with divergent ability to tolerate waterlogging. Tree Physiol. 2012, 32, 355–368. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Lyu, D.; Jia, L.; He, J.; Qin, S. Physiological and de novo transcriptome analysis of the fermentation mechanism of Cerasus sachalinensis roots in response to short-term waterlogging. BMC Genom. 2017, 18, 649. [Google Scholar] [CrossRef] [Green Version]
- Loreti, E.; Valeri, M.C.; Novi, G.; Perata, P. Gene regulation and survival under hypoxia requires starch availability and metabolism. Plant Physiol. 2018, 176, 1286–1298. [Google Scholar] [CrossRef]
- Tan, X.; Xu, H.; Khan, S.; Equiza, M.A.; Lee, S.H.; Vaziriyeganeh, M.; Zwiazek, J.J. Plant water transport and aquaporins in oxygen-deprived environments. J. Plant Physiol. 2018, 227, 20–30. [Google Scholar] [CrossRef]
- Aroca, R.; Porcel, R.; Ruiz-Lozano, J.M. Regulation of root water uptake under abiotic stress conditions. J. Exp. Bot. 2011, 63, 43–57. [Google Scholar] [CrossRef]
- Chaumont, F.; Tyerman, S.D. Aquaporins: Highly regulated channels controlling plant water relations. Plant Physiol. 2014, 164, 1600. [Google Scholar] [CrossRef] [Green Version]
- Pawłowicz, I.; Masajada, K. Aquaporins as a link between water relations and photosynthetic pathway in abiotic stress tolerance in plants. Gene 2019, 687, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Tournaire-Roux, C.; Sutka, M.; Javot, H.; Gout, E.; Gerbeau, P.; Luu, D.-T.; Bligny, R.; Maurel, C. Cytosolic pH regulates root water transport during anoxic stress through gating of aquaporins. Nature 2003, 425, 393–397. [Google Scholar] [CrossRef] [PubMed]
- Tylova, E.; Pecková, E.; Blascheová, Z.; Soukup, A. Casparian bands and suberin lamellae in exodermis of lateral roots: An important trait of roots system response to abiotic stress factors. Ann. Bot. 2017, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasidharan, R.; Hartman, S.; Liu, Z.; Martopawiro, S.; Sajeev, N.; Van Veen, H.; Yeung, E.; Voesenek, L.A.C.J. Signal dynamics and interactions during flooding stress. Plant Physiol. 2018, 176, 1106–1117. [Google Scholar] [CrossRef] [Green Version]
- Møller, M. Plant mitochondria and oxidative stress: Electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 561–591. [Google Scholar] [CrossRef] [Green Version]
- Asada, K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006, 141, 391–396. [Google Scholar] [CrossRef] [Green Version]
- Mignolet-Spruyt, L.; Xu, E.; Idänheimo, N.; Hoeberichts, F.A.; Mühlenbock, P.; Brosché, M.; Van Breusegem, F.; Kangasjärvi, J. Spreading the news: Subcellular and organellar reactive oxygen species production and signalling. J. Exp. Bot. 2016, 67, 3831–3844. [Google Scholar] [CrossRef] [Green Version]
- Chapman, J.M.; Muhlemann, J.K.; Gayomba, S.R.; Muday, G.K. RBOH-dependent ROS synthesis and ROS scavenging by plant specialized metabolites to modulate plant development and stress responses. Chem. Res. Toxicol. 2019, 32, 370–396. [Google Scholar] [CrossRef]
- Demidchik, V.; Shabala, S. Mechanisms of cytosolic calcium elevation in plants: The role of ion channels, calcium extrusion systems and NADPH oxidase-mediated ‘ROS-Ca2+ Hub’. Funct. Plant Biol. 2017, 45, 9–27. [Google Scholar] [CrossRef]
