Digital Absolute Gene Expression Analysis of Essential Starch-Related Genes in a Radiation Developed Amaranthus cruentus L. Variety in Comparison with Real-Time PCR
Abstract
1. Introduction
2. Results and Discussion
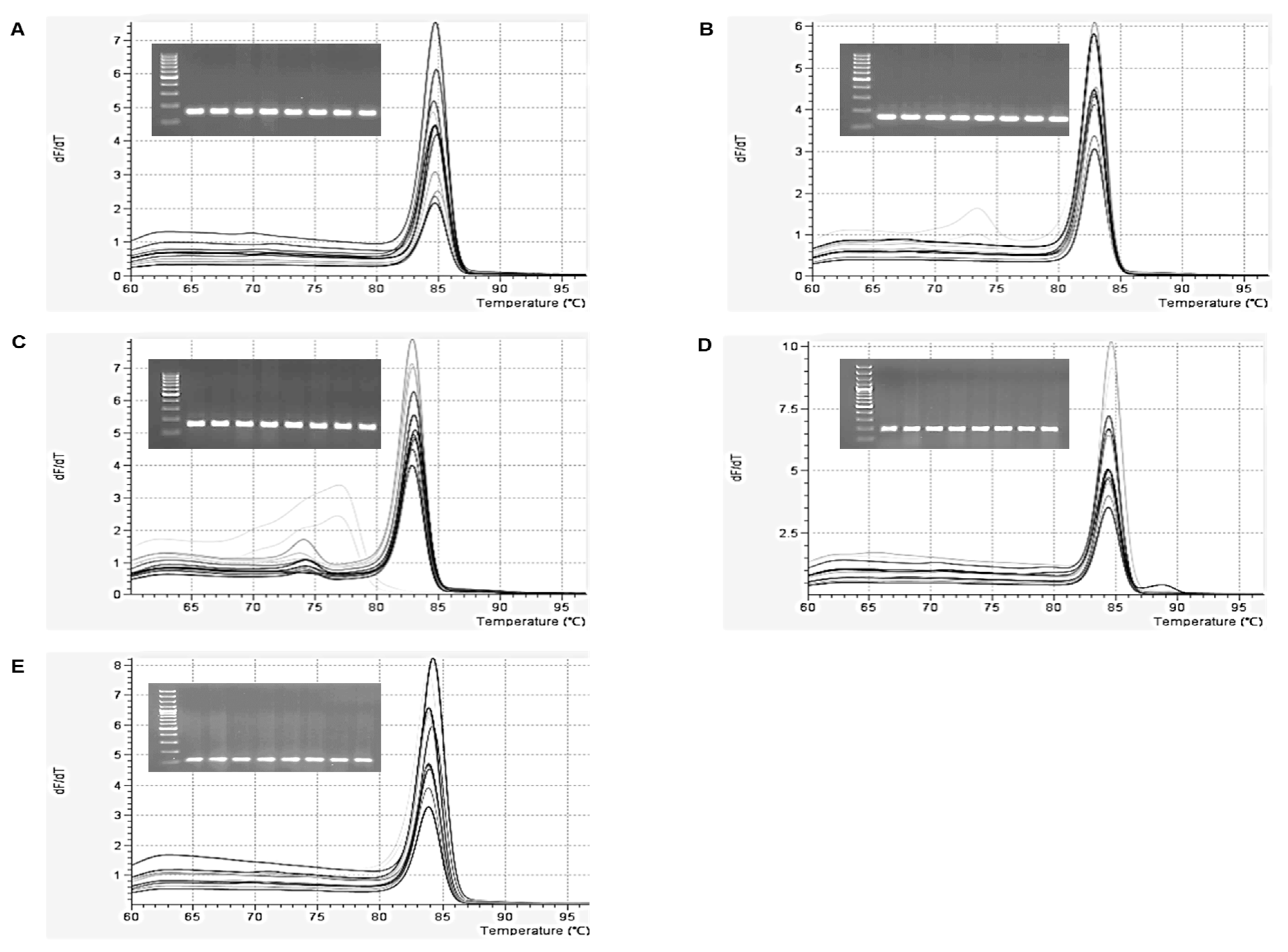
2.1. Optimization of qPCR and ddPCR Assay
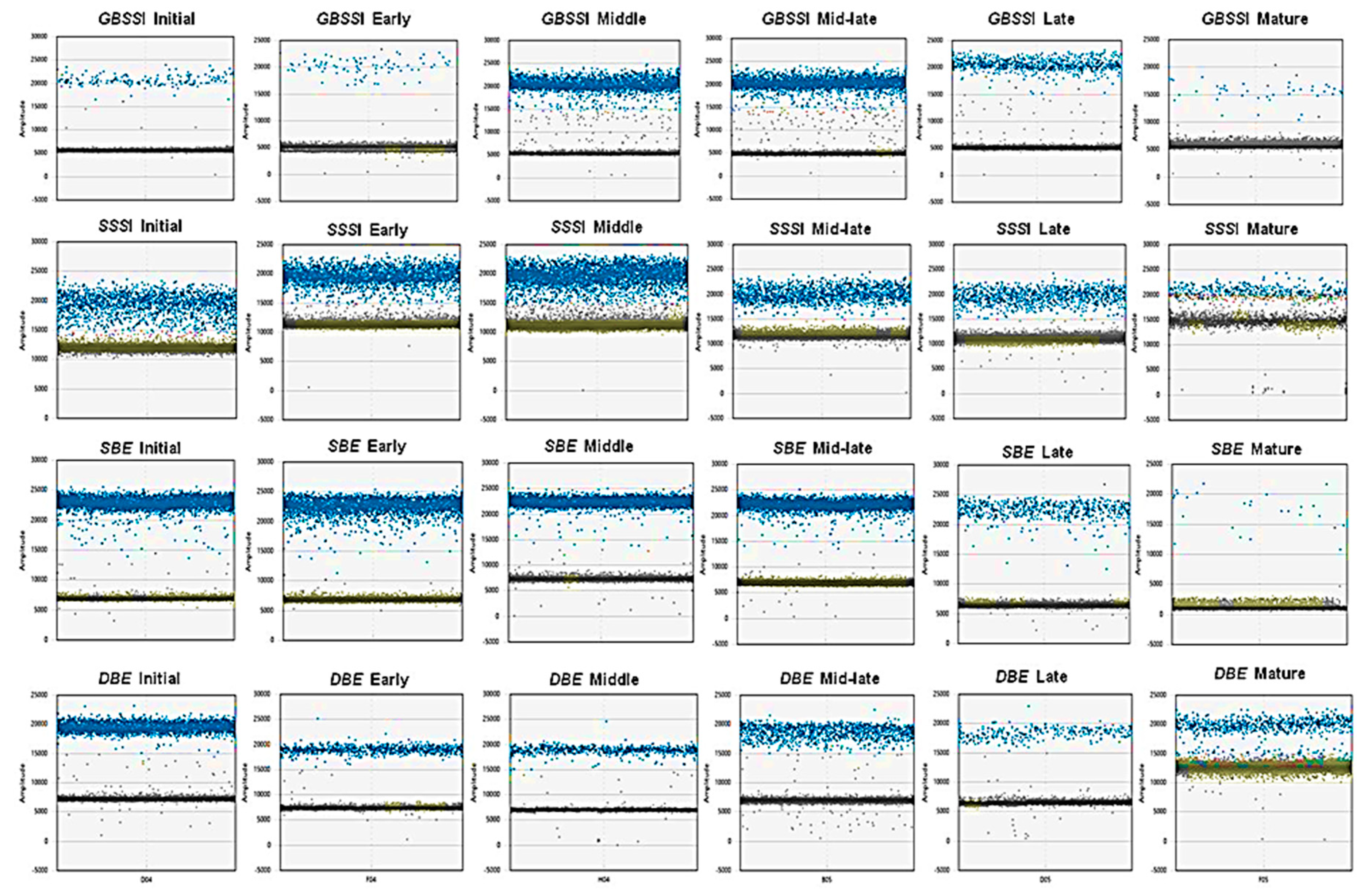
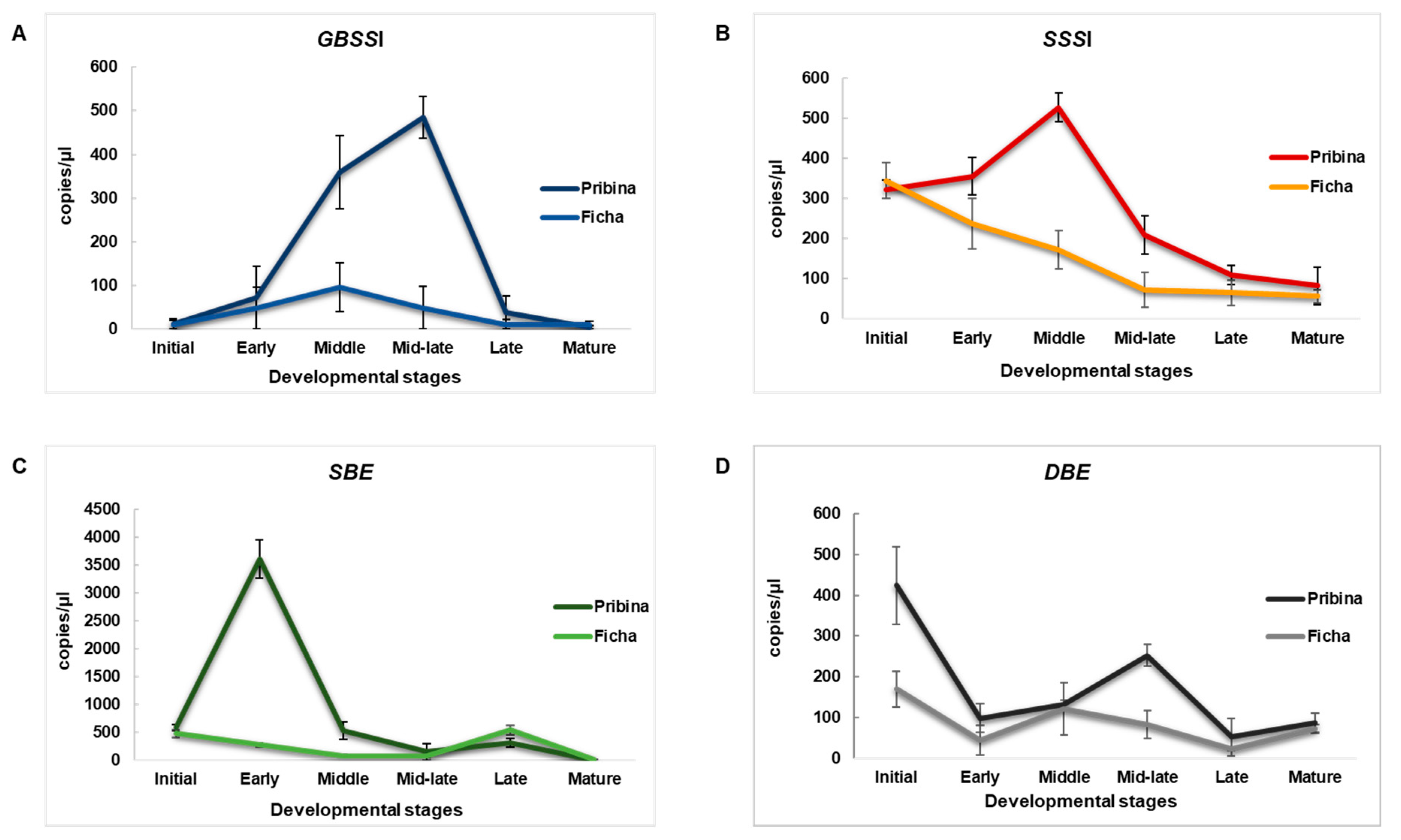
2.2. Absolute Quantification of Essential Amaranth Starch-Related Genes Using ddPCR
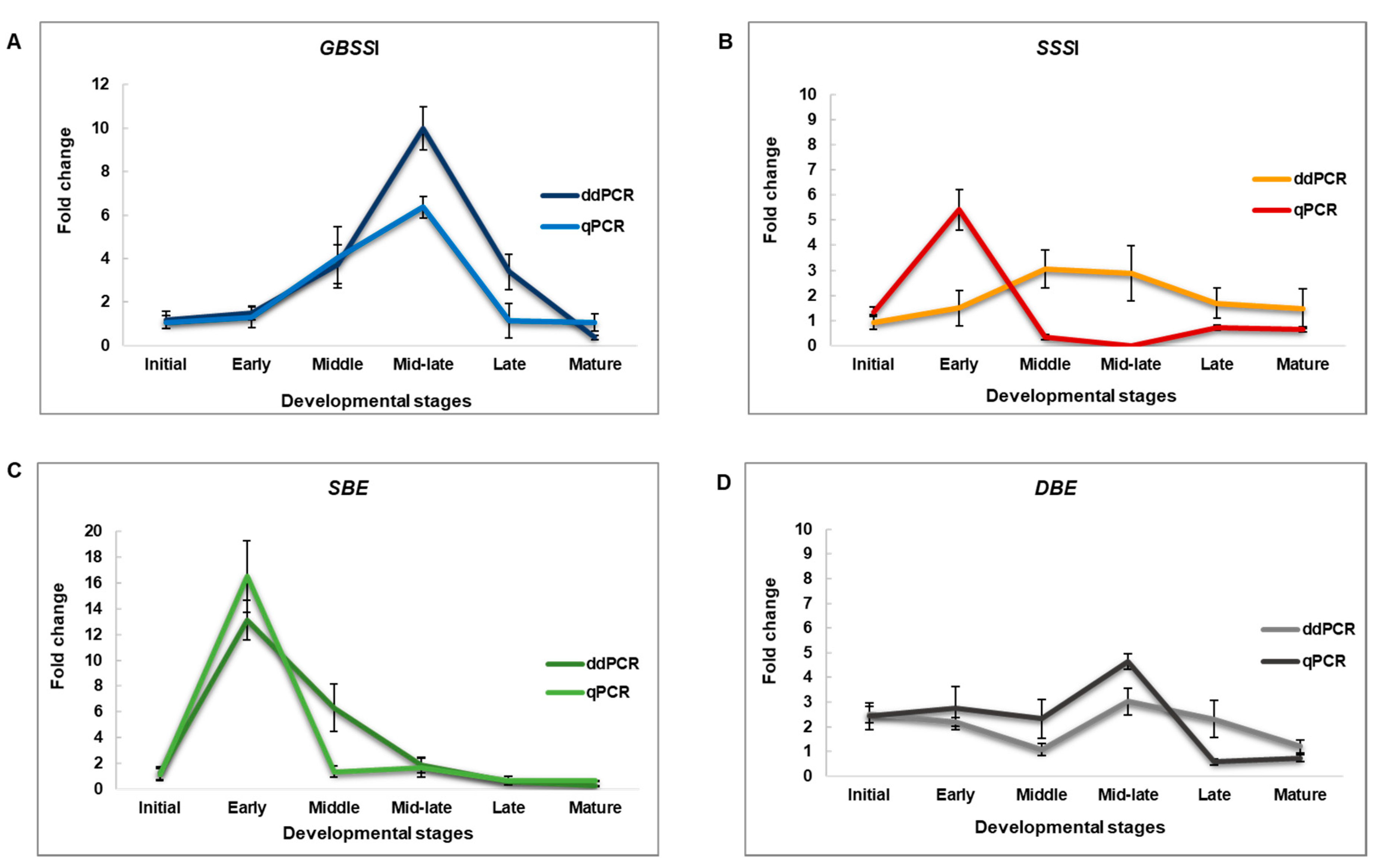
2.3. Performance of ddPCR vs. qPCR
3. Materials and Methods
3.1. Plant Material and Experimental Field
3.2. Gene-Specific Primer Design
3.3. RNA Extraction and cDNA Synthesis
3.4. Relative Gene Expression Analysis Using qRT-PCR
3.5. Absolute Gene Expression Analysis Using ddPCR
Author Contributions
Funding
Conflicts of Interest
References
- Głowacka, K.; Kromdijk, J.; Leonelli, L.; Niyogi, K.K.; Clemente, T.E.; Long, S.P. An evaluation of new and established methods to determine T-DNA copy number and homozygosity in transgenic plants. Plant Cell Environ. 2016, 39, 908–917. [Google Scholar] [CrossRef] [PubMed]
- Finke, A.; Rozhon, W.; Pecinka, A. Analysis of DNA methylation content and patterns in plants. Methods Mol. Biol. 2018, 1694, 277–298. [Google Scholar] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucl. Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Leong, D.T.; Gupta, A.; Bai, H.F.; Wan, G.; Yoong, L.F.; Too, H.P.; Chew, F.T.; Hutmacher, D.W. Absolute quantification of gene expression in biomaterials research using real-time PCR. Biomaterials 2007, 28, 203–210. [Google Scholar] [CrossRef]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef]
