The East Asian Winter Monsoon Acts as a Major Selective Factor in the Intraspecific Differentiation of Drought-Tolerant Nitraria tangutorum in Northwest China
Abstract
:1. Introduction
2. Results
2.1. Phylogenetic Analysis and Geographical Distribution of Haplotypes
2.2. Population and Phylogeographical Analysis
2.3. Estimation of the Divergence and Separation of the West and East Lineages
2.4. Regional Population Demography of N. tangutorum
2.5. Ecological Niche Modeling (ENM)
3. Discussion
3.1. Geographic Structure and Genetic Diversity of N. tangutorum
3.2. Intraspecific Divergence in N. tangutorum
3.3. Intraspecific Differentiations Triggered by the Local Monsoon Climate
3.4. Population Demography and Colonization
4. Materials and Methods
4.1. Population Sampling
4.2. DNA Extraction, PCR Amplification and Sequencing
4.3. Population Nucleotide Diversity and Phylogeographical Analysis
4.4. Phylogenetic Analyses and Molecular Dating
4.5. Inference of Demographic History and Ecological Niche Modelling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| QTP | Qinghai-Tibet Plateau |
| EAMS | East Asian monsoon system |
| EASM | East Asian summer monsoon |
| EAWM | East Asian winter monsoon |
| Ma | Million years ago |
| cpDNA | chloroplast DNA |
| GPS | global positioning system |
| AMOVA | analysis of molecular variance |
| ML | Maximum likelihood |
| RASP | reconstruct ancestral state in phylogenies |
| BSP | Bayesian skyline plot |
| MDA | mismatch distribution analysis |
| ENM | ecological niche modeling |
| LGM | Last Glacial Maximum |
| LIG | Last Inter-Glacial |
References
- Hewitt, G. The genetic legacy of the Quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, G.M. Genetic consequences of climatic oscillations in the Quaternary. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2004, 359, 183–195. [Google Scholar] [CrossRef] [Green Version]
- Meng, H.H.; Gao, X.Y.; Huang, J.F.; Zhang, M.L. Plant phylogeography in arid Northwest China: Retrospectives and perspectives. J. Syst. Evol. 2015, 53, 33–46. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, X.; Tang, N.; Liu, J.; Xu, L.; Wang, K. Phylogeography of Libanotis buchtormensis (Umbelliferae) in disjunct populations along the deserts in northwest China. PLoS ONE 2016, 11, e0159790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Yu, Q.; Zhang, Q.; Hu, X.; Hu, J.; Fan, B. Regional-scale differentiation and phylogeography of a desert plant Allium mongolicum (Liliaceae) inferred from chloroplast DNA sequence variation. Plant Syst. Evol. 2017, 303, 451–466. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, H.; Pan, B.; Zhang, M. Intraspecific divergences and phylogeography of Panzerina lanata (Lamiaceae) in northwest China. PeerJ 2019, 7, e6264. [Google Scholar] [CrossRef] [Green Version]
- Favre, A.; Päckert, M.; Pauls, S.U.; Jähnig, S.C.; Uhl, D.; Michalak, I.; Muellner-Riehl, A.N. The role of the uplift of the Qinghai-Tibetan Plateau for the evolution of Tibetan biotas. Biol. Rev. Camb. Philos. Soc. 2015, 90, 236–253. [Google Scholar] [CrossRef]