- Igamberdiev, A.U.; Hill, R.D. Elevation of cytosolic Ca2+ in response to energy deficiency in plants: The general mechanism of adaptation to low oxygen stress. Biochem. J. 2018, 475, 1411–1425. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Voesenek, L.A.C.J. Flooding stress: Acclimations and genetic diversity. Annu. Rev. Plant. Biol. 2008, 59, 313–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukao, T.; Barrera-Figueroa, B.E.; Juntawong, P.; Peña-Castro, J.M. Submergence and waterlogging stress in plants: A review highlighting research opportunities and understudied aspects. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Arismendi, M.J.; Almada, R.; Pimentel, P.; Bastias, A.; Salvatierra, A.; Rojas, P.; Hinrichsen, P.; Pinto, M.; Di Genova, A.; Travisany, D.; et al. Transcriptome sequencing of Prunus sp. rootstocks roots to identify candidate genes involved in the response to root hypoxia. Tree Genet. Genomes 2015, 11, 1–16. [Google Scholar] [CrossRef]
- Mittler, R. ROS are good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arbona, V.; Hossain, Z.; López-Climent, M.F.; Pérez-Clemente, R.M.; Gómez-Cadenas, A. Antioxidant enzymatic activity is linked to waterlogging stress tolerance in citrus. Physiol. Plant. 2008, 132, 452–466. [Google Scholar] [CrossRef]
- Bai, T.; Li, C.; Ma, F.; Feng, F.; Shu, H. Responses of growth and antioxidant system to root-zone hypoxia stress in two Malus species. Plant Soil 2010, 327, 95–105. [Google Scholar] [CrossRef]
- Toro, G.; Pinto, M.; Pimentel, P. Root respiratory components of Prunus spp. rootstocks under low oxygen: Regulation of growth, maintenance, and ion uptake respiration. Sci. Hortic. 2018, 239, 259–268. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9. [Google Scholar] [CrossRef]
- Radmann, E.; Klumb, E.; Deuner, S.; Bianchi, V. Antioxidant capacity in leaf and root tissues of Prunus spp. under flooding. J. Exp. Agric. Int. 2018, 26, 1–10. [Google Scholar] [CrossRef]
- Sauter, M. Root responses to flooding. Curr. Opin. Plant Biol. 2013, 16, 282–286. [Google Scholar] [CrossRef]
- Kozlowski, T.T. Responses of woody plants to flooding and salinity. Tree Physiol. 1997, 17, 490. [Google Scholar] [CrossRef]
- Yamauchi, T.; Shimamura, S.; Nakazono, M.; Mochizuki, T. Aerenchyma formation in crop species: A review. Field Crops Res. 2013, 152, 8–16. [Google Scholar] [CrossRef]
- Le Provost, G.; Sulmon, C.; Frigerio, J.-M.; Bodénès, C.; Kremer, A.; Plomion, C. Role of waterlogging-responsive genes in shaping interspecific differentiation between two sympatric oak species. Tree Physiol. 2011, 32, 119–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimamura, S.; Yamamoto, R.; Nakamura, T.; Shimada, S.; Komatsu, S. Stem hypertrophic lenticels and secondary aerenchyma enable oxygen transport to roots of soybean in flooded soil. Ann. Bot. 2010, 106, 277–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armstrong, W. Aeration in higher plants. In Advances in Botanical Research; Woolhouse, H.W., Ed.; Academic Press: London, UK, 1979; Volume 7, pp. 225–332. [Google Scholar]
- Colmer, T.D. Long-distance transport of gases in plants: A perspective on internal aeration and radial oxygen loss from roots. Plant Cell Environ. 2003, 26, 17–36. [Google Scholar] [CrossRef] [Green Version]
- Armstrong, J.; Armstrong, W. Rice: Sulfide-induced barriers to root radial oxygen loss, Fe2+ and water uptake, and lateral root emergence. Ann. Bot. 2005, 96, 625–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colmer, T.D.; Voesenek, L.A.C.J. Flooding tolerance: Suites of plant traits in variable environments. Funct. Plant Biol. 2009, 36, 665–681. [Google Scholar] [CrossRef]
- Yamauchi, T.; Colmer, T.D.; Pedersen, O.; Nakazono, M. Regulation of root traits for internal aeration and tolerance to soil waterlogging-flooding stress. Plant Physiol. 2018, 176, 1118–1130. [Google Scholar] [CrossRef] [Green Version]
