I-SceI Endonuclease-Mediated Plant Genome Editing by Protein Transport through a Bacterial Type III Secretion System
Abstract
1. Introduction
2. Results
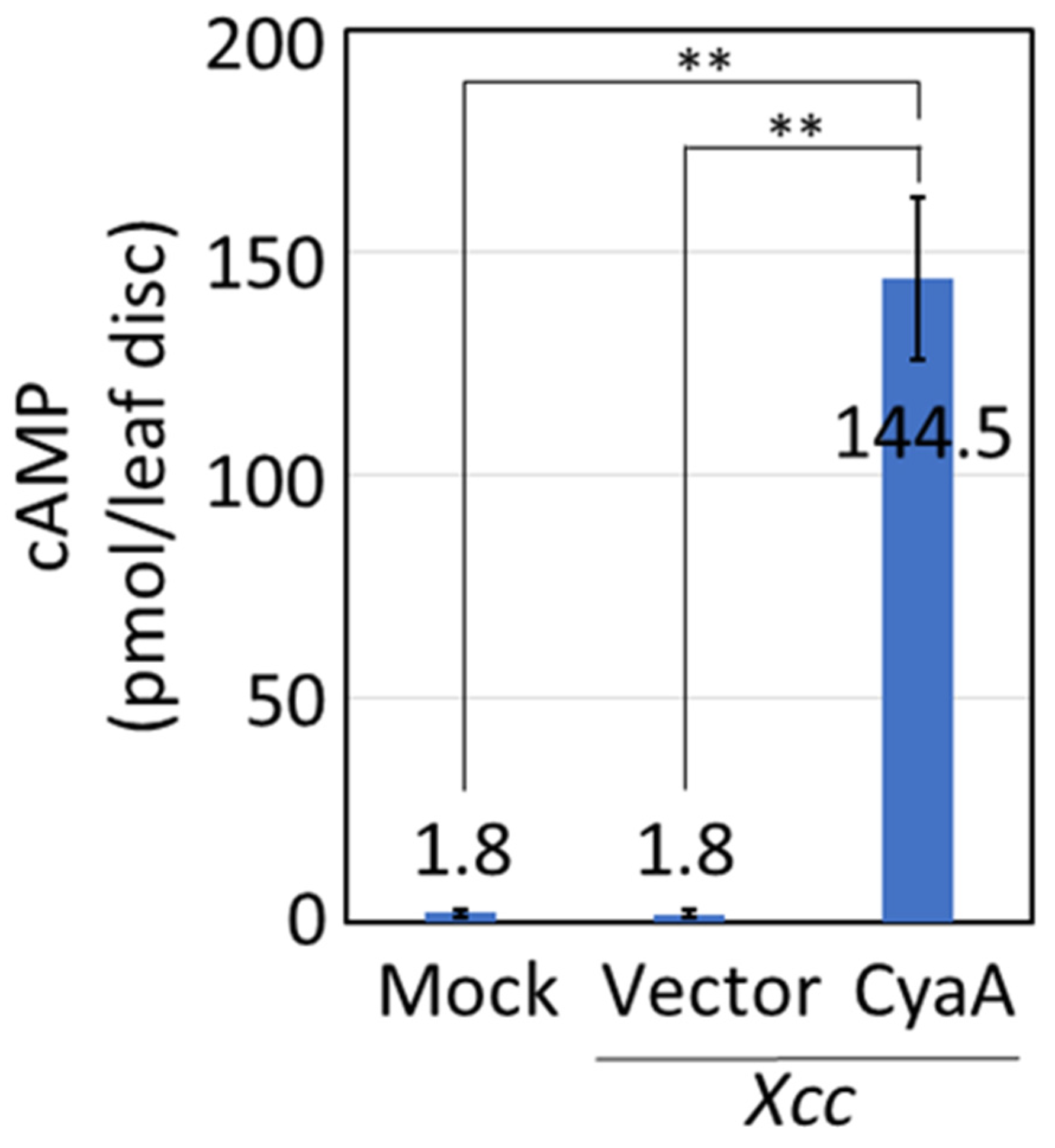
2.1. Protein Transport into Plant Leaf Cells by Xcc, and its Sterilization with Antibiotics
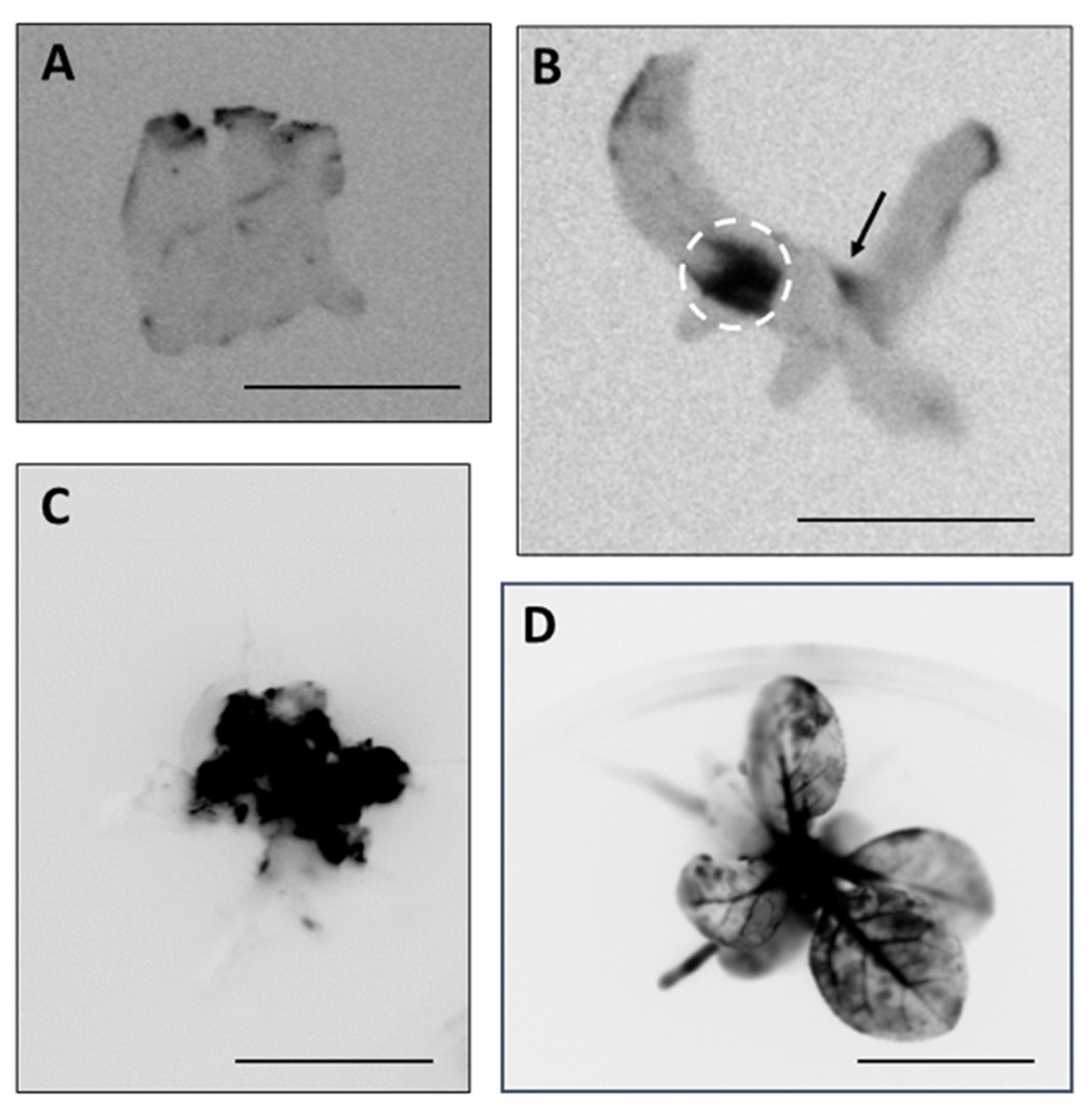
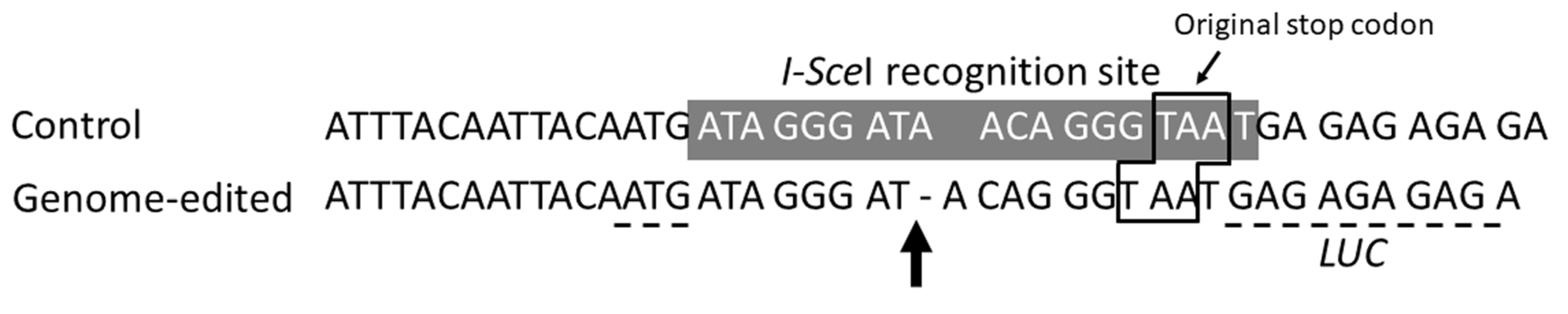
2.2. Genome Editing through I-SceI Protein Transport by Xcc into Plant Leaf Cells
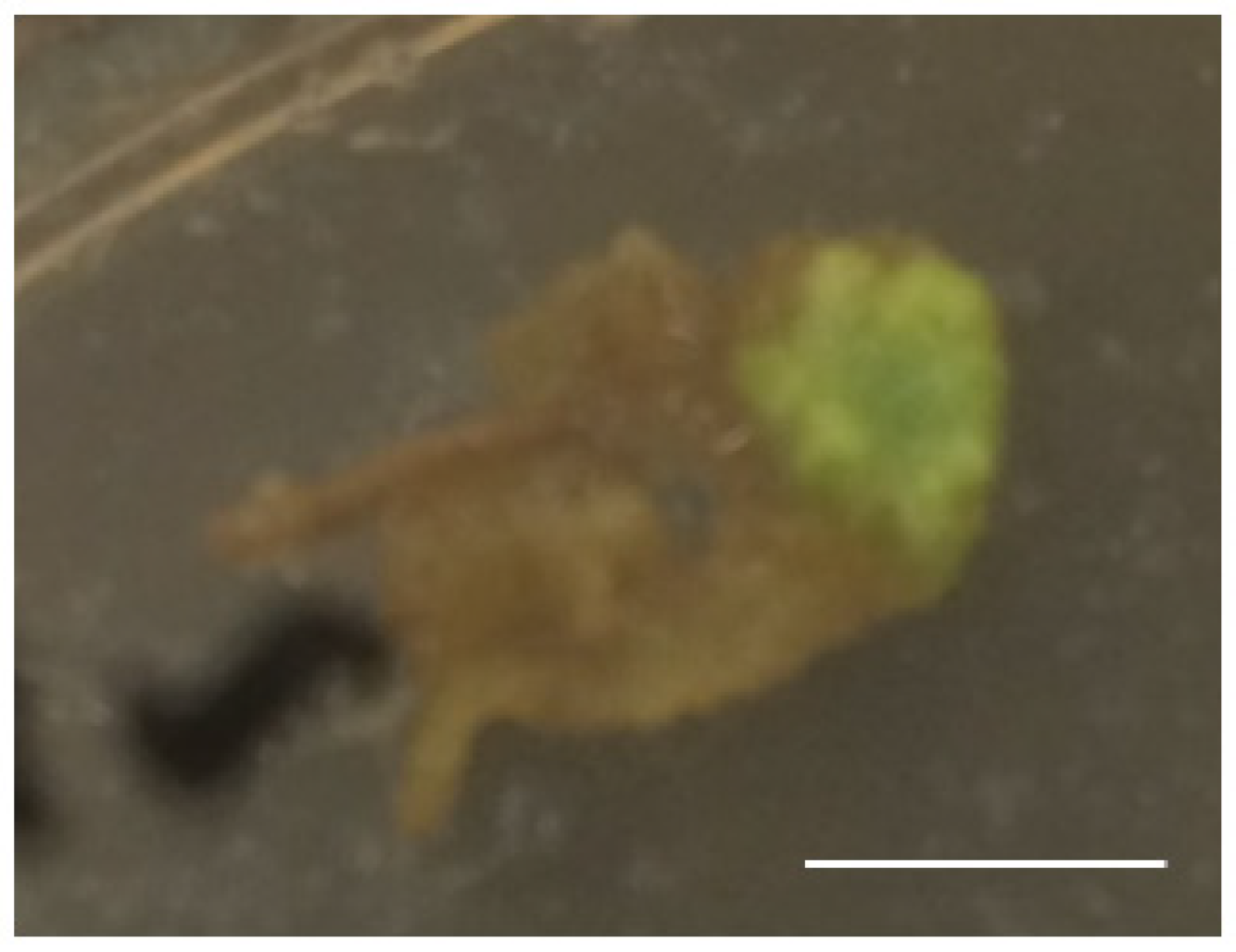
2.3. Production of Genome-Edited Tobacco Plants Through I-SceI Protein Transport by Xcc
3. Discussion
4. Materials and Methods
4.1. Plant and Bacterial Materials
4.2. Construction of Plasmids and Transformation into Bacteria
4.3. Infection of Bacteria and cAMP Enzyme Immunoassay
4.4. Genome Editing by I-SceI, Detection of LUC Chemiluminescence, and Sequencing of I-SceI Recognition Site
5. Patents
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Choo, Y.; Sanchez-Garcia, I.; Klug, A. In vivo repression by a site-specific DNA-binding protein designed against an oncogenic sequence. Nature 1994, 372, 642–645. [Google Scholar] [CrossRef] [PubMed]