- Liu, J.Q.; Duan, Y.W.; Hao, G.; Ge, X.J.; Sun, H. Evolutionary history and underlying adaptation of alpine plants on the Qinghai-Tibet Plateau. J. Syst. Evol. 2014, 52, 241–249. [Google Scholar] [CrossRef]
- Liu, J.Q.; Wang, Y.J.; Wang, A.L.; Hideaki, O.; Abbott, R.J. Radiation and diversification within the Ligularia-Cremanthodium-Parasenecio complex (Asteraceae) triggered by uplift of the Qinghai-Tibetan Plateau. Mol. Phylogenet. Evol. 2006, 38, 31–49. [Google Scholar] [CrossRef]
- Muellner-Riehl, A.N. Mountains as evolutionary arenas: Patterns, emerging approaches, paradigm shifts and their implications for plant phylogeographic research in the Tibeto-Himalayan Region. Front. Plant Sci. 2019, 10, 195. [Google Scholar] [CrossRef] [Green Version]
- Meng, H.H.; Zhang, M.L. Diversification of plant species in arid northwest China: Species-level phylogeographical history of Lagochilus Bunge ex Bentham (Lamiaceae). Mol. Phylogenet. Evol. 2013, 68, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Christmas, M.J.; Breed, M.F.; Lowe, A.J. Constraints to and conservation implications for climate change adaptation in plants. Conserv. Genet. 2016, 17, 305–320. [Google Scholar] [CrossRef] [Green Version]
- Guo, F.; Lenoir, J.; Bonebrake, T.C. Land-use change interacts with climate to determine elevational species redistribution. Nat. Commun. 2018, 9, 1315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, S.W.; Zhang, M.L. Pleistocene climate change and phylogeographic structure of the Gymnocarpos przewalskii (Caryophyllaceae) in the northwest China: Evidence from plastid DNA, ITS sequences and Microsatellite. Ecol. Evol. 2019, 9, 5219–5235. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Guo, X.; Niu, J.; Duojie, D.; Li, X.; Opgenoorth, L.; Zou, J. Molecular phylogeography and evolutionary history of the endemic species Corydalis hendersonii (Papaveraceae) on the Tibetan Plateau inferred from chloroplast DNA and ITS sequence variation. Front. Plant Sci. 2020, 11, 436. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Gao, Q.B.; Gengji, Z.M.; Jia, L.K.; Wang, Z.H.; Chen, S.L. Rapid intraspecific diversification of the alpine species Saxifraga sinomontana (Saxifragaceae) in the Qinghai-Tibetan Plateau and Himalayas. Front. Genet. 2018, 9, 381. [Google Scholar] [CrossRef] [Green Version]
- Shahzad, K.; Jia, Y.; Chen, F.L.; Zeb, U.; Li, Z.H. Effects of mountain uplift and climatic oscillations on phylogeography and species divergence in four endangered Notopterygium herbs. Front. Plant Sci. 2017, 8, 1929. [Google Scholar] [CrossRef] [Green Version]
- Renner, S.S. Available data point to a 4-km-high Tibetan Plateau by 40 Ma, but 100 molecular-clock papers have linked supposed recent uplift to young node ages. J. Biogeogr. 2016, 43, 1479–1487. [Google Scholar] [CrossRef]
- Wang, Z.M.; Meng, S.Y.; Rao, G.Y. Quaternary climate change and habitat preference shaped the genetic differentiation and phylogeography of Rhodiola sect. Prainia in the southern Qinghai-Tibetan Plateau. Ecol. Evol. 2019, 9, 8305–8319. [Google Scholar] [CrossRef] [Green Version]