- Marchioretto, L.D.R.; Rossi, A.D.; Amaral, L.O.d.; Ribeiro, A.M.A.d.S. Tolerance of apple rootstocks to short-term waterlogging. Ciência Rural 2018, 48. [Google Scholar] [CrossRef] [Green Version]
- Andersen, P.C.; Lombard, P.B.; Westwood, M.N. Effect of root anaerobiosis on the water relations of several Pyrus species. Physiol. Plant. 1984, 62, 245–252. [Google Scholar] [CrossRef]
- Vartapetian, B.B.; Andreeva, I.N.; Generozova, I.P.; Polyakova, L.I.; Maslova, I.P.; Dolgikh, Y.I.; Stepanova, A.Y. Functional electron microscopy in studies of plant response and adaptation to anaerobic stress. Ann. Bot. 2003, 91, 155–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kreuzwieser, J.; Hauberg, J.; Howell, K.A.; Carroll, A.; Rennenberg, H.; Millar, A.H.; Whelan, J. Differential response of gray poplar leaves and roots underpins stress adaptation during hypoxia. Plant Physiol. 2009, 149, 461–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, B.; Yang, Y.; Yin, Y.; Xu, M.; Li, H. De novo sequencing, assembly, and analysis of the Taxodium‘Zhongshansa’ roots and shoots transcriptome in response to short-term waterlogging. BMC Plant Biol. 2014, 14, 201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Provost, G.; Lesur, I.; Lalanne, C.; Da Silva, C.; Labadie, K.; Aury, J.M.; Leple, J.C.; Plomion, C. Implication of the suberin pathway in adaptation to waterlogging and hypertrophied lenticels formation in pedunculate oak (Quercus robur L.). Tree Physiol. 2016, 36, 1330–1342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reeksting, B.J.; Coetzer, N.; Mahomed, W.; Engelbrecht, J.; Van den Berg, N. De novo sequencing, assembly, and analysis of the root transcriptome of Persea americana (Mill.) in response to Phytophthora cinnamomi and flooding. PLoS ONE 2014, 9, e86399. [Google Scholar] [CrossRef]
- Rubio-Cabetas, M.J.; Pons, C.; Bielsa, B.; Amador, M.L.; Marti, C.; Granell, A. Preformed and induced mechanisms underlies the differential responses of Prunus rootstock to hypoxia. J. Plant Physiol. 2018, 228, 134–149. [Google Scholar] [CrossRef]
- Zhang, J.-Y.; Huang, S.-N.; Mo, Z.-H.; Xuan, J.-P.; Jia, X.-D.; Wang, G.; Guo, Z.-R. De novo transcriptome sequencing and comparative analysis of differentially expressed genes in kiwifruit under waterlogging stress. Mol. Breed. 2015, 35, 208. [Google Scholar] [CrossRef]
- Zhu, X.; Li, X.; Jiu, S.; Zhang, K.; Wang, C.; Fang, J. Analysis of the regulation networks in grapevine reveals response to waterlogging stress and candidate gene-marker selection for damage severity. R. Soc. Open Sci. 2018, 5, 172253. [Google Scholar] [CrossRef] [Green Version]
- Ferner, E.; Rennenberg, H.; Kreuzwieser, J. Effect of flooding on C metabolism of flood-tolerant (Quercus robur) and non-tolerant (Fagus sylvatica) tree species. Tree Physiol. 2012, 32, 135–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Alcántara, B.; Jover, S.; Quiñones, A.; Forner-Giner, M.Á.; Rodríguez-Gamir, J.; Legaz, F.; Primo-Millo, E.; Iglesias, D.J. Flooding affects uptake and distribution of carbon and nitrogen in citrus seedlings. J. Plant Physiol. 2012, 169, 1150–1157. [Google Scholar] [CrossRef]