- Boch, J.; Scholze, H.; Schornack, S.; Landgraf, A.; Hahn, S.; Kay, S.; Lahaye, T.; Nickstadt, A.; Bonas, U. Breaking the code of DNA binding specificity of TAL-type III eectors. Science 2009, 326, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Moscou, M.J.; Bogdanove, A.J. A simple cipher governs DNA recognition by TAL eectors. Science 2009, 326, 1501. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Jacobi, A.M.; Rettig, G.R.; Turk, R.; Collingwood, M.A.; Zeiner, S.A.; Quadros, R.M.; Harms, D.W.; Bonthuis, O.J.; Gregg, C.; Ohtsuka, M.; et al. Simplified CRISPR tools for efficient genome editing and streamlined protocols for their delivery into mammalian cells and mouse zygotes. Methods 2017, 15, 121–122. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Conboy, M.; Park, H.M.; Jiang, F.; Kim, H.J.; Dewitt, M.A.; Mackley, V.A.; Chang, K.; Rao, A.; Skinner, C.; et al. Nanoparticle delivery of Cas9 ribonucleoprotein and donar DNA in vivo induces homology-directed DNA repair. Nat. Biomed. Eng. 2017, 1, 889–901. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Rudis, M.R.; Cheplick, M.H.; Millwood, R.J.; Yang, J.