- Stewart, J.R.; Lister, A.M.; Barnes, I.; Dalén, L. Refugia revisited: Individualistic responses of species in space and time. Proc. Biol. Sci. 2010, 277, 661–671. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.Y.; Gichira, A.W.; Wang, Q.F.; Chen, J.M. Molecular phylogeography of four endemic Sagittaria species (Alismataceae) in the Sino-Japanese floristic region of east Asia. Bot. J. Linn. Soc. 2015, 180, 6–20. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.W.; Chen, S.T.; Nie, Z.L.; Zhang, J.W.; Zhou, Z.; Deng, T.; Sun, H. Climatic factors drive population divergence and demography: Insights based on the phylogeography of a riparian plant species endemic to the Hengduan mountains and adjacent regions. PLoS ONE 2015, 10, e0145014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, H.; Yan, X.; Shi, Y.; Qian, C.; Li, Z.; Zhang, W.; Wang, L.; Li, Y.; Li, X.; Chen, G.; et al. The role of East Asian monsoon system in shaping population divergence and dynamics of a constructive desert shrub Reaumuria soongarica. Sci. Rep. 2015, 5, 15823. [Google Scholar] [CrossRef] [Green Version]
- Gomez-diaz, E.; Sindaco, R.; Pupin, F.; Fasola, M.; Carranza, S. Origin and in situ diversification in Hemidactylus geckos of the Socotra Archipelago. Mol. Ecol. 2012, 21, 4074–4092. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Condamine, F.L.; Harris, A.; Chen, J.; Pan, B.; Möller, M.; Hoang, V.S.; Kang, M. Both temperature fluctuations and East Asian monsoons have driven plant diversification in the karst ecosystems from southern China. Mol. Ecol. 2017, 26, 6414–6429. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Wang, Z.; Tian, B.; Sun, H. Phylogeographic analyses of the East Asian endemic genus Prinsepia and the role of the East Asian monsoon system in shaping a north-south divergence pattern in China. Front. Genet. 2019, 10, 128. [Google Scholar] [CrossRef] [Green Version]
- Shi, C.M.; Ji, Y.J.; Liu, L.; Wang, L.; Zhang, D.X. Impact of climate changes from Middle Miocene onwards on evolutionary diversification in Eurasia: Insights from the mesobuthid scorpions. Mol. Ecol. 2013, 22, 1700–1716. [Google Scholar] [CrossRef]
- Qian, C.; Yin, H.; Shi, Y.; Zhao, J.; Yin, C.; Luo, W.; Dong, Z.; Chen, G.; Yan, X.; Wang, X.-R.; et al. Population dynamics of Agriophyllum squarrosum, a pioneer annual plant endemic to mobile sand dunes, in response to global climate change. Sci. Rep. 2016, 6, 26613. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Y.X.; Fu, C.X.; Comes, H.P. Plant molecular phylogeography in China and adjacent regions: Tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Mol. Phylogenet. Evol. 2011, 59, 225–244. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, Y.; Yang, F.; Dong, Q.; Wang, H.; Hu, N. Rapid determination of amino acids of Nitraria tangutorum Bobr. from the Qinghai–Tibet Plateau using HPLC-FLD-MS/MS and a highly selective and sensitive pre-column derivatization method. Molecules 2019, 24, 1665. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Chen, M.; Yan, L.; Yang, G.; Ma, J.; Deng, W. Quantifying threshold water tables for ecological restoration in arid northwestern China. Ground Water 2020, 58, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.J.; Lu, Q.; Wu, B.; Li, Y.H.; Yao, B.; Zhang, J.X. Effects of increased precipitation on the water use of Nitraira tangutorum at southeast edge of Baddain Jaran Desert in China. J. Appl. Ecol. 2013, 24, 41–48. [Google Scholar]