- Fukao, T.; Bailey-Serres, J. Plant responses to hypoxia—Is survival a balancing act? Trends Plant Sci. 2004, 9, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Granot, D.; Kelly, G.; Stein, O.; David-Schwartz, R. Substantial roles of hexokinase and fructokinase in the effects of sugars on plant physiology and development. J. Exp. Bot. 2013, 65, 809–819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Huang, T.; Zhou, Y.; Han, Y.; Xu, M.; Gu, J. AfterQC: Automatic filtering, trimming, error removing and quality control for fastq data. BMC Bioinform. 2017, 18, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protocols 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2018, 47, D427–D432. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-Hit: A Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araujo, F.A.; Barh, D.; Silva, A.; Guimarães, L.; Ramos, R.T.J. GO FEAT: A rapid web-based functional annotation tool for genomic and transcriptomic data. Sci. Rep. 2018, 8, 1794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Mateluna, P.; Salvatierra, A.; Solis, S.; Nuñez, G.; Pimentel, P. Involvement of aquaporin NIP1;1 in the contrasting tolerance response to root hypoxia in Prunus rootstocks. J. Plant Physiol. 2018, 228, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Toroser, D.; Plaut, Z.; Huber, S.C. Regulation of a plant SNF1-related protein kinase by glucose-6-Phosphate. Plant Physiol. 2000, 123, 403–412. [Google Scholar] [CrossRef] [Green Version]
- Rocha, M.; Licausi, F.; Araújo, W.L.; Nunes-Nesi, A.; Sodek, L.; Fernie, A.R.; Van Dongen, J.T. Glycolysis and the tricarboxylic acid cycle are linked by alanine aminotransferase during hypoxia induced by waterlogging of Lotus japonicus. Plant Physiol. 2010, 152, 1501–1513. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Bai, T.; Wang, Y.; Wu, T.; Zhang, X.; Xu, X.; Han, Z. Morpholoical and enzymatic responses to waterlogging in three Prunus species. Sci. Hort. 2017, 221, 62–67. [Google Scholar] [CrossRef]
- Xia, J.-H.; Saglio, P.H. Lactic acid efflux as a mechanism of hypoxic acclimation of maize root tips to anoxia. Plant Physiol. 1992, 100, 40–46. [Google Scholar] [CrossRef] [Green Version]
- Rivoal, J.; Hanson, A.D. Evidence for a large and sustained glycolytic flux to lactate in anoxic roots of some members of the halophytic genus Limonium. Plant Physiol. 1993, 101, 553–560. [Google Scholar] [CrossRef] [Green Version]
- O’Carra, P.; Mulcahy, P. Plant lactate dehydrogenase: NADH kinetics and inhibition by ATP. Phytochemistry 1997, 45, 897–902. [Google Scholar] [CrossRef]
- Patel, M.S.; Roche, T.E. Molecular biology and biochemistry of pyruvate dehydrogenase complexes1. FASEB J. 1990, 4, 3224–3233. [Google Scholar] [CrossRef] [PubMed]
- De Sousa, C.A.F.; Sodek, L. Alanine metabolism and alanine aminotransferase activity in soybean (Glycine max) during hypoxia of the root system and subsequent return to normoxia. Environ. Exp. Bot. 2003, 50, 1–8. [Google Scholar] [CrossRef]
- Limami, A.M.; Glévarec, G.; Ricoult, C.; Cliquet, J.-B.; Planchet, E. Concerted modulation of alanine and glutamate metabolism in young Medicago truncatula seedlings under hypoxic stress. J. Exp. Bot. 2008, 59, 2325–2335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, S.M.; Di Leo, R.; Dhanoa, P.K.; Van Cauwenberghe, O.R.; Mullen, R.T.; Shelp, B.J. Biochemical characterization, mitochondrial localization, expression, and potential functions for an Arabidopsis γ-aminobutyrate transaminase that utilizes both pyruvate and glyoxylate. J. Exp. Bot. 2009, 60, 1743–1757. [Google Scholar] [CrossRef] [Green Version]
- Clark, S.M.; Di Leo, R.; Van Cauwenberghe, O.R.; Mullen, R.T.; Shelp, B.J. Subcellular localization and expression of multiple tomato γ-aminobutyrate transaminases that utilize both pyruvate and glyoxylate. J. Exp. Bot. 2009, 60, 3255–3267. [Google Scholar] [CrossRef] [Green Version]