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005. [Google Scholar] [CrossRef]
- Rački, N.; Dreo, T.; Gutierrez-Aguirre, I.; Blejec, A.; Ravnikar, M. Reverse transcriptase droplet digital PCR shows high resilience to PCR inhibitors from plant, soil and water samples. Plant Methods 2014, 10, 42. [Google Scholar] [CrossRef]
- Quan, P.L.; Sauzade, M.; Brouzes, E. dPCR: A technology review. Sensors 2018, 18, 1271. [Google Scholar] [CrossRef]
- Morisset, D.; Stebih, D.; Milavec, M.; Gruden, K.; Zel, J. Quantitative analysis of food and feed samples with droplet digital PCR. PLoS ONE 2013, 8, e62583. [Google Scholar] [CrossRef]
- Dreo, T.; Pirc, M.; Ramšak, Ž.; Pavšič, J.; Milavec, M.; Žel, J.; Gruden, K. Optimising droplet digital PCR analysis approaches for detection and quantification of bacteria: A case study of fire blight and potato brown rot. Anal. Bioanal. Chem. 2014, 406, 6513–6528. [Google Scholar] [CrossRef]
- Taylor, C.; Laperriere, G.; Germain, H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: From variable nonsense to publication quality data. Sci. Rep. 2017, 7, 2409. [Google Scholar] [CrossRef] [PubMed]
- Nathan, L.M.; Simmons, M.; Wegleitner, B.J.; Jerde, C.L.; Mahon, A.R. Quantifying environmental DNA signals for aquatic invasive species across multiple detection platforms. Environ. Sci. Technol. 2014, 248, 12800–12806. [Google Scholar] [CrossRef] [PubMed]