- Ni, W.; Gao, T.; Wang, H.; Du, Y.; Li, J.; Li, C.; Wei, L.; Bi, H. Anti-fatigue activity of polysaccharides from the fruits of four Tibetan plateau indigenous medicinal plants. J. Ethnopharmacol. 2013, 150, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.Q.; Wang, Y.M.; Yang, Y.L.; Zeng, Y.; Wang, Q.L.; Shao, Y.; Mei, L.J.; Shi, Y.P.; Tao, Y.D. Isolation and identification of antioxidant and α-glucosidase inhibitory compounds from fruit juice of Nitraria tangutorum. Food Chem. 2017, 227, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Petit, R.J.; Duminil, J.; Fineschi, S.; Hampe, A.; Salvini, D.; Vendramin, G.G. Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol. Ecol. 2005, 14, 689–701. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.L.; Temirbayeva, K.; Sanderson, S.C.; Chen, X. Young dispersal of xerophil Nitraria lineages in intercontinental disjunctions of the Old World. Sci. Rep. 2015, 5, 13840. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Zhang, J.Q.; Nie, Z.L.; Zhong, Y.; Sun, H. Evolutionary diversifications of plants on the Qinghai-Tibetan Plateau. Front. Genet. 2014, 5, 4. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Y.F.; Zhang, J.G.; Abuduhamiti, B.; Wang, W.T.; Jia, Z.Q. Phylogeographic patterns of the desert poplar in Northwest China shaped by both geology and climatic oscillations. BMC Evol. Biol. 2018, 18, 75. [Google Scholar] [CrossRef] [Green Version]
- Xia, M.; Tian, Z.; Zhang, F.; Khan, G.; Gao, Q.; Xing, R.; Zhang, Y.; Yu, J.; Chen, S. Deep intraspecific divergence in the endemic herb Lancea tibetica (Mazaceae) distributed over the Qinghai-Tibetan Plateau. Front. Genet. 2018, 9, 492. [Google Scholar] [CrossRef]
- Polato, N.R.; Gray, M.M.; Gill, B.A.; Becker, C.G.; Casner, K.L.; Flecker, A.S.; Kondratieff, B.C.; Encalada, A.C.; Poff, N.L.; Funk, W.C.; et al. Genetic diversity and gene flow decline with elevation in montane mayflies. Heredity 2017, 119, 107–116. [Google Scholar] [CrossRef]
- Garot, E.; Joët, T.; Combes, M.C.; Lashermes, P. Genetic diversity and population divergences of an indigenous tree (Coffea mauritiana) in Reunion Island: Role of climatic and geographical factors. Heredity 2019, 122, 833–847. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Dietrich, C.H.; Wei, C. Genetic divergence, population differentiation and phylogeography of the cicada Subpsaltria yangi based on molecular and acoustic data: an example of the early stage of speciation? BMC Evol. Biol. 2019, 19, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cun, Y.Z.; Wang, X.Q. Plant recolonization in the Himalaya from the southeastern Qinghai-Tibetan Plateau: Geographical isolation contributed to high population differentiation. Mol. Phylogenet. Evol. 2010, 56, 972–982. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Zhang, M. Evolutionary response to Quaternary climate aridification and oscillations in north-western China revealed by chloroplast phylogeography of the desert shrub Nitraria sphaerocarpa (Nitrariaceae). Biol. J. Linn. Soc. Lond. 2013, 109, 757–770. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.H.; Chen, J.; Zhao, G.F.; Guo, Y.P.; Kou, Y.X.; Ma, Y.Z.; Wang, G.; Ma, X.F. Response of a desert shrub to past geological and climatic change: A phylogeographic study of Reaumuria soongarica (Tamaricaceae) in western China. J. Syst. Evol. 2012, 50, 351–361. [Google Scholar] [CrossRef]
- Yin, H.; Yan, X.; Zhang, W.; Shi, Y.; Qian, C.; Yin, C.; Tian, F.; Wang, X.; Ma, X.F. Geographical or ecological divergence between the parapatric species Ephedra sinica and E. intermedia? Plant Syst. Evol. 2016, 302, 1157–1170. [Google Scholar] [CrossRef]
- Bai, W.N.; Wang, W.T.; Zhang, D.Y. Phylogeographic breaks within Asian butternuts indicate the existence of a phytogeographic divide in East Asia. New Phytol. 2016, 209, 1757–1772. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.L.; He, Y.L.; López-Pujol, J.; Jia, Y.; Li, Z.H. Complex population evolutionary history of four cold-tolerant Notopterygium herb species in the Qinghai-Tibetan Plateau and adjacent areas. Heredity (Edinb) 2019, 123, 242–263. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Zhu, R.W.; Zhong, D.L.; Zhang, J.Q. Nunataks or massif de refuge? A phylogeographic study of Rhodiola crenulata (Crassulaceae) on the world’s highest sky islands. BMC Evol. Biol. 2018, 18, 154. [Google Scholar] [CrossRef] [Green Version]
- Ding, L.; Liao, J. Phylogeography of the Tibetan hamster Cricetulus kamensis in response to uplift and environmental change in the Qinghai-Tibet Plateau. Ecol. Evol. 2019, 9, 7291–7306. [Google Scholar] [CrossRef] [Green Version]
- Ding, L.; Liao, J.; Liu, N. The uplift of the Qinghai-Tibet Plateau and glacial oscillations triggered the diversification of Tetraogallus (Galliformes, Phasianidae). Ecol. Evol. 2020, 10, 1722–1736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Song, G.; Liu, N.; Chang, Y.; Bao, X. Deep south-north genetic divergence in Godlewski’s bunting (Emberiza godlewskii) related to uplift of the Qinghai-Tibet Plateau and habitat preferences. BMC Evol. Biol. 2019, 19, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rehfeld, K.; Münch, T.; Ho, S.L.; Laepple, T. Global patterns of declining temperature variability from the last glacial maximum to the holocene. Nature 2018, 554, 356–359. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.L.; Fritsch, P.W. Evolutionary response of Caragana (Fabaceae) to Qinghai–Tibetan Plateau uplift and Asian interior aridification. Plant Syst. Evol. 2010, 288, 191–199. [Google Scholar] [CrossRef]
- Yang, F.S.; Qin, A.L.; Li, Y.F.; Wang, X.Q. Great genetic differentiation among populations of Meconopsis integrifolia and its implication for plant speciation in the Qinghai-Tibetan Plateau. PLoS ONE 2012, 7, e37196. [Google Scholar] [CrossRef] [Green Version]