- Michaeli, S.; Fromm, H. Closing the loop on the GABA shunt in plants: Are GABA metabolism and signaling entwined? Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Lea, P.J.; Miflin, B.J. Glutamate synthase and the synthesis of glutamate in plants. Plant Physiol. Biochem. 2003, 41, 555–564. [Google Scholar] [CrossRef]
- Horchani, F.; Aschi-Smiti, S. Prolonged root hypoxia effects on enzymes involved in nitrogen assimilation pathway in tomato plants. Plant Signal. Behav. 2010, 5, 1583–1589. [Google Scholar] [CrossRef]
- Canuel, E.A.; Hardison, A.K. Carbon Cycle. In Encyclopedia of Geochemistry: A Comprehensive Reference Source on the Chemistry of the Earth; White, W.M., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 191–194. [Google Scholar] [CrossRef]
- Gifford, R.M. Plant respiration in productivity models: Conceptualisation, representation and issues for global terrestrial carbon-cycle research. Funct. Plant Biol. 2003, 30, 171–186. [Google Scholar] [CrossRef]
- Van Dongen, J.T.; Gupta, K.J.; Ramírez-Aguilar, S.J.; Araújo, W.L.; Nunez-Nesi, A.; Fernie, A.R. Regulation of respiration in plants: A role for alternative metabolic pathways. J. Plant Physiol. 2011, 168, 1434–1443. [Google Scholar] [CrossRef]
- Millar, A.H.; Whelan, J.; Soole, K.L.; Day, D.A. Organization and Regulation of Mitochondrial Respiration in Plants. Ann. Rev. Plant Biol. 2011, 62, 79–104. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.J.; Zabalza, A.; Van Dongen, J.T. Regulation of respiration when the oxygen availability changes. Physiol. Plant. 2009, 137, 383–391. [Google Scholar] [CrossRef]
- Martínez, F.; Lazo, Y.O.; Fernández-Galiano, J.M.; Merino, J.A. Chemical composition and construction cost for roots of Mediterranean trees, shrub species and grassland communities. Plant Cell Environ. 2002, 25, 601–608. [Google Scholar] [CrossRef]
- Rachmilevitch, S.; Lambers, H.; Huang, B. Root respiratory characteristics associated with plant adaptation to high soil temperature for geothermal and turf-type Agrostis species. J. Exp. Bot. 2006, 57, 623–631. [Google Scholar] [CrossRef] [Green Version]
- Rewald, B.; Shelef, O.; Ephrath, J.E.; Rachmilevitch, S. Adaptive Plasticity of Salt-Stressed Root Systems. In Ecophysiology and Responses of Plants under Salt Stress; Ahmad, P., Azooz, M.M., Prasad, M.N.V., Eds.; Springer: New York, NY, USA, 2013; pp. 169–201. [Google Scholar] [CrossRef]
- Gibbs, J.; Greenway, H. Review: Mechanisms of anoxia tolerance in plants. I. Growth, survival and anaerobic catabolism. Funct. Plant Biol. 2003, 30, 1–47. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, W.; Beckett, P.M.; Colmer, T.D.; Setter, T.L.; Greenway, H. Tolerance of roots to low oxygen: ‘Anoxic’ cores, the phytoglobin-nitric oxide cycle, and energy or oxygen sensing. J. Plant Physiol. 2019, 239, 92–108. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Nakamura, M. Root respiratory costs of ion uptake, root growth, and root maintenance in wetland plants: Efficiency and strategy of O2 use for adaptation to hypoxia. Oecologia 2016, 182, 667–678. [Google Scholar] [CrossRef]
- Greenway, H.; Gibbs, J. Review: Mechanisms of anoxia tolerance in plants. II. Energy requirements for maintenance and energy distribution to essential processes. Funct. Plant Biol. 2003, 30, 999–1036. [Google Scholar] [CrossRef]