- Doi, H.; Takahara, T.; Minamoto, T.; Matsuhashi, S.; Uchii, K.; Yamanak, H. Droplet digital polymerase chain reaction (PCR) outperforms Real-Time PCR in the detection of environmental DNA from an invasive fish species. Environ. Sci. Technol. 2015, 49, 5601–5608. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Kim, H.M.; Min, J.; Kim, K.; Park, M.G.; Jeong, H.J.; Kim, K.Y. An advanced tool, droplet digital PCR (ddPCR), for absolute quantification of the red-tide dinoflagellate, Cochlodinium polykrikoides Margalef (Dinophyceae). Algae 2017, 32, 189–197. [Google Scholar] [CrossRef]
- Dupas, E.; Legendre, B.; Olivier, V.; Poliakoff, F.; Manceau, C.; Cunty, A. Comparison of real-time PCR and droplet digital PCR for the detection of Xylella fastidiosa in plants. J. Microbiol. Methods 2019, 162, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Santander, R.D.; Meredith, C.L.; Aćimović, S.G. Development of a viability digital PCR protocol for the selective detection and quantification of live Erwinia amylovora cells in cankers. Sci. Rep. 2019, 9, 11530. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhang, H.; Zhao, Z.; Wen, C.; Wu, P.; Song, S.; Yu, S.; Luo, L.; Xu, X. Application of droplet digital PCR in detection of seed-transmitted pathogen Acidovorax citrulli. J. Integr. Agric. 2020, 19, 561–569. [Google Scholar] [CrossRef]
- Gerdes, L.; Iwobi, A.; Busch, U.; Pecoraco, S. Optimization of digital droplet polymerase chain reaction for quantification of genetically modified organisms. Biomol. Detect. Quantif. 2016, 7, 9–20. [Google Scholar] [CrossRef]
- Demeke, T.; Dobnik, D. Critical assessment of digital PCR for the detection and quantification of genetically modified organisms. Anal. Bioanal. Chem. 2018, 410, 4039–4050. [Google Scholar] [CrossRef]