-P.; Ondzighi-Assoume, C.A.; Montgomery, G.A.; Burris, K.P.; Mazarei, M.; Chesnut, J.D.; et al. Lipofection-mediated genome editing using DNA-free delivery of theCas9/gRNA ribonucleoprotein into plant cells. Plant Cell Rep. 2020, 39, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Itami, J.; Oikawa, K.; Motoda, Y.; Kigawa, Y.; Numata, K. Native protein delivery into rice callus using ionic complexes of proteins and cell-penetrating peptides. PLoS ONE 2019, 14, e0214033. [Google Scholar] [CrossRef]
- Yanagawa, Y.; Kawano, H.; Kobayashi, T.; Miyahara, H.; Okino, A.; Mitshuhara, I. Direct protein introduction into plant cells using a multi-gas plasma jet. PLoS ONE 2017, 12, e0171942. [Google Scholar] [CrossRef]
- Frank, F.; Potnis, N.; Jones, J.B.; Jiebbuj, R. The type III effectors of Xanthomonas. Mol. Plant Pathol. 2009, 10, 749–766. [Google Scholar]
- Büttner, D. Behind the lines-actions of bacterial type III effector protein in plant cells. FEMS Microbiol. Rev. 2016, 40, 894–937. [Google Scholar] [CrossRef] [PubMed]