- Sorenson, R.; Bailey-Serres, J. Selective mRNA Translation Tailors Low Oxygen Energetics. In Low-Oxygen Stress in Plants: Oxygen Sensing and Adaptive Responses to Hypoxia; Van Dongen, J.T., Licausi, F., Eds.; Springer: Vienna, Austria, 2014; pp. 95–115. [Google Scholar] [CrossRef]
- Thornley, J.H. Plant growth and respiration re-visited: Maintenance respiration defined—It is an emergent property of, not a separate process within, the system - and why the respiration: Photosynthesis ratio is conservative. Ann. Bot. 2011, 108, 1365–1380. [Google Scholar] [CrossRef]
- Penning de Vries, F.W.T. The Cost of Maintenance Processes in Plant Cells. Ann. Bot. 1975, 39, 77–92. [Google Scholar] [CrossRef]
- Penning de Vries, F.W.T.; Brunsting, A.H.; Van Laar, H.H. Products, requirements and efficiency of biosynthesis: A quantitative approach. J. Theor. Biol. 1974, 45, 339–377. [Google Scholar] [CrossRef]
- Noctor, G.; De Paepe, R.; Foyer, C.H. Mitochondrial redox biology and homeostasis in plants. Trends Plant Sci. 2007, 12, 125–134. [Google Scholar] [CrossRef]
- Dudkina, N.V.; Heinemeyer, J.; Sunderhaus, S.; Boekema, E.J.; Braun, H.-P. Respiratory chain supercomplexes in the plant mitochondrial membrane. Trends Plant Sci. 2006, 11, 232–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey-Serres, J.; Fukao, T.; Gibbs, D.J.; Holdsworth, M.J.; Lee, S.C.; Licausi, F.; Perata, P.; Voesenek, L.A.C.J.; Van Dongen, J.T. Making sense of low oxygen sensing. Trends Plant Sci. 2012, 17, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Blokhina, O.; Virolainen, E.; Fagerstedt, K.V. Antioxidants, Oxidative Damage and Oxygen Deprivation Stress: A Review. Ann. Bot. 2003, 91, 179–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toro, G.; Pinto, M. Plant respiration under low oxygen. Chil. J. Agric. Res. 2015, 75, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Caretto, S.; Linsalata, V.; Colella, G.; Mita, G.; Lattanzio, V. Carbon Fluxes between Primary Metabolism and Phenolic Pathway in Plant Tissues under Stress. Int. J. Mol. Sci. 2015, 16, 26378–26394. [Google Scholar] [CrossRef] [Green Version]
- Møller, I.M.; Jensen, P.E.; Hansson, A. Oxidative Modifications to Cellular Components in Plants. Annu. Rev. Plant Biol. 2007, 58, 459–481. [Google Scholar] [CrossRef] [Green Version]
- Gupta, K.J.; Mur, L.A.J.; Wany, A.; Kumari, A.; Fernie, A.R.; Ratcliffe, R.G. The role of nitrite and nitric oxide under low oxygen conditions in plants. New Phytol. 2020, 225, 1143–1151. [Google Scholar] [CrossRef]
- Igamberdiev, A.U.; Bykova, N.V.; Shah, J.K.; Hill, R.D. Anoxic nitric oxide cycling in plants: Participating reactions and possible mechanisms. Physiol. Plant. 2010, 138, 393–404. [Google Scholar] [CrossRef]
- Gupta, K.J.; Lee, C.P.; Ratcliffe, R.G. Nitrite Protects Mitochondrial Structure and Function under Hypoxia. Plant Cell Physiol. 2017, 58, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Vishwakarma, A.; Kumari, A.; Mur, L.A.J.; Gupta, K.J. A discrete role for alternative oxidase under hypoxia to increase nitric oxide and drive energy production. Free Radic. Biol. Med. 2018, 122, 40–51. [Google Scholar] [CrossRef] [Green Version]
- Jayawardhane, J.; Cochrane, D.W.; Vyas, P.; Bykova, N.V.; Vanlerberghe, G.C.; Igamberdiev, A.U. Roles for Plant Mitochondrial Alternative Oxidase Under Normoxia, Hypoxia, and Reoxygenation Conditions. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Stoimenova, M.; Igamberdiev, A.; Gupta, K.; Hill, R. Nitrite-driven anaerobic ATP synthesis in barley and rice root mitochondria. Planta 2007, 226, 465–474. [Google Scholar] [CrossRef]