- Grelewska-Novotko, K.; Żurawska-Zajfert, E.; Sowa, S. Optimization and verification of droplet Digital PCR even-specific methods for the quantification of GM maize DAS1507 and NK603. Appl. Biochem. Biotechnol. 2018, 185, 207–220. [Google Scholar] [CrossRef]
- Giraldo, P.A.; Cogan, N.O.I.; Spangenberg, G.C.; Smith, K.F.; Shinozuka, H. Development and application of droplet digital PCR tools for the detection of transgenes in pastures and pasture-based products. Front. Plant Sci. 2019, 9, 1923. [Google Scholar] [CrossRef]
- Park, Y.J.; Nemoto, K.; Nishikawa, T.; Matsushima, K.; Minami, M.; Kawase, M. Genetic diversity and expression analysis of granule bound starch synthase I gene in the new world grain amaranth (Amaranthus cruentus L.). J. Cereal Sci. 2011, 53, 298–305. [Google Scholar] [CrossRef]
- Park, Y.J.; Nishikawa, T. Characterization and expression analysis of the starch synthase gene family in grain amaranth (Amaranthus cruentus L.). Genes Genet. Syst. 2012, 87, 281–289. [Google Scholar] [CrossRef]
- Park, Y.J.; Nishikawa, T.; Tomooka, N.; Nemoto, K. Molecular cloning and expression analysis of a gene encoding soluble starch synthase I from grain amaranth (Amaranthus cruentus L.). Mol. Breed. 2012, 30, 1065–1107. [Google Scholar] [CrossRef]
- Park, Y.J.; Nishikawa, T.; Matsushima, K.; Minami, M.; Tomooka, N.; Nemoto, K. Moleular characterization and genetic diversity of the starch branching enzyme (SBE) gene from Amaranthus. Mol. Breed. 2014, 34, 1975–1985. [Google Scholar] [CrossRef]
- Park, Y.J.; Nishikawa, T.; Tomooka, N.; Nemoto, K. Molecular characterization of an isoamylase 1-type starch debranching enzyme (DBEI) in grain amaranth (Amaranthus cruentus L.). Mol. Biol. Rep. 2014, 41, 7857–7864. [Google Scholar] [CrossRef] [PubMed]