- Qin, A.L.; Wang, M.M.; Cun, Y.Z.; Yang, F.S.; Wang, S.S.; Ran, J.H.; Wang, X.Q. Phylogeographic evidence for a link of species divergence of Ephedra in the Qinghai-Tibetan Plateau and adjacent regions to the Miocene Asian aridification. PLoS ONE 2013, 8, e56243. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.L.; Kang, Y.; Zhong, Y.; Sanderson, S.C. Intense uplift of the Qinghai-Tibetan Plateau triggered rapid diversification of Phyllolobium (Leguminosae) in the Late Cenozoic. Plant Ecol. Divers. 2012, 5, 491–499. [Google Scholar] [CrossRef]
- Wan, S.; Li, A.; Clift, P.D.; Jiang, H. Development of the East Asian summer monsoon: Evidence from the sediment record in the South China Sea since 8.5 Ma. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2006, 241, 139–159. [Google Scholar] [CrossRef]
- Ao, H.; Roberts, A.P.; Dekkers, M.J.; Liu, X.; Rohling, E.J.; Shi, Z.; An, Z.; Zhao, X. Late Miocene–Pliocene Asian monsoon intensification linked to Antarctic ice-sheet growth. Earth Planet Sci. Lett. 2016, 444, 75–87. [Google Scholar] [CrossRef] [Green Version]
- Guo, Z.T.; Ruddiman, W.F.; Hao, Q.Z.; Wu, H.B.; Qiao, Y.S.; Zhu, R.X.; Peng, S.Z.; Wei, J.J.; Yuan, B.Y.; Liu, T.S. Onset of Asian desertification by 22 Myr ago inferred from loess deposits in China. Nature 2002, 416, 159–163. [Google Scholar] [CrossRef]
- Miao, Y.; Herrmann, M.; Wu, F.; Yan, X.; Yang, S. What controlled Mid–Late Miocene long-term aridification in Central Asia?—Global cooling or Tibetan Plateau uplift: A review. Earth Sci. Rev. 2012, 112, 155–172. [Google Scholar] [CrossRef]
- Lu, H.; Guo, Z. Evolution of the monsoon and dry climate in East Asia during late Cenozoic: A review. Sci. China Earth Sci. 2014, 57, 70–79. [Google Scholar] [CrossRef]
- Sun, J.; Alloway, B.; Fang, X.; Windley, B.F. Refuting the evidence for an earlier birth of the Taklimakan Desert. Proc. Natl. Acad. Sci. USA 2015, 112, E5556–E5557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, Z.H.; Zhang, M.L. Evolutionary history of a desert shrub Ephedra przewalskii (Ephedraceae): Allopatric divergence and range shifts in northwestern China. PLoS ONE 2016, 11, e0158284. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.X.; Zhang, M.-L.; Williams, D.M. Genetic evidence and species distribution modelling reveal the response of Larix sibirica and its related species to Quaternary climatic and ancient historical events. Biochem. Syst. Ecol. 2014, 54, 316–325. [Google Scholar] [CrossRef]
- Hallatschek, O.; Hersen, P.; Ramanathan, S.; Nelson, D.R. Genetic drift at expanding frontiers promotes gene segregation. Proc. Natl. Acad. Sci. USA 2007, 104, 19926–19930. [Google Scholar] [CrossRef] [Green Version]
- Excoffier, L.; Ray, N. Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol. Evol. 2008, 23, 347–351. [Google Scholar] [CrossRef]
- Meng, X.; Xia, P.; Zheng, J.; Wang, X. Evolution of the East Asian monsoon and its response to uplift of the Tibetan Plateau since 1.8 Ma recorded by major elements in sediments of the South China Sea. Chin. Sci. Bull. 2011, 56, 547–551. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Zhang, R.; Pryor, S.C.; Li, X.; Wang, H. Influence of wintertime surface sensible heat flux variability over the central and eastern Tibetan Plateau on the East Asian winter monsoon. Clim. Dynam. 2020, 54, 4589–4603. [Google Scholar] [CrossRef]