- Almada, R.; Arismendi, M.J.; Pimentel, P.; Rojas, P.; Hinrichsen, P.; Pinto, M.; Sagredo, B. Class 1 non-symbiotic and class 3 truncated hemoglobin-like genes are differentially expressed in stone fruit rootstocks (Prunus L.) with different degrees of tolerance to root hypoxia. Tree Genet. Genomes 2013, 9, 1051–1063. [Google Scholar] [CrossRef]
- Parent, C.; Crévecoeur, M.; Capelli, N.; Dat, J.F. Contrasting growth and adaptive responses of two oak species to flooding stress: Role of non-symbiotic haemoglobin. Plant Cell Environ. 2011, 34, 1113–1126. [Google Scholar] [CrossRef]
- Gazizova, N.; Rakhmatullina, D.; Minibayeva, F. Effect of respiratory inhibitors on mitochondrial complexes and ADP/ATP translocators in the Triticum aestivum roots. Plant Physiol. Biochem. 2020, 151, 601–607. [Google Scholar] [CrossRef]
- Millenaar, F.F.; Benschop, J.J.; Wagner, A.M.; Lambers, H. The role of the alternative oxidase in stabilizing the in vivo reduction state of the ubiquinone pool and the activation state of the alternative oxidase. Plant Physiol. 1998, 118, 599–607. [Google Scholar] [CrossRef] [Green Version]
- Arru, L.; Fornaciari, S. Root Oxygen Deprivation and Leaf Biochemistry in Trees. In Waterlogging Signalling and Tolerance in Plants; Mancuso, S., Shabala, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 181–195. [Google Scholar] [CrossRef]
- Veen, B.W. Energy cost of ion transport. In Genetic Engineering of Osmoregulation. Impact on Plant Productivity for Food, Chemicals and Energy; Rains, D.W., Valentine, R.C., Hollander, A., Eds.; Springer: New York, NY, USA, 1980; pp. 187–195. [Google Scholar]
- Bouma, T.J.; De Visser, R.; Janssen, J.; De Kock, M.; Van Leeuwen, P.; Lambers, H. Respiratory energy requirements and rate of protein turnover in vivo determined by the use of an inhibitor of protein synthesis and a probe to assess its effect. Physiol. Plant. 1994, 92, 585–594. [Google Scholar] [CrossRef]
- Hirst, J. Open questions: Respiratory chain supercomplexes—Why are they there and what do they do? BMC Biol. 2018, 16, 111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eubel, H.; Jänsch, L.; Braun, H.-P. New insights into the respiratory chain of plant mitochondria. Supercomplexes and a unique composition of complex II. Plant Physiol. 2003, 133, 274–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Römpler, K.; Müller, T.; Juris, L.; Wissel, M.; Vukotic, M.; Hofmann, K.; Deckers, M. Overlapping Role of Respiratory Supercomplex Factor Rcf2 and Its N-terminal Homolog Rcf3 in Saccharomyces cerevisiae. J. Biol. Chem. 2016, 291, 23769–23778. [Google Scholar] [CrossRef] [Green Version]
- Lobo-Jarne, T.; Ugalde, C. Respiratory chain supercomplexes: Structures, function and biogenesis. Semin. Cell Dev. Biol. 2018, 76, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Strogolova, V.; Furness, A.; Robb-McGrath, M.; Garlich, J.; Stuart, R.A. Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex. Mol. Cell. Biol. 2012, 32, 1363–1373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, A.-Y.; Koo, N.; Kim, S.; Sim, Y.M.; Choi, D.; Kim, Y.-M.; Kwon, S.-Y. Draft genome sequences of two oriental melons, Cucumis melo L. var. makuwa. Sci. Data 2019, 6, 220. [Google Scholar] [CrossRef] [Green Version]
- Schertl, P.; Braun, H.-P. Respiratory electron transfer pathways in plant mitochondria. Front. Plant Sci. 2014, 5, 163. [Google Scholar] [CrossRef] [Green Version]
- Virolainen, E.; Blokhina, O.; Fagerstedt, K. Ca2+-induced High Amplitude Swelling and Cytochrome c Release from Wheat (Triticum aestivum L.) Mitochondria Under Anoxic Stress. Ann. Bot. 2002, 90, 509–516. [Google Scholar] [CrossRef]