- Baker, M. Digital PCR hits its stride. Nat. Methods 2012, 9, 541–544. [Google Scholar] [CrossRef]
- Sidstedt, M.; Romsos, E.L.; Hedell, R.; Ansell, R.; Steffen, C.R.; Vallone, P.M.; Rådström, P. Accurate digital polymerase chain reaction quantification of challenging samples applying inhibitor-tolerant DNA polymerases. Anal. Chem. 2017, 89, 1642–1649. [Google Scholar] [CrossRef] [PubMed]
- Maheshwari, Y.; Selvaraj, V.; Hajeri, S.; Yokomi, R. Application of droplet digital PCR for quantitative detection of Spiroplasma citri in comparison with real time PCR. PLoS ONE 2017, 12, e0184751. [Google Scholar] [CrossRef]
- Coudray-Meunier, C.; Fraisse, A.; Martin-latil, S.; Guillier, L.; Dellanoy, S.; Fach, P.; Perelle, S. A comparative study of digital RT-PCR and RT-qPCR for quantification of hepatitis A virus and norovirus in lettuce and water samples. Int. J. Food Microbiol. 2015, 18, 17–26. [Google Scholar] [CrossRef]
- Gajdošová, A.; Libiaková, G.; Fejér, J. Improvement of selected Amaranthus cultivars by means of mutation induction and biotechnological approaches. In Breeding of Neglected and Under-Utilized Crops, Spices and Herbs; Science Publishers: Enfield, NH, USA, 2007; pp. 151–169. [Google Scholar]
- Hricová, A.; Fejér, J.; Libiaková, G.; Szabóová, M.; Gažo, J.; Gajdošová, A. Characterization of phenotypic and nutritional properties of valuable Amaranthus cruentus L. mutants. Turk. J. Agric. For. 2016, 40, 761–771. [Google Scholar] [CrossRef]
- Mohapatra, P.K.; Sarkar, R.K.; Kuanar, S.R. Starch synthesizing enzymes and sink strength of grains of contrasting rice cultivars. Plant Sci. 2009, 176, 256–263. [Google Scholar] [CrossRef]