- Liu, Y.X. A Study on origin and formation of the chinese desert floras. Acta Phytotax. Sin. 1995, 33, 131–143. [Google Scholar]
- Su, Z.; Lu, W.; Zhang, M. Phylogeographical patterns of two closely related desert shrubs, Nitraria roborowskii and N. sphaerocarpa (Nitrariaceae), from arid north-western China. Bot. J. Linn. Soc. 2016, 180, 334–347. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.J.; Zhang, M.L. Phylogeographical structure inferred from cpDNA sequence variation of Zygophyllum xanthoxylon across north-west China. J. Plant Res. 2015, 128, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Zhang, M.L.; Cohen, J.I. Phylogeographic history of Atraphaxis plants in arid northern China and the origin of A. bracteata in the Loess Plateau. PLoS ONE 2016, 11, e0163243. [Google Scholar] [CrossRef] [PubMed]

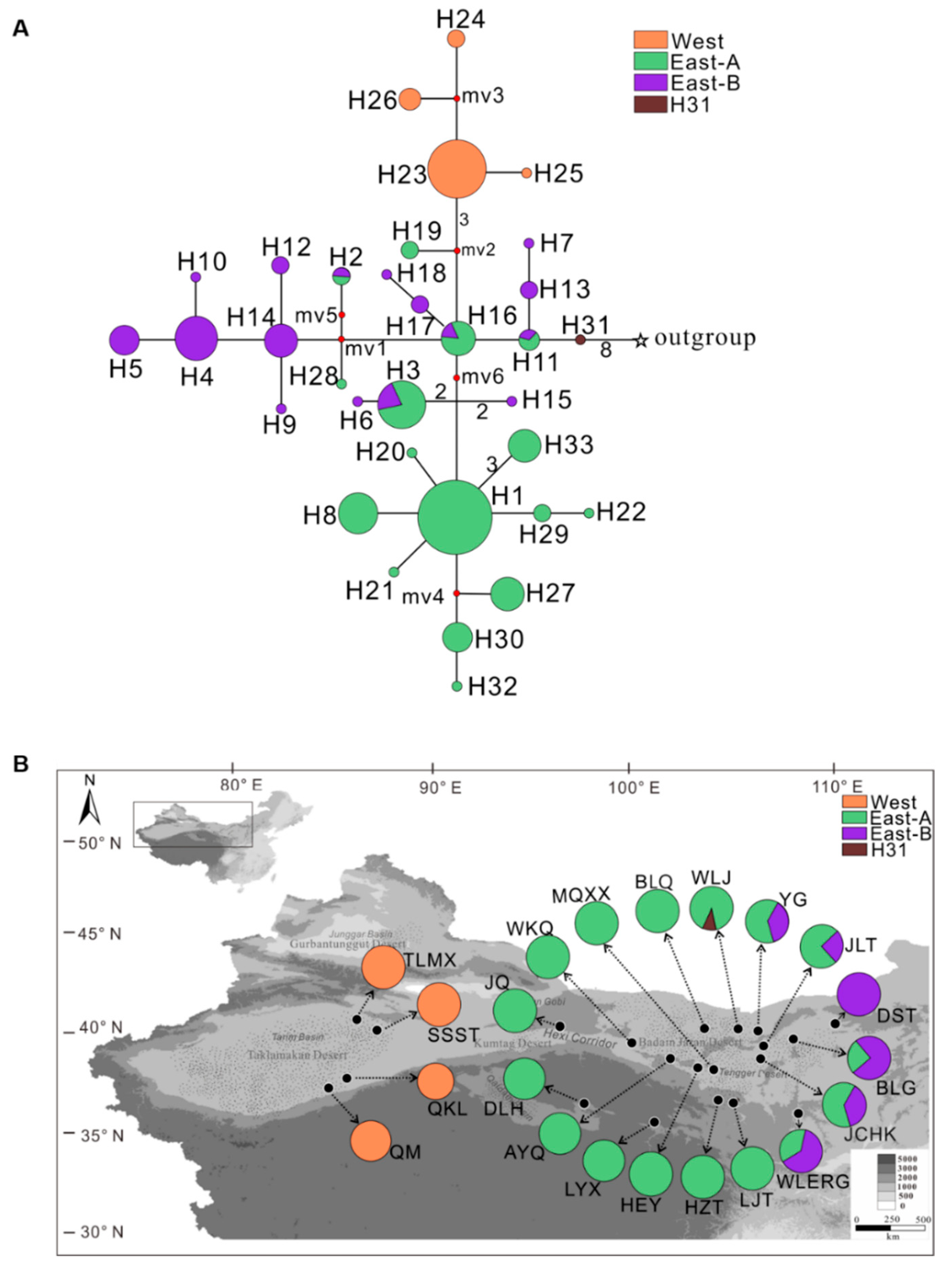
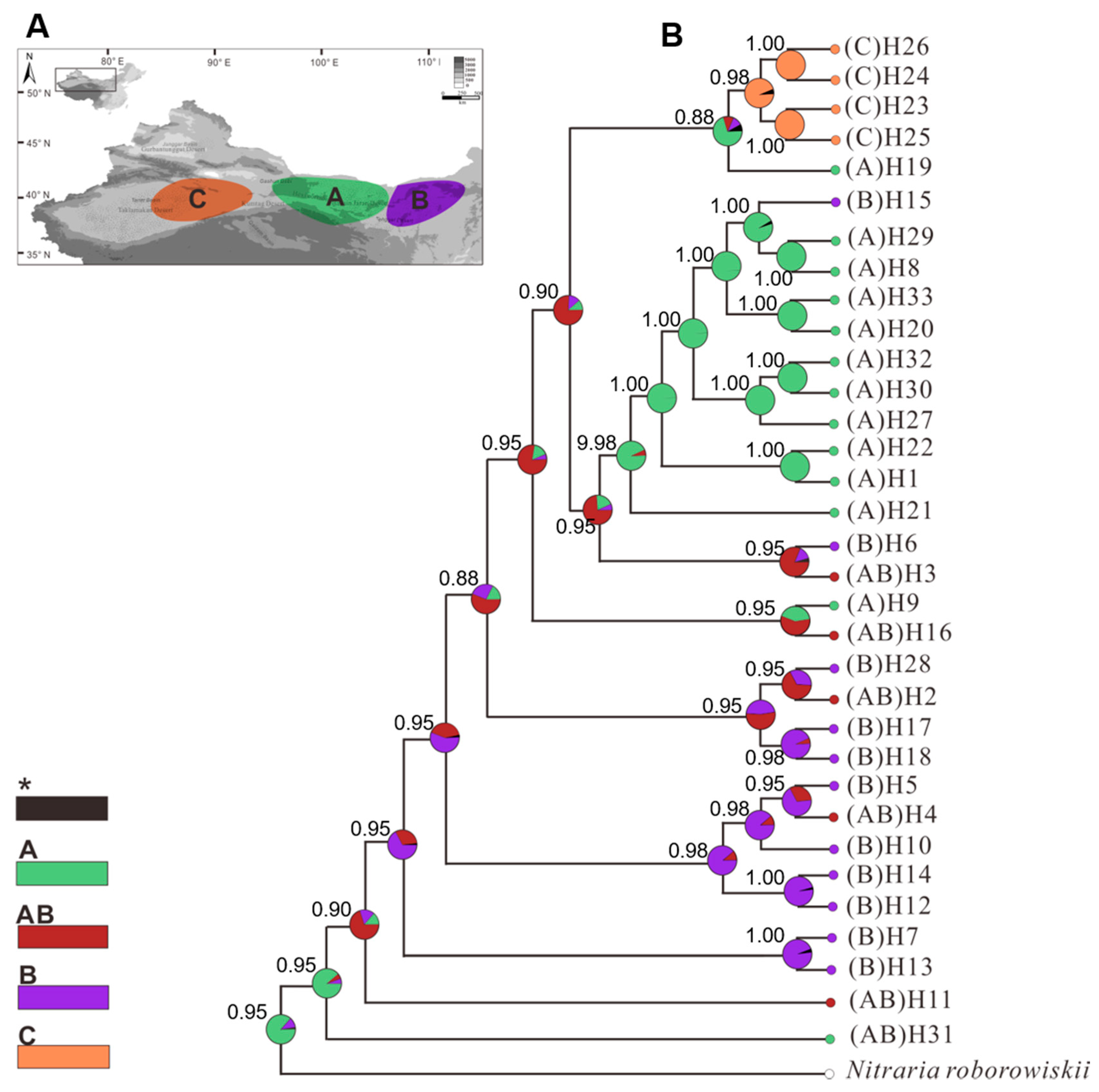

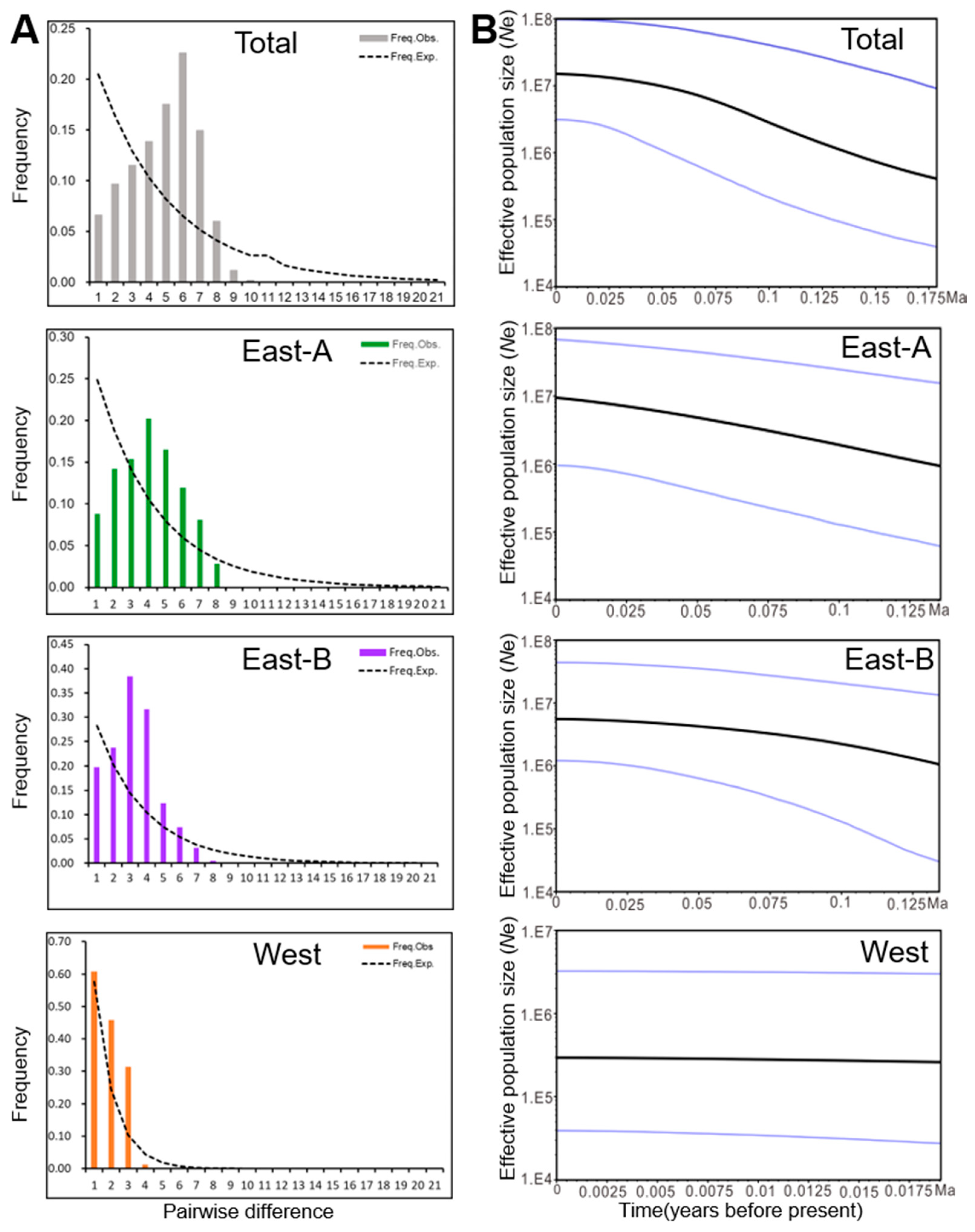
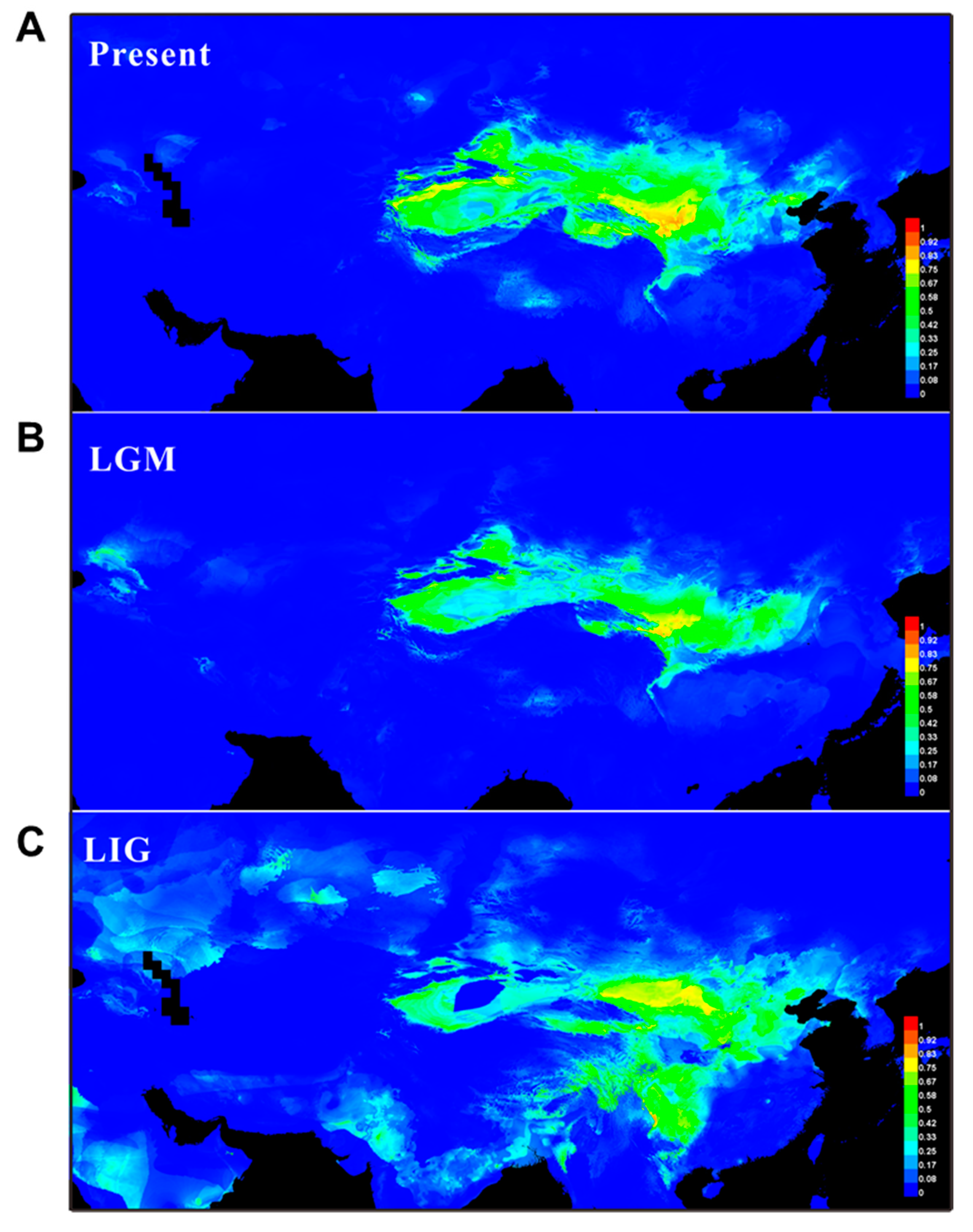

| Population (Code) | Location (All in China) | Group | Number of Individuals | Haplotypes (Individuals) | s | h | Hd | π |
|---|---|---|---|---|---|---|---|---|
| TLMX | Talimuxiang, Sinkiang | West | 8 | H23(2) H24(2) H25(1) H26(3) | 4 | 4 | 0.8210 | 0.0004 |
| SSST | Sanshisituan, Sinkiang | West | 8 | H23(8) | 0 | 1 | 0.0000 | 0.0000 |
| QKL | Qiongkule, Sinkiang | West | 5 | H23(5) | 0 | 1 | 0.0000 | 0.0000 |
| QM | Qiemo, Sinkiang | West | 6 | H23(6) | 0 | 1 | 0.0000 | 0.0000 |
| JQ | Jiuquan, Gansu | East-A | 8 | H8(3) H16(3) H19(2) | 5 | 3 | 0.7500 | 0.0006 |
| WKQ | Weikengquan, Gansu | East-A | 8 | H1(1) H8(1) H27(6) | 4 | 3 | 0.4640 | 0.0003 |
| AYQ | Ayouqi, IMG | East-A | 8 | H1(8) | 0 | 1 | 0.0000 | 0.0000 |
| HEY | Huaeryuan, Gansu | East-A | 8 | H1(8) | 0 | 1 | 0.0000 | 0.0000 |
| MQXX | Minqinxingxi, Gansu | East-A | 8 | H1(8) | 0 | 1 | 0.0000 | 0.0000 |
| HZT | Haizitan, Gansu | East-A | 8 | H1(4) H3(3) H11(1) | 5 | 2 | 0.2500 | 0.0003 |
| LJT | Luanjintan, Ningxia | East-A | 8 | H1(2) H3(2) H11(1) H20(1) H21(1) H22(1) | 10 | 6 | 0.9280 | 0.0007 |
| WLJ | Wuliji, IMG | East-A | 8 | H16(2) H29(1) H30(4) H31(1) | 6 | 4 | 0.7500 | 0.0006 |
| BLQ | Buliqi, IMG | East-A | 8 | H2(1) H3(6) H32(1) | 6 | 3 | 0.4640 | 0.0003 |
| DLH | Delingha, Qinghai | East-A | 8 | H8(7) H33(1) | 5 | 2 | 0.2500 | 0.0003 |
| LYX | Longyangxia, Qinghai | East-A | 8 | H1(8) | 0 | 1 | 0.0000 | 0.0000 |
| DST | Dashetai, IMG | East-B | 7 | H4(6) H10(1) | 5 | 4 | 0.7140 | 0.0004 |
| WLERG | Wulanerige, IMG | East-B | 8 | H3(2) H4(3) H14(2) H28(1) | 6 | 4 | 0.8210 | 0.0005 |
| YG | Yingen, IMG | East-B | 8 | H9(1) H4(2) H33(5) | 4 | 4 | 0.6420 | 0.0004 |
| BLG | Balagong, IMG | East-B | 8 | H4(2) H5(4) H6(1) H7(1) | 6 | 4 | 0.7500 | 0.0004 |
| JLT | Jilanta, IMG | East-B | 8 | H11(1) H14(2) H15(1) H16(1) H17(2) H18(1) | 8 | 6 | 0.9280 | 0.0007 |