- Bernardi, P.; Scorrano, L.; Colonna, R.; Petronilli, V.; Di Lisa, F. Mitochondria and cell death. Eur. J. Biochem. 1999, 264, 687–701. [Google Scholar] [CrossRef]
- Bouranis, D.L.; Chorianopoulou, S.N.; Siyiannis, V.F.; Protonotarios, V.E.; Hawkesford, M.J. Lysigenous aerenchyma development in roots–triggers and cross-talks for a cell elimination program. Int. J. Plant. Dev. Biol. 2007, 1, 127–140. [Google Scholar]
- Pimenta, J.A.; Bianchini, E.; Medri, M.E. Adaptations to flooding by tropical trees: Morphological and anatomical modifications. Oecol. Aust. 2010, 4, 157–176. [Google Scholar] [CrossRef]
- Hancock, J.T.; Desikan, R.; Neill, S.J. Cytochrome c, Glutathione, and the Possible Role of Redox Potentials in Apoptosis. Ann. N. Y. Acad. Sci. 2003, 1010, 446–448. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, D.M.; Umbach, A.L.; Subbaiah, C.C.; Siedow, J.N. Mitochondrial Reactive Oxygen Species. Contribution to Oxidative Stress and Interorganellar Signaling. Plant Physiol. 2006, 141, 357. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Log2 FC | |||||||
|---|---|---|---|---|---|---|---|
| ‘Mariana 2624’ | ‘Mazzard F12/1’ | ||||||
| Gene | 6 h | 24 h | 72 h | 6 h | 24 h | 72 h | |
| RBOHA | Prupe.6G321500 | 4.751 | 4.196 | 3.828 | 3.940 | 3.656 | 3.112 |
| RBOHC/RHD2 | Prupe.1G211000 | −0.300 | −1.902 | −1.240 | 0.944 | 0.935 | 1.138 |
| RBOHE | Prupe.5G107400 | 0.398 | 0.309 | 0.618 | 0.224 | 0.429 | 0.770 |
| Cu Zn SOD1 | Prupe.2G269400 | −0.451 | −1.268 | −2.900 | −0.454 | −0.695 | −0.863 |
| Cu Zn SOD2 | Prupe.1G347200 | 0.911 | 2.752 | 2.557 | −0.713 | −1.446 | −0.485 |
| Cu Zn SOD3 | Prupe.2G262400 | 1.022 | 3.112 | 2.748 | 0.930 | 3.460 | 2.933 |
| Fe SOD1 | Prupe.6G042300 | 0.512 | 1.369 | 0.869 | 1.058 | 1.402 | 0.840 |
| CAT1 | Prupe.5G011300 | 0.174 | −0.134 | −0.818 | nd | nd | nd |
| CAT2 | Prupe.5G011400 | 0.498 | −0.115 | −0.526 | 0.376 | −0.739 | −0.580 |
| APX 1 | Prupe.1G481000 | 0.681 | 0.724 | 0.792 | 0.685 | 0.873 | 0.525 |
| APX 2 | Prupe.1G493900 | −0.383 | −0.638 | −0.547 | −0.017 | −0.156 | −0.395 |
| APX 3 | Prupe.6G091600 | 0.719 | −1.498 | −2.527 | 0.555 | −1.351 | −2.630 |
| APX 5 | Prupe.6G242200 | 6.538 | 7.284 | 6.864 | 3.323 | 3.948 | 4.591 |
| APX 6 | Prupe.7G171200 | 0.718 | 0.106 | −0.252 | 1.158 | 0.502 | 0.740 |
| APX S | Prupe.8G164400 | −0.421 | −2.284 | −2.494 | −0.414 | −1.564 | −2.650 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salvatierra, A.; Toro, G.; Mateluna, P.; Opazo, I.; Ortiz, M.; Pimentel, P. Keep Calm and Survive: Adaptation Strategies to Energy Crisis in Fruit Trees under Root Hypoxia. Plants 2020, 9, 1108. https://doi.org/10.3390/plants9091108
Salvatierra A, Toro G, Mateluna P, Opazo I, Ortiz M, Pimentel P. Keep Calm and Survive: Adaptation Strategies to Energy Crisis in Fruit Trees under Root Hypoxia. Plants. 2020; 9(9):1108. https://doi.org/10.3390/plants9091108
Chicago/Turabian StyleSalvatierra, Ariel, Guillermo Toro, Patricio Mateluna, Ismael Opazo, Mauricio Ortiz, and Paula Pimentel. 2020. "Keep Calm and Survive: Adaptation Strategies to Energy Crisis in Fruit Trees under Root Hypoxia" Plants 9, no. 9: 1108. https://doi.org/10.3390/plants9091108
APA StyleSalvatierra, A., Toro, G., Mateluna, P., Opazo, I., Ortiz, M., & Pimentel, P. (2020). Keep Calm and Survive: Adaptation Strategies to Energy Crisis in Fruit Trees under Root Hypoxia. Plants, 9(9), 1108. https://doi.org/10.3390/plants9091108