- Xie, G.; Li, Z.; Ran, Q.; Wang, H.; Zhang, J. Over-expression of mutated ZmDA1 or ZmDAR1 gene improves maize kernel yield by enhancing starch synthesis. Plant Biotechnol. J. 2018, 6, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Zi, Y.; Ding, J.; Song, J.; Humphreys, G.; Peng, Y.; Li, C.; Zhu, X.; Guo, W. Grain yield, starch content and activities of key enzymes of waxy and non-waxy wheat (Triticum aestivum L.). Sci. Rep. 2018, 8, 4548. [Google Scholar] [CrossRef]
- Park, Y.J.; Nishikawa, T.; Matsushima, K.; Nemoto, K. Characterization of a new granule-bound starch synthase gene found in amaranth grains (Amaranthus cruentus L.). Mol. Breed. 2017, 37, 111. [Google Scholar] [CrossRef]
- Hirose, T.; Terao, T. A comprehensive expression analysis of the starch synthasegene family in rice (Oryza sativa L.). Planta 2004, 220, 9–16. [Google Scholar] [CrossRef]
- Caoa, Y.N.; Hub, W.G.; Wang, C.S. Expression profiles of genes involved in starch synthesis in nonwaxy and waxy wheat. Russ. Plant J. Physl. 2012, 59, 632–639. [Google Scholar] [CrossRef]
- James, M.G.; Denyer, K.; Myers, A.M. Starch synthesis in the cereal endosperm. Curr. Opin. Plant Biol. 2003, 6, 215–222. [Google Scholar] [CrossRef]
- Tsai, C.Y. The function of the waxy locus in starch synthesis in maize endosperm. Biochem. Genet. 1974, 11, 83–96. [Google Scholar] [CrossRef]
- Fujita, N.; Hasegawa, H.; Taira, T. The isolation and characterization of a waxy mutant of diploid wheat (Triticum monococcum L.). Plant Sci. 2001, 160, 595–602. [Google Scholar] [CrossRef]