- Furutani, A.; Takaoka, M.; Sanada, H.; Noguchi, Y.; Oku, T.; Tsuno, K.; Ochiai, H.; Tsuge, S. Identification of novel type III secretion effectors in Xanthomonas oryzae pv. oryzae. Mol. Plant Microbe Interact. 2009, 22, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Mitsuhara, I. Complete genome sequences of two strains of Xanthomonas campestris pv. campestris isolated in Japan. Microbiol. Resour. Announc. 2020, 9, e0123919. [Google Scholar] [CrossRef] [PubMed]
- Chujo, T.; Yoshikawa, M.; Ariga, H.; Endo, M.; Toki, S.; Ishibashi, K. A removable virus vector suitable for plant genome editing. Plant J. 2017, 91, 558–561. [Google Scholar] [CrossRef] [PubMed]
- Toki, S.; Hara, N.; Ono, K.; Onodera, H.; Tagiri, A.; Oka, S.; Tanaka, H. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 2006, 47, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Kawazu, K.; Wasano, N.; Konno, K.; Ohashi, Y.; Mochizuki, A.; Mitsuhara, I. Evaluation of anti-herbivory genes using an Agrobacterium-mediated transient expression system. Plant Biotech. 2012, 29, 495–499. [Google Scholar] [CrossRef][Green Version]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yanagawa, Y.; Takeuchi, K.; Endo, M.; Furutani, A.; Ochiai, H.; Toki, S.; Mitsuhara, I. I-SceI Endonuclease-Mediated Plant Genome Editing by Protein Transport through a Bacterial Type III Secretion System. Plants 2020, 9, 1070. https://doi.org/10.3390/plants9091070
Yanagawa Y, Takeuchi K, Endo M, Furutani A, Ochiai H, Toki S, Mitsuhara I. I-SceI Endonuclease-Mediated Plant Genome Editing by Protein Transport through a Bacterial Type III Secretion System. Plants. 2020; 9(9):1070. https://doi.org/10.3390/plants9091070
Chicago/Turabian StyleYanagawa, Yuki, Kasumi Takeuchi, Masaki Endo, Ayako Furutani, Hirokazu Ochiai, Seiichi Toki, and Ichiro Mitsuhara. 2020. "I-SceI Endonuclease-Mediated Plant Genome Editing by Protein Transport through a Bacterial Type III Secretion System" Plants 9, no. 9: 1070. https://doi.org/10.3390/plants9091070
APA StyleYanagawa, Y., Takeuchi, K., Endo, M., Furutani, A., Ochiai, H., Toki, S., & Mitsuhara, I. (2020). I-SceI Endonuclease-Mediated Plant Genome Editing by Protein Transport through a Bacterial Type III Secretion System. Plants, 9(9), 1070. https://doi.org/10.3390/plants9091070