| JCHK | Jichakou, IMG | East-B | 7 | H2(1) H3(1) H12(1) H13(2) H14(2) | 8 | 5 | 0.9040 | 0.0006 |
| Regions | HS | HT | GST | NST | Neutrality Tests | |
|---|---|---|---|---|---|---|
| Fu’s Fs | Tajiam’s D | |||||
| East-A | 0.472 (0.116) | 0.843 (0.095) | 0.440 (0.099) | 0.691 (0.136) | −8.024 ** (0.004) | −1.306 (0.067) |
| East-B | 0.824 * (0.041) | 0.930 * (0.043) | 0.114 * (0.022) | 0.220 (NC) | −6.468 ** (0.008) | −0.077 (0.527) |
| West | 0.205 * (0.020) | 0.375 * (0.027) | 0.452 (NC) | 0.364 (NC) | −0.645 (0.359) | −1.589 * (0.046) |
| Total | 0.470 (0.081) | 0.922 * (0.034) | 0.490 (0.046) | 0.647 (0.041) | −13.614 ** 0.002 | −0.624 * (0.034) |
| Regions | Source of Variation | d.f. | SS | VC | PV (%) | Fixation Index |
|---|---|---|---|---|---|---|
| East-A | Among populations | 10 | 56.705 | 0.623 | 47.770 | FST = 0.478 * |
| Within-populations | 77 | 52.500 | 0.681 | 52.230 | ||
| Total | 87 | 109.205 | 1.305 | |||
| East-B | Among populations | 5 | 38.049 | 0.849 | 43.500 | FST = 0.435 * |
| Within-populations | 40 | 44.125 | 1.103 | 56.500 | ||
| Total | 45 | 82.174 | 1.952 | |||
| West | Among populations | 3 | 3.431 | 0.132 | 33.070 | FST = 0.331 * |
| Within-populations | 23 | 6.125 | 0.266 | 66.930 | ||
| Total | 26 | 9.556 | 0.398 | |||
| West, East-A and East-B in comparison | Among groups | 2 | 121.013 | 1.314 | 51.960 | FCT = 0.520 * |
| Among populations within groups | 16 | 66.398 | 0.441 | 17.450 | FSC = 0.363 * | |
| Within populations | 126 | 97.500 | 0.774 | 30.590 | FST = 0.694 * | |
| Total | 144 | 284.910 | 2.530 |
| East-A | East-B | West | |
|---|---|---|---|
| East-A | 0.000 * | ||
| East-B | 0.388 * | 0.000 * | |
| West | 0.538 * | 0.553 ** | 0.000 * |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, H.; Wang, L.; Shi, Y.; Qian, C.; Zhou, H.; Wang, W.; Ma, X.-F.; Tran, L.-S.P.; Zhang, B. The East Asian Winter Monsoon Acts as a Major Selective Factor in the Intraspecific Differentiation of Drought-Tolerant Nitraria tangutorum in Northwest China. Plants 2020, 9, 1100. https://doi.org/10.3390/plants9091100
Yin H, Wang L, Shi Y, Qian C, Zhou H, Wang W, Ma X-F, Tran L-SP, Zhang B. The East Asian Winter Monsoon Acts as a Major Selective Factor in the Intraspecific Differentiation of Drought-Tolerant Nitraria tangutorum in Northwest China. Plants. 2020; 9(9):1100. https://doi.org/10.3390/plants9091100
Chicago/Turabian StyleYin, Hengxia, Lirong Wang, Yong Shi, Chaoju Qian, Huakun Zhou, Wenying Wang, Xiao-Fei Ma, Lam-Son Phan Tran, and Benyin Zhang. 2020. "The East Asian Winter Monsoon Acts as a Major Selective Factor in the Intraspecific Differentiation of Drought-Tolerant Nitraria tangutorum in Northwest China" Plants 9, no. 9: 1100. https://doi.org/10.3390/plants9091100
APA StyleYin, H., Wang, L., Shi, Y., Qian, C., Zhou, H., Wang, W., Ma, X.-F., Tran, L.-S. P., & Zhang, B. (2020). The East Asian Winter Monsoon Acts as a Major Selective Factor in the Intraspecific Differentiation of Drought-Tolerant Nitraria tangutorum in Northwest China. Plants, 9(9), 1100. https://doi.org/10.3390/plants9091100