- Chen, L.; Lu, D.; Wang, T.; Li, Z.; Zhao, Y.; Jiang, Y.; Zhang, Q.; Cao, Q.; Fang, K.; Xing, Y.; et al. Identification and expression analysis of starch branching enzymes involved in starch synthesis during the development of chestnut (Castanea mollissima Blume) cotyledons. PLoS ONE 2017, 12, e0177792. [Google Scholar] [CrossRef]
- Wang, J.; Hu, P.; Chen, Z.; Liu, Q.; Wei, C. Progress in high-amylose cereal crops through inactivation of starch branching enzymes. Front. Plant Sci. 2017, 8, 469. [Google Scholar] [CrossRef] [PubMed]
- Ohdan, T.; Francisco, P.B.; Sawada, T.; Hirose, T.; Terao, T.; Satoh, H.; Nakamura, Y. Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J. Exp. Bot. 2005, 56, 3229–3244. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Xia, Q.; Yin, Y.; Wang, Z. Comparison of droplet digital PCR and quantitative PCR assays for quantitative detection of Xanthomonas citri Subsp. citri. PLoS ONE 2016, 11, e0159004. [Google Scholar] [CrossRef]
- Dong, L.; Wang, S.; Fu, B.; Wang, J. Evaluation of droplet digital PCR and next generation sequencing for characterizing DNA reference material fro KRAS mutation detection. Sci. Rep. 2018, 8, 9650. [Google Scholar] [CrossRef]
- Wan, J.; Song, L.; Wu, Y.; Brzoska, P.; Keys, D.; Chen, C.; Babu, V.; Shannon, J.G.; Nguyen, H.T. Application of digital PCR in the analysis of transgenic soybean plants. Adv. Biosci. Biotechnol. 2016, 7, 403–417. [Google Scholar] [CrossRef]
- Vera Hernández, F.P.; Martínez Núñez, M.; Ruiz Rivas, M.; Vázquez Portillo, R.E.; Bibbins Martínez, M.D.; Luna Suárez, S.; Rosas Cárdenas, F.D.F. Reference genes fro RT-qPCR normalization in different tissues, developmental stages ans tress conditions of amaranth. Plant Biol. 2018, 20, 713–721. [Google Scholar] [CrossRef]
- Kečkešová, M.; Gálová, Z.; Hricová, A. Changes of protein profiles in amaranth mutant lines. J. Microbiol. Biotechnol. Food Sci. 2012, 1, 1129–1135. [Google Scholar]
- Kečkešová, M.; Palenčárová, E.; Gálová, Z.; Gažo, J.; Hricová, A. Nutritional quality of grain amaranths (Amaranthus L.) compared to putative mutant lines. J. Microbiol. Biotechnol. Food Sci. 2013, 2, 1716–1724. [Google Scholar]
- Wang, G.; Wang, G.; Zhang, X.; Wang, F.; Song, R. Isolation of high quality RNA from cereal seeds containing high levels of starch. Phytochem. Anal. 2012, 23, 159–163. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lancíková, V.; Hricová, A. Digital Absolute Gene Expression Analysis of Essential Starch-Related Genes in a Radiation Developed Amaranthus cruentus L. Variety in Comparison with Real-Time PCR. Plants 2020, 9, 966. https://doi.org/10.3390/plants9080966
Lancíková V, Hricová A. Digital Absolute Gene Expression Analysis of Essential Starch-Related Genes in a Radiation Developed Amaranthus cruentus L. Variety in Comparison with Real-Time PCR. Plants. 2020; 9(8):966. https://doi.org/10.3390/plants9080966
Chicago/Turabian StyleLancíková, Veronika, and Andrea Hricová. 2020. "Digital Absolute Gene Expression Analysis of Essential Starch-Related Genes in a Radiation Developed Amaranthus cruentus L. Variety in Comparison with Real-Time PCR" Plants 9, no. 8: 966. https://doi.org/10.3390/plants9080966
APA StyleLancíková, V., & Hricová, A. (2020). Digital Absolute Gene Expression Analysis of Essential Starch-Related Genes in a Radiation Developed Amaranthus cruentus L. Variety in Comparison with Real-Time PCR. Plants, 9(8), 966. https://doi.org/10.3390/plants9080966



